Background: De novo DNA methyltransferase 3A is essential for gene regulation, however its regulation is poorly understood.
Results: DNMT3A dimers are fully functional but eliminate methylation on clustered CpG sites (processive catalysis) that are observed with the wild-type homotetramers.
Conclusion: Protein binding by DNMT3A or DNMT3L at the tetramer interface controls the catalytic properties of DNMT3A.
Significance: Provides a structural mechanism for the regulation of methylation patterning.
Keywords: DNA methylation, Epigenetics, Protein complexes, Protein-DNA interaction, Protein-protein interaction, DNMT3A, DNMT3L, Oligomerization, Processivity
Abstract
DNMT3A is one of two human de novo DNA methyltransferases essential for regulating gene expression through cellular development and differentiation. Here we describe the consequences of single amino acid mutations, including those implicated in the development of acute myeloid leukemia (AML) and myelodysplastic syndromes, at the DNMT3A·DNMT3A homotetramer and DNMT3A·DNMT3L heterotetramer interfaces. A model for the DNMT3A homotetramer was developed via computational interface scanning and tested using light scattering and electrophoretic mobility shift assays. Distinct oligomeric states were functionally characterized using fluorescence anisotropy and steady-state kinetics. Replacement of residues that result in DNMT3A dimers, including those identified in AML patients, show minor changes in methylation activity but lose the capacity for processive catalysis on multisite DNA substrates, unlike the highly processive wild-type enzyme. Our results are consistent with the bimodal distribution of DNA methylation in vivo and the loss of clustered methylation in AML patients. Tetramerization with the known interacting partner DNMT3L rescues processive catalysis, demonstrating that protein binding at the DNMT3A tetramer interface can modulate methylation patterning. Our results provide a structural mechanism for the regulation of DNMT3A activity and epigenetic imprinting.
Introduction
DNA methylation is a critical epigenetic modification controlling chromatin stability, genomic imprinting, cellular differentiation, transcriptional regulation, and memory formation in mammals (1–3). Initial (de novo) 5-methylcytosine patterning is established by DNMT3A2 and the closely related DNMT3B (4, 5), and aberrant 5-methylcytosine patterning is highly correlated with oncogenesis (6). The mechanism by which these enzymes are directed to methylate-specific CpG sites in the genome remains unclear, but methylation patterning is likely mediated by the formation of higher order complexes. One known example is the DNMT-like protein, DNMT3L, a processivity factor (7, 8), that forms heterotetramers with DNMT3A to modulate the catalytic mechanism of DNMT3A (9–11). During early development, de novo methylation patterning is dependent on the presence of DNMT3L (12). Further characterization of the oligomeric states of DNMT3A is necessary for understanding how DNA methylation is established in mammalian genomes.
The recent crystal structure of the catalytic domain of DNMT3A with the catalytically inactive DNMT3L shows a DNMT3L·DNMT3A·DNMT3A·DNMT3L heterotetramer complex (9). The complex shows specific contacts at the DNMT3A homodimer interface (dimer interface), and dimerization brings two enzyme active sites separated by approximately one helical turn, in B-form DNA (9). Still unclear is the physical basis of the DNMT3A homotetramer that is observed in gel filtration experiments in the absence of DNMT3L. Several lines of evidence suggest that the DNMT3A homotetramer is formed by DNMT3A monomers assuming the position occupied by DNMT3L in the heterotetramer crystal structure (tetramer interface) (13). Although most characterized C5 methyltransferases function as monomers (14), the disruption of tetramerization and loss of activity with the Phe-728 → Ala DNMT3A mutant led to the proposal that tetramerization is essential for methylation (9). However, homotetramerization does not provide additional DNA binding surfaces, nor does it bring more active sites into contact with the substrate, and it is not clear why tetramerization should be essential for catalysis.
Recent studies have demonstrated that more aggressive forms of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) are correlated with mutations in DNMT3A (15–17), including three at the tetramer interface (9). Patients with DNMT3A mutations have altered methylation patterns without a decrease in global DNA methylation levels (16). We hypothesized that DNMT3A is functional in the homodimeric form, and mutations at the tetramer interface disrupt the ability of regulatory proteins to bind to the DNMT3A homodimer, resulting in aberrant methylation patterning. We sought to evaluate the functional significance of DNMT3A oligomerization at the tetramer interface and understand how the regulatory protein DNMT3L modulates DNMT3A function.
A homotetramer DNMT3A model was developed for the design of side-chain mutations that disrupt tetramerization while minimizing changes in catalysis. Mutations at five individual residues are shown by light scattering and electrophoretic mobility assays to disrupt homotetramerization. Four mutants are dimeric on DNA and have activity comparable to the wild-type homotetramer but with altered catalytic properties. Homotetramers carry out multiple cycles of methylation on the same piece of DNA (processive catalysis), whereas disruption of the tetramer interface results in faster product release, resulting in non-processive catalysis. Dimeric mutants are able to bind DNMT3L to form heterotetramers, decreasing the rate of product release, and thus restoring processive catalysis. Here we demonstrate the functional consequence of DNMT3A oligomerization and offer insights into the control of this critical epigenetic regulator.
EXPERIMENTAL PROCEDURES
Protein Cloning and Purification
The catalytic domain of DNMT3A and full-length DNMT3L were purified as stated in Holz-Schietinger et al. (8), and M.HhaI was purified as stated in Matje et al. (18). Plasmids used for protein expression and site-directed mutagenesis include codon-optimized pET28a-hDNMT3A_CD (Δ1–611) (19), pET28a-M.HhaI, and pTYB1–3L for hDNMT3L (13). Briefly, protein expression occurred in Escherichia coli Rosetta2 (DE3) for hDNMT3L, M.HhaI, and wild-type DNMT3A catalytic domain and their mutants were expressed in T7Iq cells, induction occurred at 0.6 A600 at room temperate for 5 h with 1 mm IPTG. DNMT3A and M.HhaI were purified from BioRex and nickel affinity columns, DNMT3L was purified from nickel affinity and chitin columns, and all were purified to >95%. EcoRV was prepared as described in Hiller et al. (20). Three different wild-type DNMT3A preparations were compared and showed <5% difference in kcat. The catalytic domain of DNMT3A has similar kinetic parameters as the full-length enzyme, including kcat, KmDNA, KmAdoMet, processivity, and DNMT3L activation (8, 13), and was used for the DNMT3A/DNMT3L co-crystal structure (9). Both the homo- and DNMT3L hetero-oligomerization interfaces are located on the catalytic domain.
