Background: A variant of α1-antitrypsin with an E342K mutation (ATZ) is retained in the endoplasmic reticulum (ER) of hepatocytes.
Results: In hepatoma cells, activation of a branch of the unfolded protein response (UPR) increases ER-dependent degradation of ATZ.
Conclusion: ATZ degradation can be accelerated by induction of a protective branch of the UPR.
Significance: UPR-dependent ATZ disposal is a potential target for therapy.
Keywords: Autophagy, Endoplasmic Reticulum (ER), ER-associated Degradation, Ubiquitin Ligase, Unfolded Protein Response, HRD1, Activating Transcription Factor 6, α1-Antitrypsin
Abstract
α1-Antitrypsin is a serine protease inhibitor secreted by hepatocytes. A variant of α1-antitrypsin with an E342K (Z) mutation (ATZ) has propensity to form polymers, is retained in the endoplasmic reticulum (ER), is degraded by both ER-associated degradation and autophagy, and causes hepatocyte loss. Constant features in hepatocytes of PiZZ individuals and in PiZ transgenic mice expressing ATZ are the formation of membrane-limited globular inclusions containing ATZ and mitochondrial damage. Expression of ATZ in the liver does not induce the unfolded protein response (UPR), a protective mechanism aimed to maintain ER homeostasis in the face of an increased load of proteins. Here we found that in hepatoma cells the ER E3 ligase HRD1 functioned to degrade most of the ATZ before globular inclusions are formed. Activation of the activating transcription factor 6 (ATF6) branch of the UPR by expression of spliced ATF6(1–373) decreased intracellular accumulation of ATZ and the formation of globular inclusions by a pathway that required HRD1 and the proteasome. Expression of ATF6(1–373) in ATZ-expressing hepatoma cells did not induce autophagy and increased the level of the proapoptotic factor CCAAT/enhancer-binding protein (C/EBP) homologous protein (CHOP) but did not lead to apoptotic DNA fragmentation. Expression of ATF6(1–373) did not cause inhibition of protein synthesis and prevented mitochondrial damage induced by ATZ expression. It was concluded that activation of the ATF6 pathway of the UPR limits ATZ-dependent cell toxicity by selectively promoting ER-associated degradation of ATZ and is thereby a potential target to prevent hepatocyte loss in addition to autophagy-enhancing drugs.
Introduction
A variant of α1-antitrypsin with an E342K (Z) mutation (ATZ)3 has a greatly increased tendency to form polymers as compared with wild-type α1-antitrypsin (AAT) (1, 2). PiZZ individuals (homozygotes for ATZ) develop liver disease due to intracellular retention of the mutated protein (3). Unlike other misfolded proteins retained in the ER, expression of ATZ in cultured cells and in transgenic PiZ mice does not activate the unfolded protein response (UPR) even if this pathway is still functional (4). When the ER faces an increased load of unfolded proteins, it expands by enhancing lipid synthesis and the abundance of ER chaperones, and it decreases protein transcription and translation (5, 6). If homeostasis cannot be achieved, the ER will initiate pathways that lead to cell death. The mammalian UPR includes three signaling pathways initiated by three ER transmembrane proteins: protein kinase RNA-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1 (IRE1). The pathway initiated by PERK leads to phosphorylation of eukaryotic translation initiation factor 2a (eIF2a). This leads to a general decrease in protein translation except of that encoded by transcripts such as activating transcription factor 4 (ATF4) and the proapoptotic factor CCAAT/enhancer-binding protein (C/EBP) homologous protein (CHOP), which are increased (7–9). The signaling pathway initiated by IRE1 leads to splicing of XBP mRNA in the cytosol. The spliced XBP1 (XBP1s) protein is translocated to the nucleus where it promotes transcription of chaperones, ERAD components, and enzymes that control biosynthesis of lipids such as phosphatidylcholine (10). This ultimately leads to expansion of the ER by increasing both the lipid and the protein content of the organelles. ATF6 is an ER transmembrane component that binds to the chaperone GRP78/BiP. Upon an increased load of misfolded proteins in the ER, ATF6 dissociates from GRP78/BiP and traffics to the Golgi where it is cleaved to its active form, ATF6(1–373). ATF6(1–373) then migrates to the nucleus where it increases transcription of different ER chaperones, including GRP78/BiP, and induces ER expansion independently of XBP1 (11, 12). Thus, it appears that increased ER capacity can be artificially induced by overexpression of the active, nuclear form of ATF6 (ATF6(1–373)) (13, 14)). Expression of ATZ in the liver of transgenic mice does not induce the UPR (4), suggesting the possibility that induction of a protective branch of the pathway such as that initiated by ATF6 may promote ATZ degradation by ERAD. In this work, we found that most of the ATZ synthesized in hepatoma cells was degraded by an HRD1-dependent pathway and that artificial induction of the ATF6 branch of the UPR led to increased HRD1-dependent disposal of ATZ and protected cells from ATZ-dependent toxicity.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Lipofectamine 2000 was purchased from Invitrogen. Rat monoclonal anti-HA antibody (3F10), mouse monoclonal anti-GFP antibody, peroxidase-conjugated anti-hemagglutinin (HA) antibody, peroxidase-conjugated anti-mouse IgG, protease inhibitor (Complete Mini), and the terminal deoxynucleotidyltransferase dUTP nick end labeling (TUNEL) assay were from Roche Applied Science. Rabbit polyclonal anti-HA antibodies were from Sigma-Aldrich. Rabbit polyclonal antibodies against calnexin and GRP78/BiP and mouse monoclonal anti-KDEL were from Stressgen (San Diego, CA). Rabbit polyclonal and mouse monoclonal anti-Myc antibodies and rabbit polyclonal anti-CHOP antibodies were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Mouse monoclonal antibodies against AAT and actin were from Chemicon International (Temecula, CA). Rabbit polyclonal antibodies against synovial apoptosis inhibitor 1 (HRD1) were from Abgent (San Diego, CA). Mouse monoclonal antibodies against cleaved caspase 3 were from Cell Signaling Technology (Boston, MA). Rabbit polyclonal antibodies against prohibitin and Rab5 and rabbit monoclonal antibodies against GM130 were from Abcam (Cambridge, MA). Mouse monoclonal antibodies against calreticulin were from BD Biosciences. Cy3-conjugated anti-rat IgG was from Jackson ImmunoResearch Laboratories (West Grove, PA). Peroxidase-conjugated anti-rabbit IgG, peroxidase-conjugated anti-goat IgG, and protein A/G-agarose beads were purchased from Pierce. Cy3-conjugated anti-rat, Cy5-conjugated anti-mouse IgG, Cy3-conjugated anti-rabbit IgG, and Cy5-conjugated anti-rabbit IgG were from Jackson ImmunoResearch Laboratories. Enhanced chemiluminescence detection kits and NEG-072 Expres35S35S were purchased from PerkinElmer Life Sciences. Dithiothreitol (DTT), MG132, carbonyl cyanide 3-chlorophenylhydrazone (CCCP), oligomycin, and tunicamycin were from Sigma. Lactacystin was purchased from Calbiochem, and l-methionine and l-cysteine were from Acros Organics (Morris Plains, NJ). Amplify, Hyperfilm, and OptiPrep® (iodixanol) were purchased from GE Healthcare.
cDNAs and Constructs
The enhanced green fluorescent protein (EGFP) expression vector pEGFP-N2 was purchased from Clontech. The cDNAs for human AAT and proinsulin were obtained from Origene Technologies Inc. (Rockville, MD). Construction of the plasmids AAT-myc-pcDNA3.1, ATZ-myc-pcDNA3.1, ATZ-pEGFP-N2, and proinsulin-pcDNA3.1 have been described before (15). AAT-pEGFP-N2 was constructed using site-directed mutagenesis of the ATZ-pEGFP-N2 construct with the following primers: forward, 5′-CTGACCATCGACGAGAAAGGGACTG-3′; and reverse, 5′-CAGTCCCTTTCTCGTCGATGGTCAG-3′. ATF6-HA-pCGN and ATF6(1–373)-HA-pCGN plasmids were a kind gift from Dr. Ron Prywes (Department of Biological Sciences, Columbia University, New York, NY), and LC3-pEGFPC1 plasmid was a kind gift from Dr. Tamotsu Yoshimori (Department of Cell Genetics, National Institute of Genetics, Mishima, Japan).
Cell Culture and Transfection
Mouse hepatoma Hepa 1-6 cells were cultured in DMEM with 10% fetal bovine serum (FBS) and 5% penicillin/streptomycin. Cells were transiently transfected with the indicated plasmids using Lipofectamine 2000 according to the manufacturer instructions. Small interfering RNAs (siRNA) specific to murine synovial apoptosis inhibitor 1 (HRD1) (identification number 83279) was purchased from Applied Biosystems/Ambion (Austin, TX). The non-targeting siRNA pool (siRNA control) was from Dharmacon, Inc. (Chicago, IL). Cells were transfected with 100 nm siRNA using Lipofectamine 2000.
Preparation of Cell Lysates and Cell-free Media
Cells were washed once with phosphate-buffered saline (PBS) and scraped from plates with lysis buffer (100 mm Tris-HCl, pH 7.4, 50 mm NaCl, 1% Triton X-100, and protease inhibitors (Complete Mini)). Samples were passed five times through a 27-gauge 0.5-inch needle and incubated for 30 min at 4 °C. Cell lysates were centrifuged at 10,000 rpm for 10 min, and the pellet was discarded. Cell lysates were used for Western blot analysis and ELISA. To measure AAT in the medium by ELISA, transfected Hepa 1-6 cells were washed with M2 medium (20 mm HEPES, pH 7.4, 150 mm NaCl, 5 mm KCl, 1 mm MgCl2, 1 mm CaCl2, and 0.1 mg/ml bovine serum albumin) and incubated at 37 °C for 1 h in 1 ml of M2 medium with 5 mm glucose. The medium was collected and centrifuged at 1,000 rpm for 3 min to remove cell debris.
