Background: Inhibition of pathogenic protein aggregation by small molecules is poorly understood.
Results: Aggregation inhibitors alter the free energy landscape of a relevant fragment of Alzheimer amyloid-β peptide in subtle but complex fashion.
Conclusion: Intrinsic disorder of disease proteins persists at the level of binding small molecules.
Significance: Hallmark characteristics and efficacy of inhibitors can be reconciled with lack of specificity.
Keywords: Alzheimer Disease, Computation, Intrinsically Disordered Proteins, Molecular Dynamics, Protein Aggregation, Protein Conformation, Protein Drug Interactions, Aggregation Inhibitors
Abstract
In recent years, an increasing number of small molecules and short peptides have been identified that interfere with aggregation and/or oligomerization of the Alzheimer β-amyloid peptide (Aβ). Many of them possess aromatic moieties, suggesting a dominant role for those in interacting with Aβ along various stages of the aggregation process. In this study, we attempt to elucidate whether interactions of such aromatic inhibitors with monomeric Aβ(12–28) point to a common mechanism of action by performing atomistic molecular dynamics simulations at equilibrium. Our results suggest that, independently of the presence of inhibitors, monomeric Aβ(12–28) populates a partially collapsed ensemble that is largely devoid of canonical secondary structure at 300 K and neutral pH. The small molecules have different affinities for Aβ(12–28) that can be partially rationalized by the balance of aromatic and charged moieties constituting the molecules. There are no predominant binding modes, although aggregation inhibitors preferentially interact with the N-terminal portion of the fragment (residues 13–20). Analysis of the free energy landscape of Aβ(12–28) reveals differences highlighted by altered populations of a looplike conformer in the presence of inhibitors. We conclude that intrinsic disorder of Aβ persists at the level of binding small molecules and that inhibitors can significantly alter properties of monomeric Aβ via multiple routes of differing specificity.
Introduction
Alzheimer disease (AD)3 is the most common form of dementia in the elderly. Strong genetic, physiological and biochemical evidence suggests that the β-amyloid peptide (Aβ) plays a key role in the pathogenesis of AD (1). Neuropathological changes in the brain of AD patients include neuronal death in the regions related to memory and cognition as well as the abnormal presence of intra- and extracellular protein aggregates (2, 3) known as neurofibrillary tangles and amyloid plaques. They are the final results of a complex series of oligomerization and polymerization events that typically follow a nucleation-dependent mechanism (4). The left-hand side of Fig. 1 shows a schematic illustrating a few steps along the pathway of Aβ fibrillization. The nucleus is typically assumed to be a larger oligomer (4–6), and the nucleation event itself may be linked to a critical structural transition involving tertiary and quaternary contacts within such an oligomer or protofibril (5). Subsequent monomer addition appears to be the dominant mode of fibril elongation (4). Peptide aggregation processes have been studied in depth with several experimental (7, 8) and computational techniques (9–12) but often remain poorly understood. Although little is known about the link between the aggregation mechanism and neurotoxicity (13), experimental evidence indicates that soluble oligomers and fibrillar precursors of Aβ may be the dominant neurotoxic species (14).
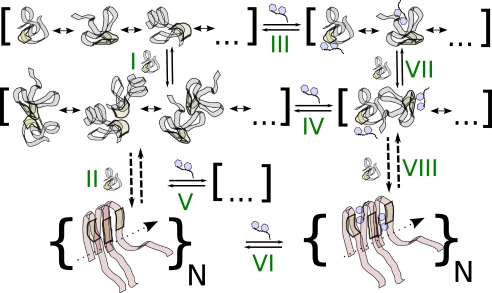
FIGURE 1.
Schematic depicting the coupled equilibria that could be involved in inhibition of fibrillization and/or oligomerization of Aβ by small molecules. Largely unstructured peptides are shown as gray ribbons with the central hydrophobic cluster highlighted in yellow. Peptides in fibril-compatible conformations are shown in light red, and inhibitor molecules are shown in blue. Square brackets indicate conformational equilibria at different assembly levels, and curly brackets indicate a repeating unit replicated along the indicated axis. Steps I and II pertain to the unperturbed, nucleation-dependent aggregation pathway. On- and off-pathway assembly steps beyond the dimer are not shown explicitly. Steps VII and VIII are the analogous steps with inhibitor bound to the aggregating peptides. Finally, steps III–VI describe binding equilibria to various peptide species. Preferential interactions of the central hydrophobic cluster with inhibitor molecules containing aromatic moieties have been postulated (81). See “Results and Discussion” for further details.