Mutagenesis Design and Computational Modeling
DNMT3A chain A of the PDB file 2QRV (9) was kept rigid, whereas the backbone of chain D was aligned to the backbone of DNMT3L chain B using PyMOL (DeLano Scientific LLC). Chain D was then dragged into a random orientation ∼10 Å away from the tetramer interface of chain A. The complex was submitted to the RosettaDock Server (21) to generate the initial tetramer interface model. After visual inspection, the best scoring models were re-submitted until all iterations repeatedly converged on a common model. This model was submitted to the Rosetta computational alanine scanning server (22, 23) to evaluate the contribution of individual residues to the tetramer interface. The side chains of residues Arg-771, Glu-733, and Arg-720 were manually optimized by choosing backbone-dependent rotamer conformations that gave the highest scores using the computational alanine-scanning protocol.
DNA Sequences
The DNA used as substrates include duplex poly(dI-dC) (∼1000 bp, Sigma-Aldrich), plasmid pCpGL (24) (non-CpG substrate), and fluorescent DNA with fluorescein (6-FAM) on the 5′-end of the top strand, GCbox30 (5′/6-FAM/TGGATATCTAGGGGCGCTATGATATCT-3′) duplex and were purchased from Integrated DNA Technologies, HPLC-purified. The recognition site for DNMT3A is underlined.
Size-exclusion Chromatography Coupled to Multiangle Light Scattering
Protein samples were passed through Bio-Rad P6 resin pre-equilibrated with light scattering buffer (50 mm KH2PO4/K2HPO4, pH 7.8, with 200 mm NaCl, 1 mm DTT, 1 mm EDTA, and 0.2% azide), then 100 μl of this sample (∼30 μm of monomeric enzyme) was injected into an analytical size-exclusion silica gel chromatography column (300-Å pore size, Wyatt Technologies) with a flow rate of 0.5 ml/min. The sample was run through a Waters 996 diode array measuring A260 and A280, optilab T-rEx differential refractometer (Wyatt), and triple-angle light-scattering detector miniDaWN TREOS (Wyatt). Molecular weight was calculated using ASTRA 5.3.4 software using a dn/dc value of 0.185. Standard error was reported from at least two separate samples.
Native Gel Mobility DNA Shift Assays
DNMT3A wild-type, mutants (150 nm or varied as indicated), or size control standards (M.HhaI and EcoRV, 150 nm) were incubated at room temperature for 15 min with 200 nm duplex 5′ 6-FAM GCbox30 that contains a single-site central CG site (DNMT3A) along with two sites for EcoRV and one site for M.HhaI binding. For DNMT3L supershifting, DNMT3L was preincubated for 30 min with DNMT3A before the addition of DNA. Binding occurred in reaction buffer with 5 μm S-adenosyl-l-homocysteine. Samples were run on native 4.5% polyacrylamide gels in 0.5× Tris-Boric acid EDTA, pH 7.8, at 250 V for 35 min, visualized for fluorescein using a Typhoon scanner, and data were analyzed using ImageJ (25).
Methylation Assays
DNMT3A methylation assays measured the amount of tritiated methyl groups transferred from cofactor S-adenosyl-l-methionine (AdoMet) to the DNA by the enzyme. Reactions were carried out at 37 °C in reaction buffer (50 mm KH2PO4/K2HPO4, 1 mm EDTA, 1 mm DTT, 0.2 mg/ml BSA, and 20 mm NaCl at pH 7.8). Enzyme was at 150 nm total, which is 27 nm active enzyme (previously determined in Purdy et al. (19)). DNA was the multiple site substrate poly(dI-dC), used at saturation for kcat, KmAdoMet, and DNMT3L activation. AdoMet was used at saturation for kcat, KmDNA, DNMT3L activation, and processivity. Processivity assays and DNMT3L activation assays were carried out as described in Holz-Schietinger et al. (8). Briefly, processivity assays involve three separate reactions; positive control (20 μm base-pairs (bp) substrate), experiment (substrate followed at 20 min by chase DNA (pCpGL) at 20-fold excess over substrate concentration), and negative control (chase and substrate at the start of reaction). Varying DNMT3L 0.25- to 4-fold the concentration of DNMT3A was used to test DNMT3L activation. DNMT3A and DNMT3L were preincubated in reaction buffer with AdoMet for 1 h at 37 °C before starting the reaction with DNA. A 1:1 ratio with a 1-hour preincubation (with AdoMet) was used for all DNMT3L assays. All reactions were quenched by addition of 0.1% SDS. Samples were spotted onto Whatman DE81 filters then washed, dried, and counted as described previously. For Km values data were fit to Michaelis-Menten equation, kcat, and processivity data were fit to either linear regression or a fit to a nonlinear regression using Prism v5 (GraphPad). The fold stimulation was calculated by dividing the amount of product formed by DNMT3A with DNMT3L by product formed by DNMT3A without DNMT3L. Error bars are standard error from three reactions. Bar charts of kinetic values compared mutants to wild-type using one-way analysis of variance to determine p value using Prism. M.HhaI kchem and Kd assays carried out as described in Matje et al. (18), Km, and kcat values determined as described in Lindstrom et al. (26).
Fluorescence Anisotropy
koff values were determined with enzyme (250 nm) and 5′ 6-FAM-labeled GCbox30 duplex DNA at 20 nm, reaching maximum anisotropy, followed by adding in chase DNA (unlabeled GCbox30) at 100× concentration of labeled DNA. The decrease in anisotropy was measured with time on a PerkinElmer LS 55 fluorescence spectrometer using Fl Winlab software. Data were fit to a one-phase exponential decay (Y = span e−kt + plateau) using GraphPad, from two independent experiments.
RESULTS
Generation of a DNMT3A Homotetramer Model
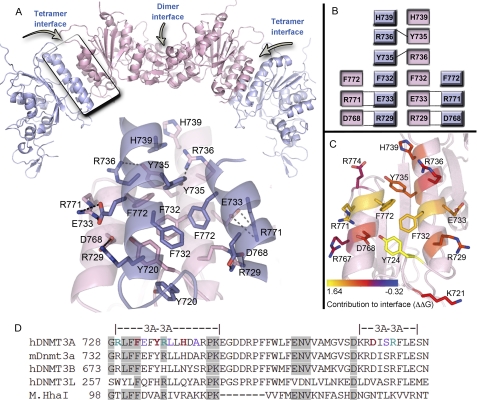
A DNMT3A homotetramer model (Fig. 1, A–C) was generated for experimental evaluation by aligning the Cα backbone of a DNMT3A monomer to the DNMT3L position in the 2QRV (9) crystal structure (root mean square deviation = 1.4 Å; supplemental Fig. S1A). This rough model was refined over several 1000 model iterations with the RosettaDock server (18) to optimize side-chain interactions at the tetramer interface. After numerous simulations converged upon the same model (Fig. 1A), the refined complex was submitted to the (22, 23) to obtain approximations for the energetic contributions of individual residues to the stabilization of this interface (ΔΔG). A summary of the homotetramer model and the contributions to tetramer stability are shown in Fig. 1, supplemental Fig. S3, and supplemental Table S4.
FIGURE 1.