Exogenous AAT and Proinsulin Measurements
Proinsulin and AAT levels in cell lysates and cell-free media were measured using the human insulin and human proinsulin ELISA kit from Linco Research (St. Charles, Missouri) and the human AAT ELISA kit from Alpco Diagnostics (Salem, NH), respectively, according to the manufacturers' instructions.
Immunoblotting
Cell lysates were electrophoresed on a 7% SDS-polyacrylamide gel using loading buffer with or without SDS and β-mercaptoethanol (denaturing and non-denaturing conditions, respectively). Separated proteins were transferred to a nitrocellulose membrane probed with the indicated primary antibodies and secondary peroxidase-conjugated antibodies. Enhanced chemiluminescence detention, densitometry, and protein determination were performed as described previously (15).
Electron Microscopy
Transmission electron microscopy of transfected Hepa 1-6 cell was carried out as described (15). Images were taken by using an FEI F20 200-keV electron microscope.
Iodixanol Gradients and Cell Fractionation
Hepa 1-6 cells were scraped in homogenization medium (0.35 m sucrose, 1 mm EDTA, and 10 mm Tris-HCl) containing protease inhibitors (Complete Mini). Human and mouse liver tissues were cut into small pieces using a scalpel and homogenized using a Teflon-glass homogenizer (Thomas Scientific, Swedesboro, NJ). Cells or liver samples were then homogenized using a ball-bearing homogenizer with a 14-μm clearance (Isobiotech, Hamburg, Germany). Next, the cell homogenate was centrifuged at 1,000 rpm for 5 min to remove nuclei or unbroken cells. The supernatant was then centrifuged at 5,000 rpm (Beckman Optima ultracentrifuge) in a fixed angle rotor (TLA 100.2) for 20 min. The pellet was resuspended in 200 μl of homogenization medium and placed on top of a 15–20% (w/v) discontinuous OptiPrep gradient. Human and mouse liver samples were placed on top of a 20–30% (w/v) discontinuous OptiPrep gradient. The gradient was then centrifuged in a swinging bucket rotor (TLS-55) at 45,000 rpm (Beckman Optima ultracentrifuge) for 2 h. Samples were collected in 130-μl fractions from the top and diluted 1:1 with homogenization medium before electrophoresis and Western blot.
Determination of Spliced XBP1 and ATF4 mRNA Levels by Quantitative Real Time PCR
Hepa 1-6 cells plated in 100-mm dishes were transiently transfected with ATZ-GFP-PEGFP-N2 and ATF6-HA-pCGN or ATF6(1–373)-HA-pCGN plasmids for 48 h. As a positive control of ER stress, cells were incubated with 1 μg/ml tunicamycin for 16 h. Total mRNA was isolated from cells using the RNeasy Plus® kit from Qiagen (Valencia, CA). cDNA was amplified using the SuperScript® First-Strand Synthesis System for RT-PCR from Invitrogen. Quantitative real time PCR was carried out using SYBR® Premix Ex TaqTM DNA polymerase (perfect real time) from Clontech in a Bio-Rad MJ Mini personal thermocycler. The primers used to measure mRNA were as follows: murine XBP1s: forward, 5′-GAGTCCGCAGCAGGTG-3′; reverse, 5′-GTGTCAGAGTCCATGGGA-3′; murine ATF4: forward, 5′-ATGGCCGGCTATGGATGAT-3′; reverse, 5′-CGAAGTCAAACTCTTTCAGATCCATT-3′; and murine β-actin: forward, 5′-GATCTGGCACCACACCTTCT-3′; reverse, 5′-GGGGTGTTGAAGGTCTCAAA-3′.
Pulse-Chase Experiments
Hepa 1-6 cells were incubated in Met/Cys-free medium for 1 h at 37 °C followed by pulse labeling with 150 μCi/ml [35S]Met and [35S]Cys mixture (NEG-072, Expres35S35S) in Met/Cys-free medium for 30 min at 37 °C. Cells were then rinsed with chase medium (Met/Cys-free medium with 30 mg/liter Met, 30 mg/liter Cys, and 10% FBS) and chased for different time periods. Cells were scraped in IP buffer (100 mm Tris-HCl, pH 7.4, 50 mm NaCl, 1% Nonidet P-40, and protease inhibitors (Complete Mini)) and incubated for 30 min at 4 °C. Cell lysates were rotated overnight at 4 °C with the indicated antibody. After addition of protein A/G-agarose beads, the samples were further incubated for 1 h at 4 °C. After four washes with IP buffer and one wash with a high salt buffer (150 mm Tris, pH 7.4, 400 mm NaCl, and 1% Nonidet P-40 at pH 8), the beads were resuspended in sample buffer, boiled for 5 min, and centrifuged. Supernatants and cell extracts were loaded onto a 7% SDS-polyacrylamide gel. After electrophoresis, the gel was fixed for 30 min in fixation buffer (isopropyl alcohol/water/acetic acid, 25:65:10) and then enhanced for fluorography with Amplify solution with 7% glycerol. The gel was dried using a gel dryer (Bio-Rad 583) under vacuum (Bio-Rad HydroTech) at 60 °C for 2 h. The fluorograph was exposed for 48 h at −80 °C using preflashed Hyperfilm.
Fluorescence Microscopy
Epifluorescence and confocal fluorescence images were captured as described previously (15).
Quantification of CHOP Expression by Immunofluorescence
Epifluorescence images of cells were obtained with constant parameters of acquisition. The region of interest was selected by manually drawing the cell margins following GFP fluorescence for each individual cell. Fluorescence pixel intensity in the Cy3 channel (CHOP) was measured using NIH ImageJ software.
TUNEL Assay
The TUNEL assay in fixed Hepa 1-6 cells was performed as recommended by the manufacturer. Epifluorescence images of cells were obtained with constant parameters of acquisition. The region of interest was selected by manually drawing the cell margins. Fluorescence pixel intensity in the Cy3 channel (TUNEL) was measured using NIH ImageJ software.
Measurement of Mitochondrial Potential
Mitochondrial potential was quantified as described previously (16). Briefly, Hepa 1-6 cells were washed twice with DMEM and incubated with 200 nm MitoTracker® Red CMXRos in DMEM for 30 min. Next, cells were washed with DMEM twice and incubated in DMEM for an additional 30 min. As a control of mitochondrial depolarization, Hepa 1-6 cells were treated with 20 μg/ml CCCP for 2 h. As a control of mitochondrial hyperpolarization, Hepa 1-6 cells were treated with 4 μg/ml oligomycin for 2 h. The region of interest (each individual cell) was selected by autosegmentation, and the fluorescence pixel intensity was measured using IPlab software (Biovision Technologies, Exton, PA). The data were expressed as the ratio fluorescence pixel intensity/number of mitochondria per cell.
Autophagy Assays
Autophagy was measured as described previously (15). Briefly, Hepa 1-6 cells transiently transfected with LC3-GFP (17) were starved for 6 h in Hanks' balanced salt solution to induce autophagy or maintained in normal growth media. To quantify the level of autophagy, the number of LC3-GFP dots per cell was determined by manual counting.
Statistical Analysis
Data are expressed as mean ± S.D. GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA) was used to perform an unpaired t test or one-way analysis of variance.