In recent years, increasing evidence points to a link between disease and disorder, specifically the functions and properties of intrinsically disordered proteins (IDPs) and polypeptide stretches within proteins (15, 16). The ensembles explored by such sequences, which are estimated to make up about 20% (17) of eukaryotic genomes, are highly diverse and devoid of long lived, “folded” conformers (18). Extensive analyses have shown that simple sequence-based classifiers, such as mean hydrophobicity or net charge, can be used to distinguish folded proteins from IDPs (19). Aβ40/42 belongs to the class of collapsed-disordered IDPs (20) on account of its low net charge and high hydrophobicity (21, 22). IDPs often attain partial order upon functional or deleterious interactions with folded proteins or with other IDPs (23). Indeed, pathogenic self-assembly can be viewed as a specific variant of the latter case. Given that collapse and aggregation are guided by the same driving forces, it is perhaps not surprising that IDPs, such as Aβ or polyglutamine, are associated with protein aggregation diseases (24–26).
Inherently, structural drug design aimed at finding compounds that interfere with an IDP-mediated process faces the challenge that structural targets emerge only later on the pathway. Nevertheless, the identification and detailed biophysical characterization of small molecules that modulate Aβ peptide self-assembly are expected to generate new lead candidates for clinical studies. Several therapeutic strategies have been suggested for blocking key steps in the amyloid aggregation process, including the direct inhibition of aggregation by using either peptides or small molecules (27–38). As an example, indole derivatives inhibited fibril formation of Aβ peptide (39, 40) and lysozyme (41). Anthraquinones were shown to be inhibitors of Tau protein (42) and Aβ40 aggregation (37), and hybrid molecules bearing both indole and quinone rings have been effective in the recovery of a fly model of AD (43). In addition, antioxidants (e.g. resveratrol (44, 45) and epigallocatechin-3-gallate (46)) and non-steroidal anti-inflammatory molecules, such as naproxen (47, 48), revealed new biological activities in the inhibition of amyloid aggregation.
Recent x-ray microcrystallography (49, 50) and solid-state nuclear magnetic resonance (NMR) spectroscopy (51) studies have provided atomistic information on the interactions between small molecule binders and amyloid fibrils. Fig. 1 illustrates why this may be less relevant than the interactions of inhibitors with soluble peptide species. In essence, compounds that specifically bind fibrils (step VI in Fig. 1) may destabilize the latter (52) but will have little impact on the association and conformational equilibria prior to nucleation (steps I and II). An alternative mechanism of inhibition could be a depletion of nucleation-competent (53) and/or toxic oligomers (54) either by stabilization of low molecular weight species, such as monomers and dimers (steps III and IV), or stabilization of larger off-pathway oligomers (step V). Given the disordered nature of the binding partner (55), it is quite reasonable to stipulate that small molecules can have differential effects for all indicated steps, including the known ability to increase the rate of fibrillization (56, 57). The complexity is exacerbated by the fact that the dominant pathways may shift in a dose-dependent manner. Fig. 1 implies that studies of the binding equilibria of monomeric Aβ with inhibitors can yield insight regarding the molecular mechanisms of inhibition.
Here, we use molecular dynamics (MD) simulations to analyze 10 different small molecule inhibitors of Aβ peptide aggregation and focus on their influence on the free energy surface of monomeric Aβ(12–28). The choice of studying a truncated construct is motivated by three reasons. First and foremost, it is important to be able to obtain statistically reliable simulation results. Conformational transitions in the full-length alloforms can occur on time scales that exceed the currently accessible regime (58). Second, residues 12–28 highlight the role of the so-called central hydrophobic cluster (59), residues 17–21, which is often assumed to be critical in mediating peptide-inhibitor (60) as well as peptide-peptide interactions (61). By discarding the hydrophobic C terminus, our simulations allow us to delineate the possible specific roles played by the residues in this stretch. Truncation of residues 1–11 is justified by experimental (62) and simulation (21) studies that show this segment to be completely unstructured. Third, segment 12–28 could be used in concentrations high enough to be suitable for solution NMR spectroscopy experiments (43) because it has lower oligomerization and fibrillization propensities than Aβ40 and Aβ42. This implies that it will be easier to derive testable hypotheses. We use the cut-based free energy profile (cFEP) method (63) to identify the metastable states of monomeric Aβ(12–28) and the change of their relative stability upon inhibitor binding.