DNMT3A homotetramer model. A, a homotetramer model was generated by aligning a DNMT3A monomer to DNMT3L (from PDB: 2QRV) followed by prediction of the lowest energy orientation using RosettaDock. Below is a close-up of the DNMT3A·DNMT3A tetramer interface showing the core residues and the predicted interactions. The interface shows an aromatic pocket in the center of the interface, with ionic interactions on each edge of the interface. B, depiction of the interactions at the DNMT3A·DNMT3A tetramer interface identified from the model. C, DNMT3A residues for one molecule (pink) of the tetramer interface. Residues are colored based upon their contribution, in ΔΔG, to the tetramer interface compared with alanine as determined using the Rosetta interface alanine-scanning module. Bright yellow residues provide the greatest contribution to the DNMT3A·DNMT3A interface. D, multiple sequence alignment of DNMT3A tetramer interface with related DNMTs and monomeric bacterial homolog (M.HhaI). Red and green residues were mutated in this study to alanine; green and purple residues are mutated in AML or MDS patients. Shaded regions show conserved residues between DNMT3A to M.HhaI.
Computational alanine screening details an aromatic core at the center of the interface providing the highest energetic contribution to tetramerization, with several salt bridges surrounding the core also having significant contributions. One residue with a high ΔΔG score, Phe-732, has previously been shown to stabilize the DNMT3A homotetramer (9). Mutation to alanine results in a large reduction in methylation activity, which was interpreted to demonstrate that tetramerization is required for catalysis by DNMT3A. However, Phe-732 resides at a structurally well conserved region of the catalytic domain. Although the DNMT3A catalytic domain (Δ1–611) has just 13% sequence identity to the well characterized homolog M.HhaI, the tertiary structures show a remarkably small backbone root mean square deviation of 2.3 Å across 220 residues of the catalytic domains (supplemental Fig. S1B). Sequence alignments of DNMT3A with monomeric bacterial C5 DNA methyltransferases reveal four conserved residues along the DNMT3A tetramer interface (Fig. 1D and supplemental Table S1) among otherwise highly divergent sequences. Of special interest is conservation of the LFF motif found at the hinge of the catalytic loop in both enzymes that includes Phe-732 (Fig. 1D).
To determine whether this conserved residue might be directly involved in catalysis in a monomeric DNA methyltransferase, we removed the side chains of the homologous residues in M.HhaI. Phe-101 and Phe-102 in the M.HhaI cognate·ternary complex (27) are nearly isomorphous with Phe-731 and Phe-732 in the DNMT3A complex (supplemental Fig. S2A). The purified F101A and F102A M.HhaI mutants showed respective decreases in kcat of 8.3- and 13.4-fold from the wild-type enzyme (supplemental Table S2). Preparations of these mutants were also found to be significantly less stable than the wild-type enzyme preparations (data not shown). Analogously, the F732A mutation in DNMT3A likely impacts critical structural elements within DNMT3A as well as tetramerization, leaving unanswered whether dimeric DNMT3A forms retain activity.
Six residues with varying ΔΔG scores and weak conservation to bacterial homologs were investigated for their contribution to tetramerization, including two residues correlated with AML and MDS syndromes (Arg-771 and Arg-729) (supplemental Table S1). Two conserved residues, Arg-736 and Phe-732, were also mutated to alanine for comparison. We found that mutation of residues with ΔΔG values of >1.0 (the combined score from the residue of the outer and inner monomers (supplemental Table S3)) resulted in the formation of homodimers on DNA, whereas residues with ΔΔG values of <1.0 remained tetramers on DNA. The correlation between the computed ΔΔG values and our experimental results (below) supports the use of the Rosetta modeling and scoring algorithms to generate the DNMT3A tetramer model.
Disruption of the Tetramer Interface Results in Monomers in Solution That Dimerize on DNA
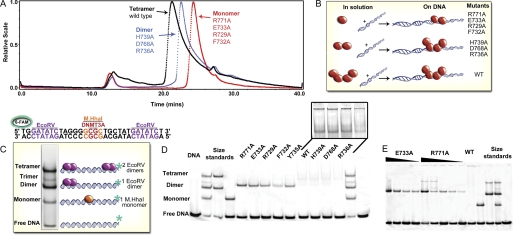
We combined size exclusion chromatography with multiangle light scattering to evaluate the oligomeric state of wild-type DNMT3A and interface mutants. Light scattering shows our preparations of wild-type DNMT3A catalytic domain to have a weight-average molecular mass of 127 kDa in solution, in agreement with four 36-kDa monomers forming a tetramer (Fig. 2A) (9). However, the broad, tailing peak suggests the tetrameric form is in equilibrium with both the dimeric and monomeric enzyme forms on the column. A cofactor product, S-adenosyl-l-homocysteine, did not affect the form of the enzyme (supplemental Fig. S4B). Surprisingly, all alanine mutations at the interface disrupted the oligomeric state of the enzyme observed by gel filtration and multiangle light scattering (Fig. 2A, supplemental Table S3, and supplemental Fig. S4). Three of the mutants had weight-average molecular masses slightly larger than the theoretical dimer mass (H739A, D768A, and R736A). Four mutants (F732A, R771A, E733A, and R729A) have approximate molecular masses close to the theoretical mass of a DNMT3A monomer (Fig. 2A). These monomeric mutants have very little tailing in their elution profiles and elute from the column slower than dimers and tetramers.
FIGURE 2.
DNMT3A biophysical characterization. A, light scattering data of DNMT3A mutations along the tetramer interface shows that mutants in solution are either mostly monomer or dimers unlike wild-type tetramers. Size-exclusion chromatography results are shown for light scattering traces of tetrameric wild-type catalytic domain (black trace), representative dimeric H739A (blue trace), and representative dimeric/monomeric R771A (red trace). Molecular weights were determined from the amount of scattered light, in relation to protein concentration determined by A280. B, diagram of oligomeric mutants with and without DNA, showing that DNA facilitates oligomerization for DNMT3A mutants via the dimer interface. C, electrophoretic mobility assay of size markers; DNA (GCbox30) has binding sites for size standards, one site for M.HhaI (37 kDa), a known monomer and two binding sites for EcoRV (29 kDa), a known dimer, which creates a dimer and ∼ tetramer band. D, DNA facilitates oligomerization for DNMT3A mutants; five mutants were dimers on DNA. The distance the fluorescently labeled duplex DNA (GCbox30) shifted with DNMT3A determined the oligomeric state. E, electrophoretic mobility assay varying the concentration of R771A and E773A (20–400 nm) with DNA at 200 nm demonstrating these mutates are dimers even at low concentrations, unlike the monomers seen in solution at 200× the concentration.