RESULTS
ATZ-GFP-containing IBs Recapitulate Features of ATZ-containing Globules in Liver from PiZZ Individuals and PiZ Mice
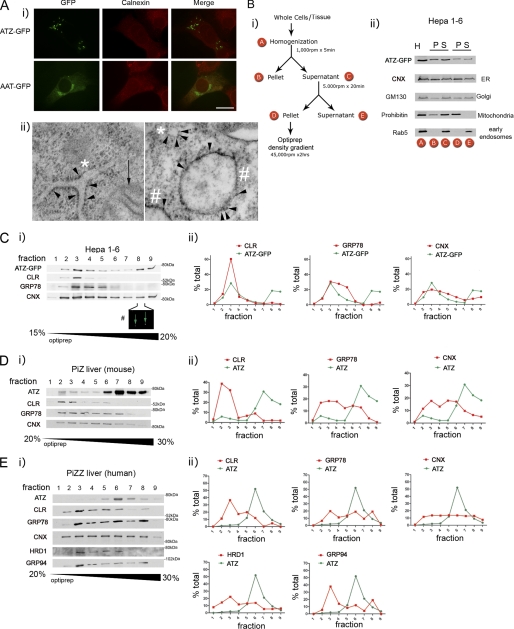
When ATZ-GFP was transiently expressed in murine hepatoma (Hepa 1-6) cells, the protein had an ER-like reticular distribution like the ER chaperone calnexin in most of the cells at 24 h after transfection (not shown) and at 48 h formed globular accumulations referred to as inclusion bodies (IBs) that appeared to reside in a subcompartment of the ER (Fig. 1A, panel i). Conversely, AAT-GFP did not form IBs at 48 h after transfection and was mostly visible in a Golgi-like perinuclear compartment. These data are similar to those obtained by transfecting Hepa 1-6 cells with HA-tagged ATZ and AAT, which localized, respectively, to IBs and to the Golgi both at ∼48 h after cell transfection (15). In the livers of PiZZ individuals and PiZ mice as well as in human fibroblasts, ATZ aggregates are included in membrane-limited globules with associated ribosomes (15, 18). Also in Hepa 1-6 cells, expression of ATZ induces dilation of the ER into globules (15). In the same Hepa 1-6 cells expressing ATZ-GFP, it is possible to visualize small round structures to which ribosomes are attached (Fig. 1A, panel ii, right panel, asterisk) and that correspond to ER in longitudinal section (Fig. 1A, panel ii, left panel, asterisks) as well as larger, ribosome-associated globules (Fig. 1A, panel ii, pound signs). This suggests that in the transfected cells (similar to the liver) ATZ concentrates into IBs that appear as subregions of the ER. Here we used density centrifugation to find further evidence that the ATZ-GFP-containing IBs of transfected cells are similar to the ATZ-containing globules in livers of PiZZ individuals and PiZ mice. Approximately 50% of ATZ-GFP from the postnuclear supernatant of Hepa 1-6 cells harvested 48 h after transfection was recovered in a pellet (fraction D) after a 5,000-rpm centrifugation step together with the ER, mitochondria, and Golgi and without early endosomes (Fig. 1B). Fraction D was then loaded onto an OptiPrep density gradient to determine the distribution of ATZ in respect to ER markers. ATZ-GFP migrated in the OptiPrep gradient as two peaks with one (Fig. 1C, fraction 3) co-migrating with the main peak of the ER proteins calreticulin, GRP78/BiP, and calnexin, indicating that a fraction of the protein localizes to the main ER. There was also another ATZ-GFP peak at fractions 8 and 9 that had higher density, had virtually no detectable calreticulin or GRP78/BiP, and retained some calnexin. Fraction 8 contained IBs of the same size as those visualized by fluorescence microscopy of the ATZ-GFP-expressing Hepa 1-6 cells (Fig. 1C, panel i). This indicates that in Hepa 1-6 cells ∼50% of ATZ-GFP is in the ER, and the rest localizes to IBs with higher density than the main ER. When fraction D from the PiZ mouse liver was prepared as outlined for the Hepa 1-6 cells and loaded onto the OptiPrep gradient, ATZ migrated in the gradient forming a single peak (Fig. 1D, fraction 7) at higher density than most of the calreticulin, GRP78/BiP, and calnexin (fractions 2–5). Thus, in the mouse liver, most of the ATZ migrates at higher density than ER chaperones, suggesting that the murine ATZ-containing globules have higher density than the rest of the ER. When PiZZ human liver was fractionated with the OptiPrep gradient, the main peak of ATZ (Fig. 1; all samples for ATZ analysis were diluted 10×) was again at higher density than most of the calreticulin and GRP94 (Fig. 1E, fraction 6), again suggesting that human ATZ-containing globules have higher density than the rest of the ER. Different from that in the mouse liver, a second peak of GRP78/BiP co-migrated with ATZ peak in fraction 6, suggesting some difference in protein composition of murine and human ATZ globules. A fraction of HRD1, an E3 ubiquitin ligase of the ER (19), also partially co-migrated with the ATZ peak. These data and the previous observation that ATZ-containing membranes from tissue and cells have attached ribosomes (15, 18) indicate that expression of ATZ leads to the formation of ER-derived globular organelles that have a higher density than the rest of the ER perhaps because they have high concentrations of ATZ aggregates.
FIGURE 1.
Fractionation of ATZ-GFP-containing IBs and ATZ-containing globules. A, panel i, Hepa 1-6 cells were transiently transfected with AAT-pEGFP-N2 or ATZ-pEGFP-N2 and analyzed by immunofluorescence microscopy 48 h after transfection. Cells were stained with rabbit polyclonal antibodies against calnexin. Secondary antibody staining was carried out using Cy3-conjugated anti-rabbit antibodies. Bar, 25 μm. Panel ii, electron microscopy of Hepa 1-6 cell transfected with ATZ-pEGFP-N2. Arrowheads, membrane-associated ribosomes; asterisks, endoplasmic reticulum; pound sign, globular structures with attached ribosomes; black arrow, mitochondrion with visible cristae. Bar, 200 nm. B, panel i, Hepa 1-6 cells transiently transfected with ATZ-pEGFP-N2 for 48 h or liver tissues from a PiZ mouse and a PiZZ individual were homogenized and fractioned as shown in the scheme. Panel ii, Western blot analysis of the fractions obtained by differential centrifugation of Hepa 1-6 cells transiently transfected with ATZ-pEGFP-N2 using the indicated antibodies. (H, homogenate; P, pellet; S, supernatant). C–E, fraction D obtained as outlined in B from Hepa 1-6 cells (C, panel i), PiZ mouse liver (D, panel i), and PiZZ human liver (E, panel i) was loaded onto OptiPrep gradients containing the indicated amount of glycerol (w/v). Fractions from the gradient were analyzed by Western blot with the indicated antibodies and quantified by densitometry (panel ii). In C, the pound sign indicates fluorescence (GFP) of IBs in fraction 8 from the OptiPrep gradient in Hepa 1-6 cells. The magnification is the same as in A, panel i. CLR, calreticulin; CNX, calnexin.
Active ATF6 Selectively Reduces Expression of ATZ and Formation of IBs
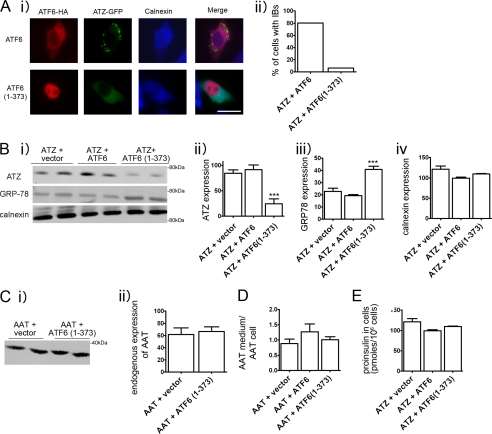
The accumulation of incorrectly folded proteins in the ER leads to induction of the UPR, a protective cell response, which results in increased protein folding and degradation. The UPR consists of three branches of which two, the ATF6 and IRE1/XBP1 branch, are protective and induce increased expression of ER chaperones and ERAD components. The third branch, activated by phosphorylation of PERK, leads to reduced protein translation and, if ER stress persists, promotes cell apoptosis (5). However, expression of ATZ does not induce the UPR (4), suggesting the possibility that induction of protective branches of the pathway may promote ATZ degradation by ERAD. ATF6 is an ER transmembrane component that binds to the chaperone GRP78/BiP. Upon an increased load of misfolded proteins in the ER, ATF6 dissociates from GRP78/BiP and traffics to the Golgi where it is cleaved to its active form, ATF6(1–373). ATF-6(1–373) then migrates to the nucleus where it increases transcription of different ER chaperones, including GRP78/BiP, and induces ER expansion (11, 12). Thus, it appears that increased ER capacity can be artificially induced by overexpression of the active, nuclear form of ATF6 (ATF6(1–373) (13, 14)). Here we tested whether exogenous expression of ATF6(1–373) in the Hepa 1-6 cells promotes ATZ disposal. By immunofluorescence analysis of transfected Hepa 1-6 cells, inactive, full-length ATF6 was distributed in an ER pattern, whereas ATF6(1–373) was localized in the nucleus (Fig. 2A) consistent with other reports (20, 21). In cells co-expressing full-length ATF6 with ATZ-GFP, ATZ-GFP was distributed to the IBs. Conversely, in cells co-transfected with ATF6(1–373) and ATZ-GFP, ATZ-GFP was distributed prevalently to the ER with a reduced population of cells containing the IBs (from 80 to 6.2%) (Fig. 2A). These results indicate that induction of the UPR inhibits the formation of the IBs. When Hepa 1-6 cells were transfected with ATF6(1–373), the amount of steady-state ATZ-GFP detected by Western blot was reduced as compared with control cells co-transfected with empty vector or full-length ATF6 (Fig. 2B, panels i and ii). This suggests that active ATF6 promotes ATZ disposal. As expected, in cells expressing active ATF6, the levels of the chaperone GRP78/BiP were increased by approximately 2-fold as compared with the controls (Fig. 2B, panels i and iii). Another ER chaperone, calnexin, remained constant in all three conditions (Fig. 2B, panels i and iv). Thus, ATF6(1–373) co-expressed with ATZ-GFP in the Hepa 1-6 cells induces transcriptional activation of specific cell chaperones that results in a reduction of ATZ-GFP protein. It is possible that induction of the UPR by expression of ATF6(1–373) in addition to promoting disposal of ATZ also promotes the disappearance of correctly folded proteins. However, when active ATF6(1–373) was expressed, there were no changes in the cell abundance of endogenous AAT (Fig. 2C). There was also no change in the ratio exogenous HA-AAT in the medium/exogenous intracellular HA-AAT when cells co-expressed either full-length ATF6 or ATF6(1–373) (Fig. 2D). This indicates that protein secretion is normally maintained in cells expressing ATF6(1–373). Moreover, intracellular levels of exogenous proinsulin were also unchanged by expression of ATF6(1–373) (Fig. 2E). This indicates that the UPR induced by active ATF6 specifically targets disposal of ATZ.
FIGURE 2.