We also investigated if binding to DNA changes the oligomeric state of wild-type and mutant enzymes. Gel filtration of the wild-type DNMT3A catalytic domain·DNA complex was attempted but resulted in large aggregates with equimolar and 2-fold excess DNA concentrations (supplemental Fig. S4B) at the micromolar concentrations needed for detectable multiangle scattering. As a control, M.HhaI was tested with and without DNA, showing distinct peaks corresponding to monomeric bound or free states, respectively (supplemental Fig. S4A).
To resolve the oligomeric states of DNMT3A with DNA at concentrations closer to cellular conditions and in vitro assay conditions, electrophoretic mobility gel shift assays using a 30-bp, fluorescently labeled deoxyoligonucleotide (GCbox30) were performed. The GCbox30 contains binding sites for two well behaved proteins; a central GCGC site bound by the monomeric M.HhaI (37 kDa) and two GATATC sites bound by the homodimeric EcoRV endonuclease (29 kDa) (28) at either end (Fig. 2B). At equimolar concentrations of protein and DNA, one or two EcoRV dimers bind to the DNA, resulting in standards for dimers and tetramers, respectively (supplemental Fig. S5A), whereas M.HhaI on DNA represents the monomer standard.
The GCbox30 has a single central CG site that is the major recognition site of DNMT3A (29). Fig. 2C shows the formation of discrete bands when wild-type and mutant DNMT3A enzymes are incubated with GCbox30. Wild-type enzyme bound to DNA as a tetramer, and smearing in the higher molecular mass region and titrating enzyme concentration from 40 to 360 nm did not change this pattern (supplemental Fig. S5B). H739A, D768A, and R736A formed tetramers or larger aggregate forms on the DNA but were dimeric by light scattering. Five mutants (R771A, E733A, R729A, F732A, and Y735A) shift the DNA closest to the dimer size standard (EcoRV dimer: 58 kDa, DNMT3A dimer: 68 kDa). Fig. 2D shows that titration of the E733A and R771A mutant enzyme concentrations down to 40 nm in the presence of 200 nm CpG sites resulted in only the presence of dimeric enzyme bound to the DNA (see also supplemental Fig. S5C). These results clearly show that DNA facilitates the dimerization of DNMT3A mutants that are monomeric in the absence of DNA even at 200-fold higher concentrations (Fig. 2A). When enzyme was present at higher concentrations than DNA, bands indicative of tetramers became observable from tetramerization of the enzyme or via multiple dimers binding to a single DNA substrate (Fig. 2D). The oligomeric states of the tetramer interface mutants are summarized in Fig. 2E.
Dimers Are Fully Active with an Altered Mechanism
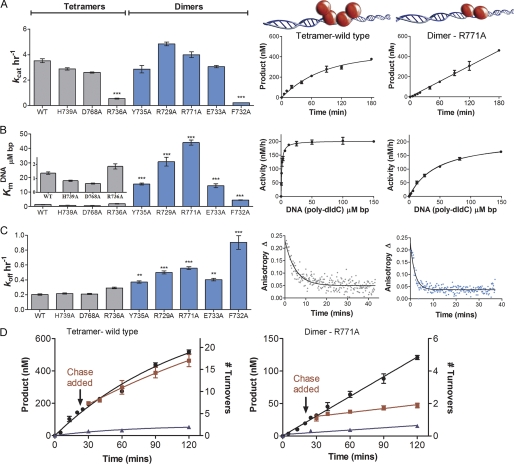
DNMT3A mutants were examined for changes in their steady-state kinetic parameters by monitoring the transfer of tritiated methyl groups from cofactor AdoMet onto DNA. kcat, KmDNA, KmAdoMet, and processivity values were obtained for all the mutants and compared with wild-type enzyme (supplemental Table S5). kcat values were obtained at saturating DNA and AdoMet concentrations after determining the respective Km values. In agreement with previous data (8), wild-type DNMT3A shows curvature in the plot of product formed over time (Fig. 3A). This is due to the slow product-release step of this processive enzyme on this multisite DNA substrate, where the enzyme partitions between migrating along the DNA to continue carrying out catalysis and dissociation from the DNA into solution (30). This same curvature was seen for the H739A and D768A mutants that did not disrupt tetramer formation, and these mutants differed < 10% in their kcat values from the wild-type value of 3.2 h−1 (Fig. 3A and supplemental Fig. S6). The other mutant that appeared tetrameric by gel shift, R736A, had a 5-fold decrease in kcat. However, Arg-736 was homologous to the previously investigated Arg-106 in M.HhaI that had a 2.5-fold decrease in kcat when mutated to alanine (supplemental Fig. S2B) (31).
FIGURE 3.
DNMT3A homodimers and homotetramers are both active with different mechanisms. A, dimers and tetramers have similar rates of reaction. Left: bar chart of kcat values for oligomeric mutants, right: representative time course for tetramer (wild type) showing curved product formation and dimer (R771A) showing linear product formation. B, eliminating tetramer formation increases KmDNA. Left: bar chart of KmDNA values for oligomeric mutants, right: representative tetramer and dimer curves. C, DNMT3A homodimers had an increase in off-rate compared with homotetramers. Left: bar chart of koff values for oligomeric mutants; koff values were determined by the enzyme being bound to fluorescein-labeled GCbox30 DNA, then adding saturating unlabeled DNA. Right: representative tetramer and dimer curves. D, DNMT3A homotetramers are processive, and dimers are non-processive. Representative tetramer and dimer processive chase assay data; ●, only substrate, 20 μm bp poly(dI-dC); ■, substrate and then 400 μm bp chase (pCpGL) at 20 min; ▴ = substrate and pCpGL at the start of the reaction. No methylation was detected after addition of chaser DNA with the dimer mutant (R771A), unlike tetramer (WT), which shows less than 10% change in activity. All error bars are at least three experiments given as ±S.E., one-way analysis of variance was used to compare wild-type values to each mutant; *, p > 0.05; **, p > 0.01; and ***, p > 0.001.
The mutants that form dimers on DNA (R771A, E733A, R729A, F732A, and Y735A) all showed linear product formation with time, unlike the curved product formation seen for the tetramers (Fig. 3A and supplemental Fig. S6A). Linear product formation with a y-intercept of zero is characteristic of an enzyme where product release is no longer the rate-limiting step on a substrate with multiple methylation sites (32). A less likely possibility is that the enzyme is 100% processive and does not dissociate from the DNA in the 3-h reaction. The kcat values for R771A and R729A were slightly greater than wild type, whereas E733A and Y735A have kcat values slightly less than the wild-type enzyme (supplemental Table S5). F732A, previously reported to have no activity (9), was found to have a 15.2-fold decrease in kcat relative to the wild-type enzyme. This reduction is very similar to the 13.4-fold kcat reduction seen in the F102A M.HhaI mutant (supplemental Table S2). Indeed, the two mutants that had significant decreases in kcat, F732A and R736A, are residues conserved in the monomeric bacterial homolog M.HhaI (supplemental Table S1 and supplemental Fig. S2). Because mutation of the Phe-102 and Arg-106 residues to alanine caused reduction in the catalytic turnover in M.HhaI (supplemental Table S2), it is unlikely the kinetic consequences of mutation of these residues in DNMT3A are a result of altered oligomeric forms. The four mutants that function as dimers have kcat values similar to wild type, demonstrating DNMT3A is a functional dimer and tetramerization is not necessary for catalysis, although the two forms have distinct kinetic mechanisms (see below).
KmAdoMet for wild-type DNMT3A is 206 nm, and similar values were found for D768A and H739A. Mutants with larger decreases in activity have a 5- to 6-fold increase in KmAdoMet (R736A and F732A) and the other dimers showed at 2- to 3-fold increase (supplemental Fig. S7, B and C). The minimal effect on cofactor binding indicates oligomerization does not result in large pleiotropic affects to catalysis.
Dimers Rapidly Dissociate from DNA
Wild-type DNMT3A has a KmDNA of 1.2 μm bp on poly(dI-dC) (19). The four mutants are fully active as dimers, and all have an increase in KmDNA from wild type between 10- and 32-fold (Fig. 3B, supplemental Table S5, and supplemental Fig. S7A). These results demonstrate that oligomerization at the tetramer interface has important consequences for DNA binding. The observed increase in KmDNA indicates a perturbation to substrate binding and/or product dissociation in the dimer mutants. To characterize the effect of oligomerization on dissociation from DNA, dissociation rates (koff) were determined by binding excess enzyme to 5′ 6-FAM-labeled GCbox30 duplex and measuring the rate of change in anisotropy upon the addition of unlabeled GCbox30 (Fig. 3C). Change in fluorescence anisotropy was plotted against time and fit to a single exponential decay to give the koff value. Wild-type DNMT3A has a koff of 0.20 min−1 (supplemental Table S5 and supplemental Fig. S8), H739A and D768A show similar rates, and R736A dissociates more quickly, 0.32 min−1. The dimeric mutants dissociate 2–5 times faster than the wild-type enzyme (Fig. 3C), indicating that the linear product formation observed for the dimer mutants (Fig. 3A) is most likely due to the increase in koff. The increase in the rate of dissociation from product also likely contributes to the observed increase in KmDNA in the dimeric mutants (Fig. 3B). Changes in processive catalysis could account for changes in in vivo methylation patterns such as those observed in AML patients with minor changes in overall methylation (28). We sought to directly relate the oligomeric form of the enzyme to changes in processivity.
The Tetramer Interface Controls Processivity
Both full-length and the catalytic domain of wild-type DNMT3A act processively on several DNA substrates, including human promoters (8). Through mathematical modeling it was determined that the enzyme carries out an average of 27 turnovers (n1/2 = 27) before 50% of the enzyme dissociates from DNA (poly(dI-dC)) (8). Tetrameric H739A and D768A have similar processivity values as wild type (n1/2 = 22 and 20, respectively (supplemental Table S6)), in agreement with their unchanged koff. The highly linear formation of product over time with the dimer mutants (Fig. 3A and supplemental Fig. S6) made fitting to the processivity equations impossible, because the multiexponential equation cannot converge to fit the linear data. Because linear product formation is observed for highly processive enzymes like M.SssI (8), we sought to qualitatively examine processivity in the dimer mutants using a chase assay (8, 30).
The processivity assay determines the length of time the enzyme stays associated with the multiple site DNA substrate and the number of turnovers using a chase experiment. The chase assay begins with DNMT3A carrying out one to two turnovers, ∼20 min, on poly(dI-dC) DNA. After this time, a 25-fold excess of pCpGL, a 3872-bp plasmid lacking the recognition site (CG) for DNMT3A, is added to capture enzyme that dissociates from the substrate after carrying out catalysis. DNMT3A shows minimal methylation activity with pCpGL (kcat 0.11 ± 0.03 h−1), and when pre-mixed at 25-fold excess poly(dI-dC), it eliminated 85% of product formation. A processive enzyme will continue to methylate the original, multisite substrate after chase DNA is added and addition of chase DNA has little, or delayed, impact on the rate of methylation (wild-type DNMT3A, Fig. 3D). A non-processive enzyme will dissociate after one methyl transfer; thus, after the addition of chase DNA, the enzyme will bind the excess pCpGL and cause an immediate decrease in product formation (see R771A, Fig. 3D).
Wild-type DNMT3A, H739A, and D768A show unaltered methylation 90 min after chase DNA is added. In contrast, the mutants that exist as dimers on DNA (supplemental Table S3) showed an immediate decrease in activity upon addition of the chase DNA. This indicates that these mutants no longer acted processively (supplemental Fig. S10), showing that tetramerization has a profound impact on the catalytic properties of DNMT3A. The enzymes with large kcat changes were not tested for processivity changes.
DNMT3L Binding to DNMT3A Dimers Restores Processivity
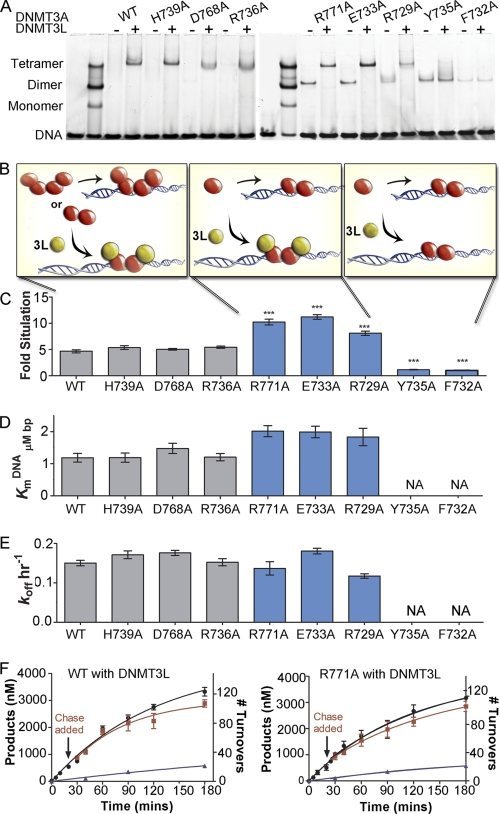
The DNMT3A·DNMT3L heterotetramer complex formed a discrete band when bound to the fluorescently labeled GCbox30 substrate in the electrophoretic mobility assay (Fig. 4A). Adding an equal concentration of DNMT3L eliminated any wild-type DNMT3A homotetramer band and formed a strong heterotetramer band (Fig. 4A). This discrete DNMT3A·DNMT3L band was seen with all interface mutants except F732A and Y735A. For dimer mutants R771A, E733A, and R729A, there was a loss of the dimer band and a gain of a heterotetramer band; gel shift results are detailed in Fig. 4B. These results are in good agreement with computational alanine screening of the heterotetramer interface that describes the central aromatic pocket (including Phe-732 and Tyr-735) as critical for the DNMT3A·DNMT3L interface, with more modest contributions from salt bridges flanking the hydrophobic core (supplemental Fig. S11). Like the homotetramer interface, disruption of residues with a ΔΔG value >1.2 resulted in loss of heterotetramerization (supplemental Tables S3 and S7).