Expression of ATF6(1–373) reduces expression of ATZ and formation of IBs. A, panel i, cells were transiently transfected with ATZ-pEGFP-N2 and ATF6-HA-pCGN or with ATZ-pEGFP-N2 and ATF6(1–373)-HA-pCGN and analyzed by immunofluorescence. Cells were stained with primary rat monoclonal antibodies against the HA tag and rabbit polyclonal antibodies against calnexin. Secondary antibody staining was carried out using Cy3-conjugated anti-rat antibodies and Cy5-conjugated anti-rabbit antibodies. Panel ii, quantification of cells with IBs. For the quantification shown in the graph, we measured the percentage of cells that had at least one IB. Averages were obtained from the experiment shown in panel i (n = 30 cells per time point). Bar, 25 μm. B, panel i, Hepa 1-6 cells were transiently transfected with ATZ-pEGFP-N2 and empty vector, ATZ-pEGFP-N2 and ATF6-HA-pCGN, or ATZ-pEGFP-N2 and ATF6(1–373)-HA-pCGN as indicated. Cell lysates were analyzed by Western blot 48 h after transfection using the indicated antibodies. Panels ii–iv, ATZ-GFP, GRP78/BiP, and calnexin expression levels from the experiment in panel i were measured by densitometry. C, panel i, Hepa 1-6 cells were transiently transfected with either ATF6-HA-pCGN or ATF6(1–373)-HA-pCGN. Endogenous AAT expression was analyzed by Western blot. Panel ii, endogenous AAT levels were measured by densitometry. D, Hepa 1-6 cells were co-transfected with AAT-pcDNA3.1 and empty vector, AAT-pcDNA3.1 and ATF6-HA-pCGN, or AAT-pcDNA3.1 and ATF6(1–373)-HA-pCGN as indicated. Data are expressed as the ratio exogenous AAT in the medium/exogenous AAT in cell lysates. E, proinsulin levels in Hepa 1-6 cells co-transfected with ATZ-pEGFP-N2, proinsulin-pcDNA3.1, and empty vector; ATZ-pEGFP-N2, proinsulin-pcDNA3.1, and ATF6-HA-pCGN; or ATZ-pEGFP-N2, proinsulin-pcDNA3.1, and ATF6(1–373)-HA-pCGN. Statistical significance is indicated by *** (p ≤ 0.0001). Error bars = standard deviation derived from 3 independent experiments.
Active ATF6 Promotes Degradation of ATZ by Proteasome-dependent Pathway
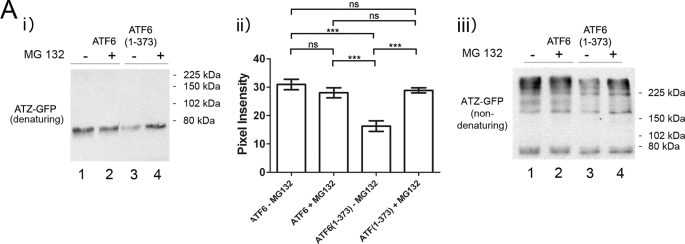
Proteins that are not properly folded are degraded by ERAD. ERAD is known to be one of the pathways involved in ATZ degradation (22–25). To determine whether ATF6(1–373)-induced ATZ disposal is dependent on increased ERAD, cells co-transfected with ATZ-GFP and ATF6 or ATF6(1–373) were treated with the proteasome inhibitor MG132. Incubation with MG132 restored the abundance of ATZ-GFP in the ATF6(1–373)-transfected cells to the same level of control cells co-transfected with full-length ATF6 (Fig. 3A, lanes 4 and 1). Conversely, MG132 did not increase the steady-state level of ATZ-GFP in Hepa 1-6 cells (Fig. 3A, lanes 1 and 2). Therefore, the expression of ATF6(1–373) activates proteasome-dependent degradation of ATZ. ATZ has a tendency to form homopolymers, and in agreement with this, hepatic globular inclusions have been found to be composed almost entirely of polymerized ATZ protein (26). It has been reported that ATZ migrates in non-denaturing gel electrophoresis as high molecular weight bands in the resolving gel with ATZ polymers also remaining in the stacking gel (26). Here we found that in non-denaturing conditions ATZ immunoreactivity appeared as high molecular weight bands both in the resolving gel and in the stacking gel (supplemental Fig. S1). Because the ratio of high molecular weight ATZ in stacking and resolving gels was equivalent, we then routinely developed the Western blot corresponding only to the resolving gel. However, quantifications of the total amount of ATZ were done using gel electrophoresis in denaturing conditions. When the same samples analyzed in Fig. 3A, panel i, were subjected to SDS-PAGE in non-denaturing conditions, only a fraction of the ATZ-GFP migrated as a band at ∼75 kDa, corresponding to the monomeric form, whereas the remaining ATZ-GFP immunoreactivity appeared as higher molecular weight species (Fig. 3A, panel iii). This suggests that most of the protein exists either in the form of polymers and/or associated to other proteins. In respect to this, we have shown that ATZ forms complexes with itself when expressed in the Hepa 1-6 cells (15). Although the amount of monomeric ATZ-GFP was similar in all samples, the high molecular weight bands of ATZ-GFP were decreased in the ATF6(1–373) samples (Fig. 3, panel iii, lanes 1 and 3) and restored by incubation with MG132. The data suggest that above a threshold level ATZ has an increased tendency to form polymers and/or associate with other proteins and that expression of active ATF6 blunts homo- and/or heterodimerization by promoting degradation of ATZ by a proteasome-dependent pathway.
FIGURE 3.
Expression of ATF6(1–373) increases degradation of ATZ by proteasome. A, Hepa 1-6 cells were transiently transfected with either ATZ-pEGFP-N2 and ATF6-HA-pCGN or ATZ-pEGFP-N2 and ATF6(1–373)-HA-pCGN. After 48 h, cells were incubated in the absence and in the presence of 1 μm MG132 for 8 h. SDS-PAGE of cell lysates was carried out in denaturing (panel i) and non-denaturing (panel iii) conditions, and Western blots were probed with mouse monoclonal antibodies against AAT. Panel ii, ATZ-GFP levels were measured by densitometry. Statistical significance is indicated by *** (p ≤ 0.0001) and ns (p > 0.05). Error bars = standard deviation derived from 3 independent experiments.
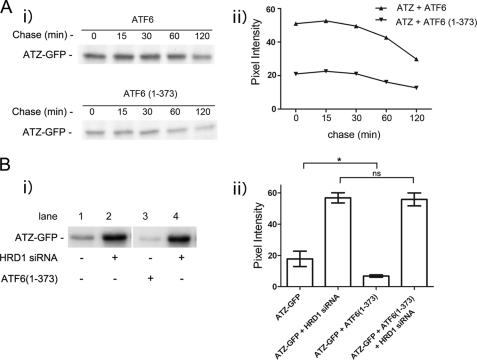
ATZ Degradation Requires HRD1 in Absence and Presence of ATF6 Activation
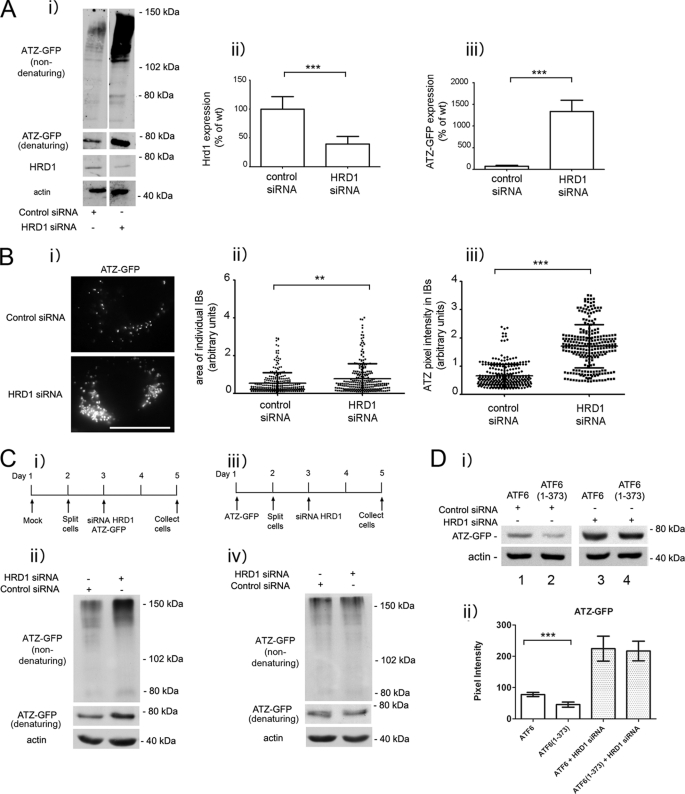
It has been reported recently that soluble, non-transmembrane proteins of the ER are degraded by an ERAD pathway that includes the E3 ubiquitin ligase HRD1 (27). ATZ does not classically belong to the group of soluble ER proteins because it often forms insoluble polymers in the ER and in globular inclusions (26), leaving open the question as to whether HRD1 contributes to ATZ disposal. When HRD1 protein expression was reduced by siRNA, ATZ-GFP levels were significantly increased (> 3-fold; Fig. 4A), indicating that an ERAD pathway that includes HRD1 degrades most of the ATZ produced in the cell. If formation of IBs in the ATZ-expressing Hepa 1-6 cells (Fig. 1A) depended on increased intracellular accumulation of ATZ, then HRD1 silencing may lead to proliferation of the IBs. In this respect, silencing of HRD1 increased significantly both the size and the brightness of ATZ-GFP-containing IBs, indicating that the formation of IBs is dependent on the level of intracellular ATZ (Fig. 4B). We have shown that at early stages after hepatoma cell transfection (24 h) most of the cell ATZ localizes in the reticular ER, and later on (48 h) it appears in IBs (15). These observations raise the question as to whether the HRD1-dependent degradation of ATZ occurs in the ER, in the IBs, or in both. In this respect, HRD1 silencing was able to increase the intracellular level of ATZ-GFP when the siRNA was co-transfected with ATZ-GFP (Fig. 4C, panels i and ii). Conversely, this was not the case when the siRNA was introduced into cells 48 h after transfection with ATZ-GFP when the protein was already localized in the IBs (Fig. 4C, panels iii and iv). Thus, HRD1-dependent degradation appears to take place at early stages after synthesis when ATZ localizes to the ER rather than to IBs. Having established that HRD1 is essential for degradation of ATZ-GFP, we asked whether the ATF6(1–373)-dependent degradation of ATZ shares the same pathway as that occurring in the absence of the active transcription factor. In cell populations expressing either ATF6 or ATF6(1–373), HRD1 silencing led to the same level of increased intracellular accumulation of ATZ-GFP (Fig. 4D). This suggests that the endogenous pathway of ATZ degradation as well as that induced by activation of the ATF6 branch of the UPR is dependent on HRD1 function.