FIGURE 4.
DNMT3L heterotetramers restores processivity in DNMT3A dimer mutants. A, DNMT3L binds to DNMT3A tetramers, forming defined heterotetramer. Three dimer mutants become heterotetramers with DNMT3L resulting in the loss of dimers; Y735A and F732A do not bind DNMT3L. B, schematic of the three forms that occurs with the addition of DNMT3L, how the forms change function is below. C, DNMT3L (1:1 ratio) actives DNMT3A tetramers ∼5-fold. Homodimers that bind DNMT3L were activated ∼10-fold; Y735A and F732A saw no activation by DNMT3L. D, heterotetramer formation decreased KmDNA for dimer mutants, and no change for homotetramers. E, DNMT3A heterotetramers had a decrease in off-rate compared with homotetramers and homodimers. All heterotetramers have similar rates. F, all DNMT3A heterotetramers are processive as demonstrated by the processive chase assay; ●, only substrate, 20 μm bp poly(dI-dC); ■, substrate and then 400 μm bp chase (pCpGL) at 20 min; ▴, substrate and pCpGL at the start of the reaction. Data indicates 100 turnovers occur before the enzyme dissociates from the DNA. All error bars are at least three experiments given as ±S.E., one-way analysis of variance was used to compare wild-type enzyme to mutants; *, p > 0.05; **, p > 0.01; and ***, p > 0.001.
DNMT3L activates DNMT3A in vitro (13) and in vivo (10) and increases processivity 3-fold (8). DNMT3L activates DNMT3A ∼5-fold at a 1:1 ratio with preincubation (8, 13) (supplemental Fig. S9A). Preincubation of DNMT3L with wild-type DNMT3A is needed before DNA is added for heterotetramer formation and activation, presumably to allow exchange of the outer two DNMT3A monomers for DNMT3L (13). Homotetramers H739A, D768A, and R736A also showed a 5-fold increase in activity when incubated with equimolar or greater concentrations of DNMT3L. Dimer mutants partitioned into two groups; activation close to 10-fold (E733A, R771A, and R729A) or no activation (F732A and Y735A) (Fig. 4C and supplemental Fig. S9). F732A and Y735A are the two residues predicted to be the most important for the DNMT3L interface (supplemental Table S3) and experimentally are the only mutants that showed no activation and no tetramerization. In addition E733A, R771A, and R729A did not require preincubation to be activated by DNMT3L and could be preincubated with DNA and were still activated unlike the tetramers (data not shown).
The addition of DNMT3L to wild-type DNMT3A enhances kcat, and KmAdoMet and leaves KmDNA unchanged (19). In contrast, DNMT3L regulates the KmDNA for the functional dimers, which in the absence of DNMT3L showed a 15- to 30-fold increase compared with wild type, and heterotetramerization restored the KmDNA to only 1.2- to 1.6-fold greater than wild-type enzyme (Fig. 4D, supplemental Fig. S9B, and supplemental Table S8). These data show that the changes in KmDNA did not result from these residues being directly involved in DNA binding, but rather through the disruption of homo- or heterotetramerization.
As shown above, DNMT3A dimer mutants tested in this study had an increase in koff relative to wild type (Fig. 3C and supplemental Fig. S8). The addition of DNMT3L to wild-type DNMT3A caused koff to be decreased 25% from 0.2 min−1 to 0.15 min−1 (Fig. 4E and supplemental Table S8) on a single site substrate. Previous kinetic modeling showed that DNMT3L decreases DNMT3A off-rates more substantially on a multiple site substrate (8). DNMT3L decreased the dissociation rate for R771A, R729A, and E733A to wild-type levels.
Dimer mutants showed linear product formation with time, most likely because of an increase in the off-rate, resulting in kchemistry being the rate-limiting step. Because DNMT3L decreased the off-rate for DNMT3A dimer mutants (Fig. 4F and supplemental Fig. S8), we predicted that DNMT3L would rescue the curved product formation of a processive enzyme, like the homotetramers. As expected, dimer mutants with DNMT3L (E733A, R771A, and R729A) all showed curved product formation with time like wild-type DNMT3A (Fig. 4F and supplemental Fig. S12).
The chase assay was also used on DNMT3A mutants with DNMT3L for further qualitative assessment of processivity, with DNMT3A wild-type and mutants all showing an increase in processivity. Dimer mutants R771A, E733A, and R729A were all non-processive without DNMT3L. When DNMT3L was added, these three mutants become highly processive like the wild-type DNMT3A·DNMT3L complex. As shown in Fig. 5E the addition of chase DNA had little effect on product formation even over 100 methylation cycles for all DNMT3A·DNMT3L complexes (supplemental Fig. S12).
FIGURE 5.
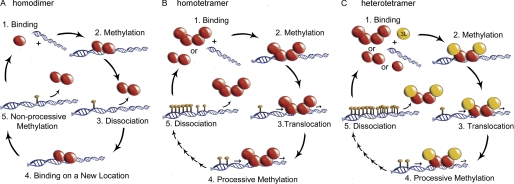
Diagram of DNMT3A oligomeric states altering methylation patterns through changes in processivity. A, homodimers (R771A, R729A, E733A, Y735A, and F732A) formed by disrupting the tetramer interface resulted in enzymes that bind DNA, methylation, fast dissociation, and rebinding a new piece of DNA (non-processive catalysis). B, homotetramers (WT, H739A, D768A, and R736A) bind DNA followed by methylation then translocation along the same piece of DNA to a new site that the enzyme methylates, thus carrying out processive catalysis multiple times before slow dissociation. C, DNMT3A·DNMT3L heterotetramers (WT, H739A, D768A, R736A, R771A, E733A, and R729A) have increased processive catalysis compared with homotetramers and restores processive in the homodimer, thus carrying out processive catalysis multiple times before slow dissociation.
DISCUSSION
The regulatory mechanisms controlling DNMT3A targeting and activity remain poorly understood, but there is increasing evidence for the role of protein-protein interactions in establishment of 5-methylcytosine patterning (33). We investigated how oligomerization contributes to DNMT3A function by testing distinct oligomers, created by mutating several residues identified via computational modeling as critical toward formation of the homotetramer complex. The DNMT3A dimer is fully functional but dissociates from DNA more rapidly, eliminating methylation on clustered CpG sites (processive catalysis) that is observed with the wild-type homotetramer. Dimeric DNMT3A mutants that form heterotetramers with DNMT3L exhibit processivity like the wild-type heterotetramer complex (diagramed in Fig. 5). Our results demonstrate protein binding by DNMT3A, DNMT3L, or other regulators of DNMT3A at the tetramer interface is important for methylation patterning. Recently, mutations in DNMT3A were found in AML (16, 17) or MDS (15) patients along the tetramer interface (Fig. 1D and supplemental Table S10), and we showed that two of these residues (Arg-771 and Arg-729) are necessary for tetramerization.