FIGURE 4.
ATZ degradation requires HRD1 in absence and presence of ATF6 activation. A, panel i, Hepa 1-6 cells were transiently transfected with ATZ-pEGFP-N2 and either siRNA negative control or siRNA against mouse HRD1. Cell lysates were analyzed by Western blot in non-denaturing or denaturing conditions as indicated. HRD1 (panel ii) and ATZ-GFP (panel iii) levels were measured by densitometry. Error bars = standard deviation derived from 3 independent experiments. B, panel i, cells were transfected as in A, fixed after 48 h, and visualized by fluorescence microscopy. Measurements of the size of individual IBs (panel ii) and pixel intensity of ATZ-GFP in IBs (panel iii) (control siRNA IB, n = 269; HRD1 siRNA IB, n = 251 in ∼10 cells). Bar, 25 μm. Panels ii and iii, Error bars = standard deviations. C, panel i, Hepa 1-6 cells were transfected as indicated in the diagram. Panel ii, cell lysates from cells treated as in panel i were analyzed by Western blot in non-denaturing or denaturing conditions as indicated. Panel iii, Hepa 1-6 cells were transfected as indicated in the diagram. Panel iv, cell lysates treated as in panel iii were analyzed by Western blot in non-denaturing or denaturing conditions as indicated. D, panel i, Hepa 1-6 cells were transiently transfected with control siRNA, ATZ-pEGFP-N2, and either ATF6-HA-pCGN (lane 1) or ATF6(1–373)-HA-pCGN (lane 2) or HRD1 siRNA, ATZ-pEGFP-N2, and either ATF6-HA-pCGN (lane 3) or ATF6(1–373)-HA-pCGN (lane 4) as indicated. Cell lysates were analyzed by Western blot 48 h after transfection using the indicated antibodies. Panel ii, ATZ-GFP levels were measured by densitometry. Averages and S.D. were determined from six independent samples, including those shown in the left panel. Statistical significance is indicated by ** (p ≤ 0.001) and *** (p ≤ 0.0001). Error bars are as in panel A.
HRD1-dependent Degradation of ATZ Occurs within 30 min of Protein Synthesis
When transiently transfected Hepa 1-6 cells were metabolically labeled with 35S-labeled Cys and Met in a 30-min pulse followed up by a 2-h chase, ∼50% of ATZ-GFP was already decreased immediately after the pulse in the cells co-transfected with ATF6(1–373) as compared with cells co-transfected with full-length ATF6 (Fig. 5A, time 0 after the pulse), suggesting that most of the ATF6(1–373)-dependent disposal of ATZ indeed occurs shortly after protein synthesis. When data were expressed as the percentage of initial ATZ-GFP that remained at each time point, the difference between the two samples was less than 10% (data not shown), again suggesting that most of the ATF6-dependent degradation occurs immediately after synthesis. To further investigate this possibility, we co-expressed ATZ-GFP with and without ATF6(1–373) in the presence and absence of HRD1 silencing. After a 30-min pulse, the abundance of metabolically labeled ATZ-GFP was reduced by more than 50% in cells co-expressing ATF6(1–373) as compared with the control without active ATF6 (Fig. 5B) consistent with previous experiments. However, when HRD1 was silenced, the level of ATZ-GFP was increased by ∼3-fold, and there was no difference in abundance between samples with and without ATF6(1–373) expression (Fig. 5B). This experiment indicates that most of ATZ is degraded within 30 min of synthesis by an HRD1-dependent mechanism. This same pathway of ATZ degradation normally occurring in the cell is further enhanced by expression of active ATF6.
FIGURE 5.
ATZ is degraded shortly after synthesis. A, panel i, Hepa 1-6 cells transiently transfected with ATZ-pEGFP-N2 and ATF6-HA-pCGN or ATZ-pEGFP-N2 and ATF6(1–373)-HA-pCGN were pulsed with 150 μCi/ml [35S]Met and [35S]Cys. Cells were chased for the indicated time intervals, and cell lysates were immunoprecipitated using mouse monoclonal anti-AAT antibodies. Panel ii, quantification of the experiments in panel i. B, panel i, Hepa 1-6 cells transiently transfected with ATZ-pEGFP-N2, pcDNA3.1, and either control siRNA (lane 1) or HRD1 siRNA (lane 2) or ATZ-pEGFP-N2, ATF6(1–373)-pCGN, and either control siRNA (lane 3) or HRD1 siRNA (lane 4). Cells were pulsed for 30 min with 150 μCi/ml [35S]Met and [35S]Cys. Cell lysates were immunoprecipitated using mouse monoclonal anti-AAT antibodies. Panel ii, quantification of the experiments in panel i (for each condition, n = 3). Statistical significance is indicated by * (p ≤ 0.01) and ns (p > 0.05). Error bars = standard deviation derived from 3 independent experiments.
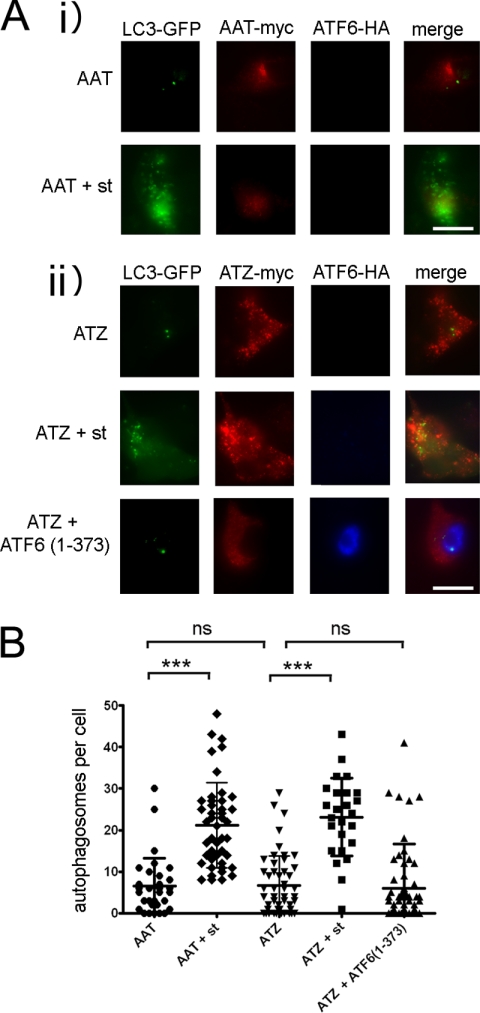
Active ATF6 Does Not Increase Autophagy
Degradation of ATZ has been proposed to occur by two pathways, ERAD and autophagy (17, 22, 24, 25, 28, 29). In addition, ER stress can activate autophagy, which in turn may facilitate ATZ degradation (30). These observations raise the question as to whether activation of the ATF6 branch of the UPR induces the autophagy pathway. LC3 is an established marker of autophagosomes (31, 32). In untreated Hepa 1-6 cells expressing either AAT-Myc or ATZ-Myc, there were very few puncta of LC3-GFP (Fig. 6, A and B). Starvation increased significantly the number of LC3-GFP-labeled autophagosomes in both cell populations, indicating that autophagy was induced. In the same experiment, expression of ATF6(1–373) did not increase the number of autophagosomes, indicating that active ATF6 does not increase autophagy.
FIGURE 6.
Expression of ATF6(1–373) does not induce autophagy. A, Hepa 1-6 cells were transfected with LC3-pEGFP-C1, AAT-myc-pcDNA3.1, and empty vector (panel i) or LC3-pEGFP-C1, ATZ-myc-pcDNA3.1, and either empty vector or ATF6(1–373)-HA-pCGN (panel ii). Where indicated, autophagy was induced by starvation (st) 48 h after transfection. Cells were fixed, and primary staining was done using the rabbit polyclonal antibody against HA to visualize ATF6 and mouse monoclonal antibody against Myc to visualize ATZ. Secondary staining was carried out using Cy5-conjugated anti-rabbit and Cy3-conjugated anti-mouse antibodies. Bar, 25 μm. B, quantification of the experiment shown in A (AAT, n = 32; AAT + starvation, n = 45; ATZ, n = 51; ATZ + starvation, n = 25; ATZ + ATF6(1–373), n = 66). Statistical significance is indicated by *** (p ≤ 0.0001) and ns (p > 0.05). Error bars = standard deviations.
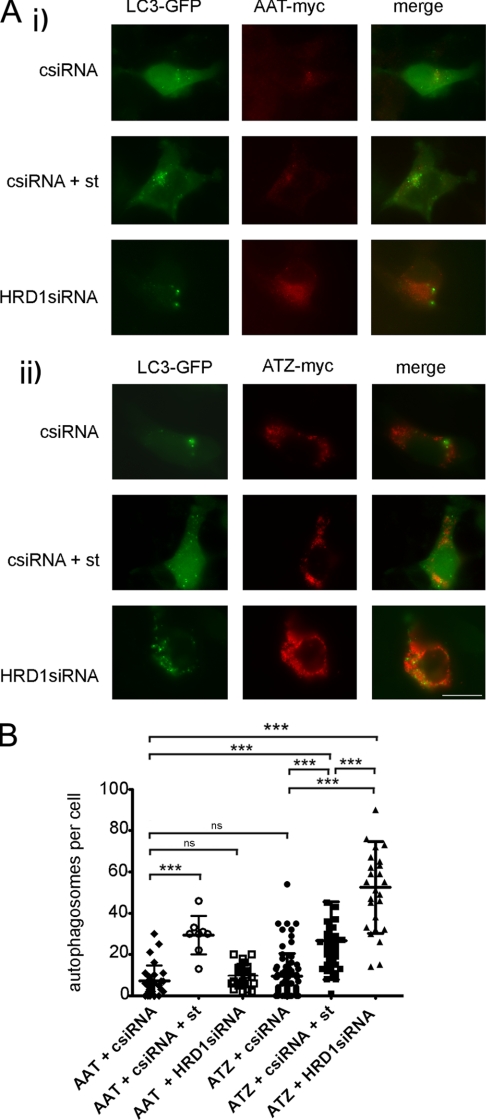
Silencing of HRD1 Induces Autophagy in ATZ-expressing Hepa 1-6 Cells
The data in Fig. 4 indicate that activation of the ATF6 pathway promotes degradation of ATZ by an HRD1-dependent mechanism. This HRD1-dependent mechanism already degrades a large fraction of the ATZ protein synthesized by the cell in the absence of the exogenous transcription factor. In transiently transfected Hepa 1-6 cells expressing ATZ, autophagy is not induced (15). This is different from that in the hepatocytes of PiZZ individuals and of PiZ mice (33). It is possible that induction of autophagy is a consequence of a relative insufficiency of ERAD to degrade the ATZ synthesized in the hepatocyte. Silencing of HRD1 in the Hepa 1-6 cells increased the number of LC3-GFP-labeled autophagosomes per cell by ∼3-fold (Fig. 7). Conversely, HRD1 silencing did not affect the number of autophagosomes in cells expressing AAT. The data indicate that insufficient ERAD of ATZ by the HRD1 pathway induces autophagy.
FIGURE 7.
Reduced levels of HRD1 induce autophagy in ATZ-expressing Hepa 1-6 cells. A, Hepa 1-6 cells were transfected with LC3-pEGFP-C1, AAT-myc-pcDNA3.1, and either control siRNA (csiRNA) or HRD1 siRNA (panel i) or LC3-pEGFP-C1, ATZ-myc-pcDNA3.1, and either control siRNA or HRD1 siRNA (panel ii). Where indicated, autophagy was induced by starvation (st) 48 h after transfection. Cells were fixed, and primary staining was done using the mouse monoclonal antibody against Myc to visualize ATZ. Secondary staining was carried out using Cy3-conjugated anti-mouse antibodies. Bar, 25 μm. B, quantification of the experiment shown in A (AAT + control siRNA, n = 29; AAT + control siRNA + starvation, n = 8; AAT + HRD1 siRNA, n = 23; ATZ + control siRNA, n = 80; ATZ + control siRNA + starvation, n = 40; ATZ + HRD1 siRNA, n = 26). Statistical significance is indicated by *** (p ≤ 0.0001) and ns (p > 0.05). Error bars = standard deviations.
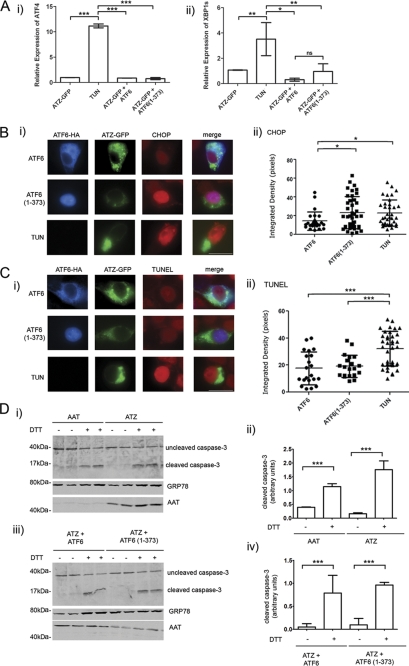
Active ATF6 Induces CHOP But Does Not Lead to Apoptosis
The UPR consists of three main branches of which two, the ATF6 and the IRE1/XBP1 pathways, are mainly protective, and the other, the PERK pathway, leads to decreased protein translation, activation of the transcription factor ATF4, and apoptosis when ER stress is unresolved (5). Because there is cross-talk between branches of the UPR, we asked whether expression of ATF6(1–373) increased components of the IRE1/XBP1 and the PERK pathway. Tunicamycin inhibits protein glycosylation and folding and induces a classical UPR with activation of the three pathways (34). Consistent with that, cell exposure to the drug increased the level of ATF4 mRNA (Fig. 8A, panel i) and induced splicing of XBP1 mRNA to the active transcription factor (Fig. 8A, panel ii) by quantitative real time PCR. When cells were co-transfected with ATZ-GFP and either ATF6 or ATF6(1–373), there was no increase of ATF4 levels or XBP1 splicing (Fig. 8A), indicating that activation of the ATF6 pathway in the Hepa 1-6 cells did not induce a concomitant increase of other UPR pathways. The absence of increased ATF4 and XBP1s mRNA was also observed by semiquantitative PCR in cells sorted by FACS to select for cells with ATZ-GFP expression (supplemental Fig. S2).
FIGURE 8.
ATF6(1–373) expression does not induce XBP1 and ATF4 or promote apoptosis. A, Hepa 1-6 cells were transiently transfected with ATZ-pEGFP-N2 and either ATF6-HA-pCGN or ATF6(1–373)-HA-pCGN. As a positive control for ER stress, cells were treated for 16 h with 1 μg/ml of tunicamycin (TUN). ATF4 (panel i) and XBP1s (panel ii) mRNA induction was measured by quantitative real time PCR 48 h after transfection. Each sample was normalized for β-actin mRNA (for each condition, n = 3). Error bars = standard deviations derived from 3 independent experiments. B, panel i, Hepa 1-6 cells were co-transfected with ATZ-pEGFP-N2 and either ATF6-HA-pCGN or ATF6(1–373)-HA-pCGN for 48 h. As a positive control, Hepa 1-6 cells were transfected with ATZ-pEGFP-N2 and treated for 16 h with 1 μg/ml tunicamycin (TUN). Cells were analyzed by immunofluorescence using rat monoclonal antibody against HA and rabbit polyclonal antibody against CHOP. Secondary staining was carried out using Cy3-conjugated anti-rat antibodies and Cy5-conjugated anti-rabbit antibodies. Panel ii, quantification of the experiment shown in panel i (ATF6, n = 25; ATF6(1–373), n = 41; tunicamycin, n = 36). C, panel i, Hepa 1-6 cells were co-transfected and treated as in B. The TUNEL assay was performed on fixed cells followed by primary staining using rat monoclonal antibody against HA. Secondary staining was carried out using of Cy5-conjugated anti-rat antibodies. Panel ii, quantification of the experiment shown in panel i (ATF6, n = 24; ATF6(1–373), n = 21; tunicamycin, n = 37). B and C, error bars = standard deviations. D, panel i, Hepa 1-6 cells were transiently transfected with AAT-myc-pcDNA3.1 or ATZ-myc-pcDNA3 and treated with or without 2 mm DTT for 16 h. Western blots were probed with the indicated antibodies. Panel ii, cleaved caspase 3 shown in panel i was measured by densitometry. Panel iii, Hepa 1-6 cells were transiently transfected with ATZ-myc-pcDNA3.1 and ATF6-HA-pCGN or ATZ-myc-pcDNA3.1 and ATF6(1–373)-HA-pCGN. Cells were incubated with or without 2 mm DTT for 16 h. Western blots were probed with the indicated antibodies. Panel iv, cleaved caspase 3 shown in panel iii was measured by densitometry. Statistical significance is indicated by * (p ≤ 0.01), ** (p ≤ 0.001), *** (p ≤ 0.0001), and ns (p > 0.05). Bars, 25 μm. Error bars as in panel A.
The data presented here indicate that induction of the ATF6 pathway leads to a specific increase of ATZ degradation without suppression of protein synthesis or perturbations of protein traffic. However, it is also known that prolonged activation of the UPR may lead to cell apoptosis. To determine whether activation of the ATF6 branch of the UPR leads to apoptosis in ATZ-expressing cells, we measured by quantitative fluorescence microscopy the expression level of a proapoptotic transcription factor, CHOP (35). Expression of ATF6(1–373) indeed increased the level of CHOP in cells co-expressing ATZ-GFP (Fig. 8B) to the same level as cell exposure to tunicamycin. To determine whether CHOP induction leads to increased apoptosis in cells expressing ATF6(1–373), we used the TUNEL assay to measure apoptotic DNA fragmentation by quantitative fluorescence microscopy. Hepa 1-6 cells exposed to tunicamycin had significantly increased apoptotic DNA fragmentation as compared with control cells transfected with inactive ATF6 (Fig. 8C). Conversely, there was no increase of TUNEL staining in cells co-expressing ATF6(1–373) and ATZ-GFP as compared with the control expressing ATZ-GFP alone (Fig. 8C). Caspase 3 activation is essential for apoptotic chromatin condensation and DNA fragmentation (36). ATZ expression by itself did not induce cleavage of caspase 3 (Fig. 8D, panels i and ii) consistent with the observation that ATZ-expressing cells do not have apoptotic DNA fragmentation (Fig. 8C). Conversely, cell exposure to DTT, which also induces ER stress, induced cleavage of caspase 3 in addition to increasing the abundance of the ER chaperone GRP78/BiP. In cells expressing both ATZ and ATF6(1–373), cleavage of caspase 3 was not induced (Fig. 8D, panels iii and iv). Thus, it appears that although in ATZ-expressing cells proapoptotic CHOP is induced as part of the ATF6 pathway (consistent with other reports (37, 38)) apoptosis itself does not take place.