The clustered loss (and gain) of promoter methylation at multiple CpG sites within CpG islands in these patients (34) is consistent with our in vitro observations involving DNMT3A mutants. Differences between oligomeric states provide a possible mechanism for how DNMT3A mutations in these cancer patients create aberrant DNA methylation patterns without global decreases in methylation levels (16).
Processive catalysis and its regulation by accessory proteins are critical to the action of several DNA-modifying enzymes (35–37). The control of DNMT3A processivity is relevant to the establishment of methylation patterning, because the degree of 5-methylcytosine content in promoter regions has important consequences for transcriptional regulation (1). Processive methylation of DNA has been demonstrated for the maintenance DNMT1 (30, 38) and both de novo DNMT3 enzymes in vitro (8, 10), with increasing evidence for this being the predominant mechanism of methylation in vivo. Recently, whole genome methylation analysis of human cells showed promoter regions to have bimodal distribution patterns, where promoters were either mostly methylated or unmethylated (39, 40). We show the major consequence of eliminating tetramerization in DNMT3A is a dramatic change in the number of processive cycles of methylation that occur each time the enzyme associates with DNA. There is also clear evidence that interactions with accessory proteins play an essential role in the establishment of methylation patterning (41, 42). DNMT3L is critical for early developmental de novo DNA methylation (2, 12), by binding DNMT3A at the tetramer interface (9), which serves to increase cofactor affinity (13) and catalytic turnover (13). This same interface is shown here to regulate DNMT3A processivity, thereby playing an essential role in coordinating genomic methylation. Because DNMT3L is only expressed in early development (12), DNMT3A either acts as a homo-oligomer or interacts with unidentified binding partners (43–46) that modulate the oligomeric state and activity in differentiated cells.
We propose that protein binding at the tetramer interface controls processivity through stabilization of the catalytic loop in the closed form. Cheng and Blumenthal suggested that binding of DNMT3L or DNMT3A at the tetramer interface locks the flexible catalytic loops residues (707–727) of the inner dimer in the closed position (47), which increases the contact area with DNA (supplemental Fig. S12). Increasing the binding surface is established to enhance processivity in other DNA-modifying enzymes (48–50) by decreasing product release and facilitating translocation along the substrate. In M.HhaI, the closure of this loop is correlated with tight binding of cofactor and DNA (18). The other human de novo methyltransferase, DNMT3B, may be regulated by a similar mechanism (10), because the tetramer interface is highly homologous with DNMT3A. Newly solved crystal structures of DNMT1 (51, 52) show a hydrophobic core at the interface analogous to the DNMT3A tetramer interface. Structures of mouse DNMT1 show that the catalytically essential CXXC domain (53) forms specific interactions across this interface, including along the catalytic loop (52). We propose that oligomerization at this structurally conserved interface is a general mechanism by which cis (CXXC domain)- and trans (DNMT3L)-acting interactions modulate the activity of the DNMT family. Further work is needed to identify and understand how binding partners (46) (including possibly regulatory RNA (54)), mutations, and/or cellular conditions drive oligomerization at the tetramer interface of DNMT3A that have important manifestations in the cellular phenotype.
Acknowledgments
We thank Frederick Dahlquist and John Perona for helpful discussions and critical reading of the manuscript and Armand Vartanian and Andrew Hadd for light scattering support. We are also thankful for Matt Purdy's work on M.HhaI F101A.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S13, Tables S1–S10, and references.
- DNMT
- DNA methyl transferase
- AML
- acute myeloid leukemia
- MDS
- myelodysplastic syndrome
- AdoMet
- S-adenosyl-l-homocysteine
- poly(dI-dC)
- poly-deoxyinosinic-deoxycytidylic
- 6-FAM
- fluorescein.
REFERENCES
- 1. Bird A. (2002) Genes Dev. 16, 6–21 [DOI] [PubMed] [Google Scholar]
- 2. Reik W., Dean W., Walter J. (2001) Science 293, 1089–1093 [DOI] [PubMed] [Google Scholar]
- 3. Wu H., Coskun V., Tao J., Xie W., Ge W., Yoshikawa K., Li E., Zhang Y., Sun Y. E. (2010) Science 329, 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Okano M., Bell D. W., Haber D. A., Li E. (1999) Cell 99, 247–257 [DOI] [PubMed] [Google Scholar]
- 5. Chen T., Ueda Y., Dodge J. E., Wang Z., Li E. (2003) Mol. Cell Biol. 23, 5594–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Robertson K. D. (2005) Nat. Rev. Genet. 6, 597–610 [DOI] [PubMed] [Google Scholar]
- 7. Van Emburgh B. O., Robertson K. D. (2011) Nucleic Acids Res. 39, 4984–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Holz-Schietinger C., Reich N. O. (2010) J. Biol. Chem. 285, 29091–29100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jia D., Jurkowska R. Z., Zhang X., Jeltsch A., Cheng X. (2007) Nature 449, 248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wienholz B. L., Kareta M. S., Moarefi A. H., Gordon C. A., Ginno P. A., Chédin F. (2010) PLoS Genet 6, pii: e1001106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chedin F., Lieber M. R., Hsieh C. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16916–16921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Webster K. E., O'Bryan M. K., Fletcher S., Crewther P. E., Aapola U., Craig J., Harrison D. K., Aung H., Phutikanit N., Lyle R., Meachem S. J., Antonarakis S. E., de Kretser D. M., Hedger M. P., Peterson P., Carroll B. J., Scott H. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4068–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kareta M. S., Botello Z. M., Ennis J. J., Chou C., Chédin F. (2006) J. Biol. Chem. 281, 25893–25902 [DOI] [PubMed] [Google Scholar]
- 14. Malygin E. G., Evdokimov A. A., Hattman S. (2009) Biol. Chem. 390, 835–844 [DOI] [PubMed] [Google Scholar]