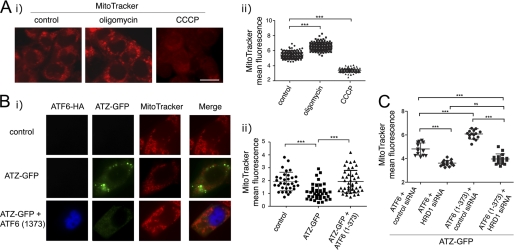
ATZ-dependent Mitochondrial Damage Is Reduced by Active ATF6 and Increased by HRD1 Silencing
A hallmark of ATZ-dependent cell toxicity in the liver of PiZZ patients as well as in cultured hepatoma cells is the appearance of damaged mitochondria with blebs in the outer membrane and dissolution of the cristae and the matrix (15, 33). We reasoned that ATZ-induced mitochondrial damage might result in the loss of mitochondrial membrane potential that could be monitored by staining the cells with the mitochondrial marker MitoTracker Red, which is sensitive to membrane potential (16). In the Hepa 1-6 cells, the uncoupler CCCP decreased the intensity of MitoTracker Red fluorescence, whereas oligomycin, an inhibitor of ATP synthase, increased it (Fig. 9A). This indicates that MitoTracker Red fluorescence indeed reflects changes in mitochondrial membrane potential in the Hepa 1-6 cells. MitoTracker Red labeling was significantly reduced in cells expressing ATZ-GFP as compared with the mock-transfected control, indicating a loss of mitochondrial membrane potential (Fig. 9B). Co-transfection with ATF6(1–373) blunted the loss of mitochondrial membrane potential induced by ATZ-GFP expression. Thus, activation of the ATF6 branch of the UPR appears to protect cells against ATZ-induced mitochondrial toxicity. Conversely, silencing of HRD1 induced a loss of mitochondrial membrane potential whether or not cells were expressing ATF6(1–373) (Fig. 9C). This and the data in Fig. 4D indicate that activation of the ATF6 pathway limits ATZ-induced mitochondrial damage by promoting HRD1-dependent degradation of ATZ.
FIGURE 9.
Expression of ATF6(1–373) reduces ATZ-dependent mitochondrial damage. A, panel i, untransfected Hepa 1-6 cells were treated either with 20 μg/ml CCCP or vehicle (DMSO; control) for 2 h or 4 μg/ml oligomycin for 2 h. Mitochondria were stained using MitoTracker Red CMXRos. Cells were fixed and visualized by fluorescence microscopy. Panel ii, quantification of the experiment shown in panel i) (control, n = 187; oligomycin, n = 185; CCCP, n = 186). B, panel i, Hepa 1-6 cells were transiently transfected with Lipofectamine alone (control), ATZ-pEGFP-N2 and empty vector, or ATZ-pEGFP-N2 and ATF6(1–373)-HA-pCGN. After 48 h, mitochondria were stained as in A. Immunofluorescence analysis was done using primary rat monoclonal antibodies against HA. Secondary staining was carried out using Cy5-conjugated anti-rat antibodies. Panel ii, quantification of the MitoTracker fluorescence of the experiment shown in panel i (control, n = 32; ATZ-GFP, n = 36; ATZ-GFP + ATF6(1–373), n = 41). C, panel i, Hepa 1-6 cells were transiently transfected with ATZ-pEGFP-N2, ATF6-HA-pCGN, and control siRNA; ATZ-pEGFP-N2, ATF6-HA-pCGN, and HRD1 siRNA; ATZ-pEGFP-N2, ATF6(1–373)-HA-pCGN, and control siRNA; or ATZ-pEGFP-N2, ATF6(1–373)-HA-pCGN, and HRD1 siRNA. Staining of mitochondria and immunofluorescence microscopy were carried out as in B, and the experiment was quantified (cells transfected with ATF6-HA-pCGN and control siRNA, n = 20; ATF6-HA-pCGN and HRD1 siRNA, n = 20; ATF6(1–373)-HA-pCGN and control siRNA, n = 20; ATF6(1–373)-HA-pCGN and HRD1 siRNA, n = 21). Statistical significance is indicated by *** (p ≤ 0.0001) and ns (p > 0.05). Bars, 25 μm. Error bars = standard deviations.
DISCUSSION
In this study, we showed that selective activation of the ATF6 branch of the UPR led to increased degradation of ATZ by an ERAD-dependent pathway that includes HRD1 and the proteasome. We also showed that the characteristic ATZ accumulations that appear in hepatoma cells as a consequence of the Z mutation, namely the IBs, were reduced by activation of the ATF6 pathway. Furthermore, it appears that selective induction of the ATF6 pathway of ERAD machinery, although promoting ATZ degradation, does not lead cells to apoptosis. This is important considering that sustained activation of the UPR and specifically of the ATF4 branch of the UPR promotes apoptosis (5). Here we found that although activation of ATF6 promoted increased expression of proapoptotic CHOP in the nucleus caspase 3 activation and chromatin condensation were not induced, indicating that apoptosis itself does not take place. Another potentially detrimental effect of activation of the UPR is phosphorylation of eIF2a and reduced protein synthesis with adverse effects on the ability of the hepatocyte to synthesize and secrete major protein components of the blood. In this respect, it appears that activation of the ATF6 pathway does not impair translation of both endogenous and exogenous proteins. The lack of increased apoptosis and of inhibition of protein synthesis in the in the ATZ-expressing hepatoma cells with induced ATF6 pathway is consistent with the observation that the ATF4 mRNA levels are not increased, thereby indicating that the PERK/eIF2a/ATF4 pathway is independent of ATF6. Another aspect of induction of the ATF6 pathway is the expansion of the ER (12), which may cause a relative inability to export proteins along the secretory pathway. However, this does not seem to be the case because secretion of AAT in the medium was maintained. It was concluded that activation of the ATF6 pathway in the ATZ-expressing cells accelerates ATZ disposal without impairing synthesis and secretion of natively folded proteins and without causing apoptosis. A characteristic feature of expression of ATZ in hepatocytes is mitochondrial damage (33). Here we found that activation of the ATF6 pathway restored mitochondrial potential to the same level as in the absence of ATZ expression. Thus, it appears that activation of the ATF6 pathway not only increases specifically the degradation of ATZ but also reduces cell toxicity. The work presented here addresses the issue of whether induction of a branch of the UPR specifically increases ATZ degradation without decreasing hepatocyte function or promoting apoptosis. In this respect, although modulation of proteostasis has been proposed as a target for therapy of misfolded proteins (39), knowledge on the consequences of such interventions on general cell function is limited.
We found that, in the absence of induction of ATF6, ERAD of ATZ by HRD1 was a major pathway of ATZ degradation. In this respect, HRD1 silencing in hepatoma cells increased by ∼3-fold the amount of intracellular ATZ, promoted the appearance of high molecular weight species of ATZ, and increased the size and ATZ content of the IBs. The IBs induced by ATZ expression in the hepatoma cells were similar to the globules in the hepatocytes of PiZZ individuals and PiZ transgenic mice not only by having a ribosome-associated limiting membrane (15, 18) but also increased density as compared with the main ER (Fig. 1). Our finding that the HRD1 pathway of degradation is a major route of ATZ disposal is consistent with the conclusion that disposal of mutant neuroserpin with propensity to polymerize is impaired by cell incubation with the proteasome inhibitor MG132 and is inhibited by HRD1 silencing (25, 40). However, incubation with MG132 did not impair bulk disposal of ATZ in the hepatoma cells and did so only when the ATF6 pathway was activated. This suggests that MG132 does not efficiently inhibit ATZ degradation by the proteasome and that this inhibition becomes apparent only when disposal of the variant is promoted by activation of ATF6. Our data suggest that a relative insufficiency of the HRD1-dependent pathway of ATZ degradation leads to formation of the hepatic globules. HRD1-dependent disposal of ATZ is likely to occur in the ER rather than in the IBs because HRD1 silencing functions to inhibit degradation of ATZ when it occurs before, rather than after, the appearance of the IBs. The data presented here converge to a model where an early step in the degradation of ATZ occurs by an HRD1-dependent mechanism in the ER that would clear up most of the protein. The fraction of ATZ still persisting in the ER would accumulate in the form of high density aggregates in subregions of the ER that appear as globular inclusions. This model is consistent with the findings that in the transfected hepatoma cells HRD1 silencing led to further increased intracellular ATZ with most of the protein appearing as high molecular weight forms and that the IBs appear as dilated sections of the ER co-existing with ER of normal appearance (Fig. 1). We have proposed that formation of IBs originating from the main ER is a protective mechanism to limit ATZ toxicity (15) (41). The new data presented here suggest that IBs may function as a protective mechanism to maintain ER homeostasis when the HRD1 pathway of ATZ degradation is overloaded. Activation of the ATF6 pathway by preventing or delaying such an overload would reduce formation of the IBs. It has been reported recently that transcriptional elevation of an ERAD component, EDEM1, increases the efficiency of degradation of ATZ function, by maintaining ER mannosidase I function (29). Moreover, a mutation in the 3′-untranslated region of ER mannosidase I may accelerate the onset of the end stage liver disease (42). More work has to be done to determine whether EDEM1 and ER mannosidase I function upstream of HRD1 to facilitate degradation of ATZ. The data presented here show that induction of the ATF6 pathway does not promote formation of autophagosomes. This and the finding that ATF6 degraded ATZ by a pathway that was both dependent on HRD1 and the proteasome are consistent with the conclusion that the transcription factor promotes degradation of ATZ by classical ERAD rather than by autophagy. On the other hand, it appears that inhibition of the HRD1 pathway of ATZ degradation induces autophagy most likely consequent to the increased amount of intracellular ATZ. It has been reported recently that autophagy-enhancing drugs such as carbamazepine and rapamycin promote ATZ degradation and reduce liver fibrosis (43, 44). These data suggest that although ERAD of ATZ would function early after ATZ synthesis, increased autophagy may function to dispose of ATZ once the globular inclusions are already formed in the hepatocytes. Because the ATF6/HRD1-dependent pathway of ATZ degradation does not involve autophagy, this pathway would be another potential target to promote disposal of ATZ by the liver in addition to the autophagy-promoting drugs.
Acknowledgments
We are grateful to Dr. Ron Prywes from the Department of Biological Sciences, Columbia University (New York, NY) for the kind gift of the plasmids for ATF6-HA and ATF6(1–373)-HA and to Jeffrey Kamykowski and Dr. Brian Storrie for help with the electron microscopy (Electron Microscopy Core Facility, Digital Microscopy Core Facility, University of Arkansas for Medical Sciences). We are also grateful to Marie Burdine and Dr. Alan Tackett in the Department of Biochemistry (University of Arkansas for Medical Sciences) for help and guidance in utilizing quantitative real time PCR techniques.
This work was supported, in whole or in part, by National Institutes of Health Grant UL1RR029884 from the National Center for Research Resources (to the University of Arkansas for Medical Sciences Translational Research Institute). This work was also supported by a grant from the Alpha-1 Foundation, the Arkansas Tobacco Settlement (to Giulia Baldini), a postdoctoral grant from the Secretaria de Estado de Universidades Del Ministerio de Educacion y Ciencia, Spain (to S. G.), and Grant BFU2009-07186 from the Ministerio de Innovación y Ciencia, Spain (to G. E.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- ATZ
- α1-antitrypsin with an E342K (Z) mutation
- ATF
- activating transcription factor
- ER
- endoplasmic reticulum
- IB
- inclusion body
- ERAD
- endoplasmic reticulum-associated degradation
- UPR
- unfolded protein response
- AAT
- wild-type α1-antitrypsin
- CHOP
- CCAAT/enhancer-binding protein (C/EBP) homologous protein
- PERK
- protein kinase RNA-like ER kinase
- IRE1
- inositol-requiring enzyme 1
- XBP
- X box-binding protein
- XBP1s
- spliced XBP1
- CCCP
- carbonyl cyanide 3-chlorophenylhydrazone
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1. Perlmutter D. H. (2011) Annu. Rev. Med. 62, 333–345 [DOI] [PubMed] [Google Scholar]
- 2. Ekeowa U. I., Freeke J., Miranda E., Gooptu B., Bush M. F., Pérez J., Teckman J., Robinson C. V., Lomas D. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 17146–17151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Teckman J. H. (2007) Semin. Liver Dis. 27, 274–281 [DOI] [PubMed] [Google Scholar]
- 4. Hidvegi T., Schmidt B. Z., Hale P., Perlmutter D. H. (2005) J. Biol. Chem. 280, 39002–39015 [DOI] [PubMed] [Google Scholar]
- 5. Malhi H., Kaufman R. J. (2011) J. Hepatol. 54, 795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaufman R. J., Back S. H., Song B., Han J., Hassler J. (2010) Diabetes Obes. Metab. 12, Suppl. 2, 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Malhotra J. D., Kaufman R. J. (2007) Semin. Cell Dev. Biol. 18, 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 9. Schröder M., Kaufman R. J. (2005) Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 10. Sriburi R., Jackowski S., Mori K., Brewer J. W. (2004) J. Cell Biol. 167, 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. (2007) Dev. Cell 13, 365–376 [DOI] [PubMed] [Google Scholar]
- 12. Bommiasamy H., Back S. H., Fagone P., Lee K., Meshinchi S., Vink E., Sriburi R., Frank M., Jackowski S., Kaufman R. J., Brewer J. W. (2009) J. Cell Sci. 122, 1626–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y., Shen J., Arenzana N., Tirasophon W., Kaufman R. J., Prywes R. (2000) J. Biol. Chem. 275, 27013–27020 [DOI] [PubMed] [Google Scholar]
- 14. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 15. Granell S., Baldini G., Mohammad S., Nicolin V., Narducci P., Storrie B., Baldini G. (2008) Mol. Biol. Cell 19, 572–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 17. Kamimoto T., Shoji S., Hidvegi T., Mizushima N., Umebayashi K., Perlmutter D. H., Yoshimori T. (2006) J. Biol. Chem. 281, 4467–4476 [DOI] [PubMed] [Google Scholar]
- 18. Teckman J. H., Perlmutter D. H. (2000) Am. J. Physiol. Gastrointest. Liver Physiol. 279, G961–G974 [DOI] [PubMed] [Google Scholar]
- 19. Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009) Nature 458, 453–460 [DOI] [PubMed] [Google Scholar]
- 20. Chen X., Shen J., Prywes R. (2002) J. Biol. Chem. 277, 13045–13052 [DOI] [PubMed] [Google Scholar]
- 21. Bobrovnikova-Marjon E., Diehl J. A. (2007) Dev. Cell 13, 322–324 [DOI] [PubMed] [Google Scholar]
- 22. Le A., Graham K. S., Sifers R. N. (1990) J. Biol. Chem. 265, 14001–14007 [PubMed] [Google Scholar]
- 23. Teckman J. H., Perlmutter D. H. (1996) J. Biol. Chem. 271, 13215–13220 [DOI] [PubMed] [Google Scholar]
- 24. Teckman J. H., Burrows J., Hidvegi T., Schmidt B., Hale P. D., Perlmutter D. H. (2001) J. Biol. Chem. 276, 44865–44872 [DOI] [PubMed] [Google Scholar]
- 25. Kroeger H., Miranda E., MacLeod I., Pérez J., Crowther D. C., Marciniak S. J., Lomas D. A. (2009) J. Biol. Chem. 284, 22793–22802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. An J. K., Blomenkamp K., Lindblad D., Teckman J. H. (2005) Hepatology 41, 160–167 [DOI] [PubMed] [Google Scholar]
- 27. Bernasconi R., Galli C., Calanca V., Nakajima T., Molinari M. (2010) J. Cell Biol. 188, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Teckman J. H., Gilmore R., Perlmutter D. H. (2000) Am. J. Physiol. Gastrointest. Liver Physiol. 278, G39–G48 [DOI] [PubMed] [Google Scholar]
- 29. Termine D. J., Moremen K. W., Sifers R. N. (2009) J. Cell Sci. 122, 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Høyer-Hansen M., Jäättelä M. (2007) Cell Death Differ. 14, 1576–1582 [DOI] [PubMed] [Google Scholar]
- 31. Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Dröge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fésüs L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., González-Estévez C., Gorski S., Gottlieb R. A., Häussinger D., He Y. W., Heidenreich K., Hill J. A., Høyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jäättelä M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovács A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., López-Otín C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Meléndez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Münz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nürnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Tallóczy Z., Tanaka K., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcátegui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Teckman J. H., An J. K., Blomenkamp K., Schmidt B., Perlmutter D. (2004) Am. J. Physiol. Gastrointest. Liver Physiol. 286, G851–G862 [DOI] [PubMed] [Google Scholar]
- 34. Okada T., Yoshida H., Akazawa R., Negishi M., Mori K. (2002) Biochem. J. 366, 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., Ron D. (1998) Genes Dev. 12, 982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kumar S. (2007) Cell Death Differ. 14, 32–43 [DOI] [PubMed] [Google Scholar]
- 37. Egawa N., Yamamoto K., Inoue H., Hikawa R., Nishi K., Mori K., Takahashi R. (2011) J. Biol. Chem. 286, 7947–7957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Senkal C. E., Ponnusamy S., Bielawski J., Hannun Y. A., Ogretmen B. (2010) FASEB J. 24, 296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. (2008) Science 319, 916–919 [DOI] [PubMed] [Google Scholar]
- 40. Ying Z., Wang H., Fan H., Wang G. (2011) J. Biol. Chem. 286, 20835–20844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Granell S., Baldini G. (2008) Autophagy 4, 375–377 [DOI] [PubMed] [Google Scholar]
- 42. Pan S., Huang L., McPherson J., Muzny D., Rouhani F., Brantly M., Gibbs R., Sifers R. N. (2009) Hepatology 50, 275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaushal S., Annamali M., Blomenkamp K., Rudnick D., Halloran D., Brunt E. M., Teckman J. H. (2010) Exp. Biol. Med. 235, 700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hidvegi T., Ewing M., Hale P., Dippold C., Beckett C., Kemp C., Maurice N., Mukherjee A., Goldbach C., Watkins S., Michalopoulos G., Perlmutter D. H. (2010) Science 329, 229–232 [DOI] [PubMed] [Google Scholar]