- 15. Walter M. J., Ding L., Shen D., Shao J., Grillot M., McLellan M., Fulton R., Schmidt H., Kalicki-Veizer J., O'Laughlin M., Kandoth C., Baty J., Westervelt P., DiPersio J. F., Mardis E. R., Wilson R. K., Ley T. J., Graubert T. A. (2011) Leukemia 25, 1153–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ley T. J., Ding L., Walter M. J., McLellan M. D., Lamprecht T., Larson D. E., Kandoth C., Payton J. E., Baty J., Welch J., Harris C. C., Lichti C. F., Townsend R. R., Fulton R. S., Dooling D. J., Koboldt D. C., Schmidt H., Zhang Q., Osborne J. R., Lin L., O'Laughlin M., McMichael J. F., Delehaunty K. D., McGrath S. D., Fulton L. A., Magrini V. J., Vickery T. L., Hundal J., Cook L. L., Conyers J. J., Swift G. W., Reed J. P., Alldredge P. A., Wylie T., Walker J., Kalicki J., Watson M. A., Heath S., Shannon W. D., Varghese N., Nagarajan R., Westervelt P., Tomasson M. H., Link D. C., Graubert T. A., DiPersio J. F., Mardis E. R., Wilson R. K. (2010) N. Engl. J. Med. 363, 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shah M. Y., Licht J. D. (2011) Nat. Genet. 43, 289–290 [DOI] [PubMed] [Google Scholar]
- 18. Matje D. M., Coughlin D. F., Connolly B. A., Dahlquist F. W., Reich N. O. (2011) Biochemistry 50, 1465–1473 [DOI] [PubMed] [Google Scholar]
- 19. Purdy M. M., Holz-Schietinger C., Reich N. O. (2010) Arch. Biochem. Biophys. 498, 13–22 [DOI] [PubMed] [Google Scholar]
- 20. Hiller D. A., Fogg J. M., Martin A. M., Beechem J. M., Reich N. O., Perona J. J. (2003) Biochemistry 42, 14375–14385 [DOI] [PubMed] [Google Scholar]
- 21. Lyskov S., Gray J. J. (2008) Nucleic Acids Res. 36, W233-W238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kortemme T., Kim D. E., Baker D. (2004) Sci. STKE 2004, pl2. [DOI] [PubMed] [Google Scholar]
- 23. Kortemme T., Baker D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14116–14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klug M., Rehli M. (2006) Epigenetics 1, 127–130 [DOI] [PubMed] [Google Scholar]
- 25. Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Biophoton. Int. 11, 36–42 [Google Scholar]
- 26. Lindstrom W. M., Jr., Flynn J., Reich N. O. (2000) J. Biol. Chem. 275, 4912–4919 [DOI] [PubMed] [Google Scholar]
- 27. Shieh F. K., Youngblood B., Reich N. O. (2006) J. Mol. Biol. 362, 516–527 [DOI] [PubMed] [Google Scholar]
- 28. Winkler F. K., Banner D. W., Oefner C., Tsernoglou D., Brown R. S., Heathman S. P., Bryan R. K., Martin P. D., Petratos K., Wilson K. S. (1993) EMBO J. 12, 1781–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goll M. G., Bestor T. H. (2005) Annu. Rev. Biochem. 74, 481–514 [DOI] [PubMed] [Google Scholar]
- 30. Svedruzi Z. M., Reich N. O. (2005) Biochemistry 44, 14977–14988 [DOI] [PubMed] [Google Scholar]
- 31. Sharma V., Youngblood B., Reich N. (2005) J. Biomol. Struct. Dyn. 22, 533–543 [DOI] [PubMed] [Google Scholar]
- 32. Fersht A. (1999) Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding, Vol. xxi, pp. 103–113, W. H. Freeman, New York [Google Scholar]
- 33. Denis H., Ndlovu M. N., Fuks F. (2011) EMBO Rep. 12, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yan X. J., Xu J., Gu Z. H., Pan C. M., Lu G., Shen Y., Shi J. Y., Zhu Y. M., Tang L., Zhang X. W., Liang W. X., Mi J. Q., Song H. D., Li K. Q., Chen Z., Chen S. J. (2011) Nat. Genet. 43, 309–315 [DOI] [PubMed] [Google Scholar]
- 35. Chelico L., Pham P., Calabrese P., Goodman M. F. (2006) Nat. Struct. Mol. Biol. 13, 392–399 [DOI] [PubMed] [Google Scholar]
- 36. Ha T., Rasnik I., Cheng W., Babcock H. P., Gauss G. H., Lohman T. M., Chu S. (2002) Nature 419, 638–641 [DOI] [PubMed] [Google Scholar]
- 37. Walstrom K. M., Dozono J. M., von Hippel P. H. (1997) Biochemistry 36, 7993–8004 [DOI] [PubMed] [Google Scholar]
- 38. Vilkaitis G., Suetake I., Klimasauskas S., Tajima S. (2005) J. Biol. Chem. 280, 64–72 [DOI] [PubMed] [Google Scholar]
- 39. Grunau C., Hindermann W., Rosenthal A. (2000) Hum. Mol. Genet. 9, 2651–2663 [DOI] [PubMed] [Google Scholar]
- 40. Strichman-Almashanu L. Z., Lee R. S., Onyango P. O., Perlman E., Flam F., Frieman M. B., Feinberg A. P. (2002) Genome Res. 12, 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eckhardt F., Lewin J., Cortese R., Rakyan V. K., Attwood J., Burger M., Burton J., Cox T. V., Davies R., Down T. A., Haefliger C., Horton R., Howe K., Jackson D. K., Kunde J., Koenig C., Liddle J., Niblett D., Otto T., Pettett R., Seemann S., Thompson C., West T., Rogers J., Olek A., Berlin K., Beck S. (2006) Nat. Genet. 38, 1378–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hata K., Okano M., Lei H., Li E. (2002) Development 129, 1983–1993 [DOI] [PubMed] [Google Scholar]
- 43. Dennis K., Fan T., Geiman T., Yan Q., Muegge K. (2001) Genes Dev. 15, 2940–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klose R. J., Bird A. P. (2006) Trends Biochem. Sci. 31, 89–97 [DOI] [PubMed] [Google Scholar]
- 45. Li Y. Q., Zhou P. Z., Zheng X. D., Walsh C. P., Xu G. L. (2007) Nucleic Acids Res. 35, 390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hervouet E., Vallette F. M., Cartron P. F. (2009) Epigenetics 4, 487–499 [DOI] [PubMed] [Google Scholar]
- 47. Cheng X., Blumenthal R. M. (2008) Structure 16, 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Serebrov V., Beran R. K., Pyle A. M. (2009) J. Biol. Chem. 284, 2512–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Symmons M. F., Jones G. H., Luisi B. F. (2000) Structure 8, 1215–1226 [DOI] [PubMed] [Google Scholar]
- 50. Nudler E., Avetissova E., Markovtsov V., Goldfarb A. (1996) Science 273, 211–217 [DOI] [PubMed] [Google Scholar]
- 51. Song J., Rechkoblit O., Bestor T. H., Patel D. J. (2011) Science 331, 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Takeshita K., Suetake I., Yamashita E., Suga M., Narita H., Nakagawa A., Tajima S. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 9055–9059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pradhan M., Estève P. O., Chin H. G., Samaranayke M., Kim G. D., Pradhan S. (2008) Biochemistry 47, 10000–10009 [DOI] [PubMed] [Google Scholar]
- 54. Aravin A. A., Bourc'his D. (2008) Genes Dev. 22, 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]