This study was inspired by the following questions. How does the free energy surface of monomeric Aβ(12–28) change in the presence of small molecules that are known to interfere with oligomerization and/or fibril formation? Do different inhibitors of Aβ peptide self-assembly share similar interaction motifs with monomeric Aβ(12–28)? Is there a major binding mode? The MD simulation results indicate that monomeric Aβ(12–28) is largely disordered with and without inhibitors. The most frequent interaction motifs are similar for different inhibitors. There is no predominant binding mode because Aβ(12–28) is highly flexible, and its plasticity is marginally influenced by the small molecule inhibitors. An analysis of binding frequency and the enhancement of a specific, otherwise transiently populated conformation of Aβ(12–28) in the presence of inhibitors suggests a complex interplay of interfacial effects, trends that can be mapped back to simple physicochemical properties of the primary sequence, and last, highly specific effects that require elucidation by atomistic simulations.
EXPERIMENTAL PROCEDURES
Implicit Solvent Simulations
Simulations were performed with the CHARMM program (64). The Aβ(12–28) peptide and inhibitors were modeled using the united atoms CHARMM PARAM19 force field with its default truncation scheme for non-bonded interactions (cut-off of 7.5 Å). Parameters for 1,4-napthoquinon-2-yl-l-tryptophan (NQTrp), anthracene, and 9,10-anthraquinone were derived as reported (37, 43). Protonation states of titratable residues were considered at neutral pH. In particular, the His side chains of Aβ(12–28) and β-Ala-His were neutral (protonated at the Nδ), whereas the charges of the Asp/Glu and Arg/Lys side chains were −1 and +1 electronic units, respectively. The net charge of the Aβ(12–28) segment is zero because there are two positively charged residues (Lys-16 and Lys-28) and two negatively charged residues (Glu-22 and Asp-23), and the N and C terminus were capped with acetyl and N-methylamide groups, respectively. The electrostatic contribution to solvation was accounted for by using FACTS (65), an efficient generalized Born implicit solvent model based on the fully analytical evaluation of the volume and spatial symmetry of the solvent that is displaced from around a solute atom by its neighboring atoms. The non-polar contribution to the total effective solvation energy was approximated by a term proportional to the solvent-accessible surface area of the solute using a surface tension-like, multiplicative parameter of 7.5 cal mol−1 Å−2. Starting conformations were prepared by placing fully extended Aβ(12–28) in the presence or absence of a single molecule of the inhibitor in the simulation box (1:1 concentration ratio). Simulations were carried out with periodic boundary conditions at a fixed peptide concentration of ∼2.5 mm (87-Å cubic simulation box) using the Langevin integrator at low friction (coefficient of 0.15 ps−1) and at a temperature of 300 K. Using a time step of 2 fs, for each system, we performed three independent runs of 5 μs each.
Explicit Solvent Simulations
Using GROMACS version 4.5.3 (66), capped Aβ(12–28) was simulated in a cubic box of 60-Å side length in the isothermal-isobaric ensemble. The velocity rescaling thermostat of Bussi et al. (67) was used to keep a constant temperature of 310 K, whereas an ambient pressure of 1 bar was maintained using the Parrinello-Rahman barostat (68). The peptide was represented with the CHARMM27 all-atom force field, including CMAP corrections (69). The bath consisted of a solution of ∼150 mm NaCl in TIP3P water (70). Electrostatic interactions were modeled by the particle mesh Ewald method (71). All real space interactions were truncated at 12 Å. Neighbor lists were recalculated every five steps. LINCS (72) was used to constrain all bonds involving hydrogen atoms to their parameter-derived values. The time step was 2 fs, and we obtained three independent simulations starting from random, extended structures that each are 380 ns in length, the first 20 ns of which we discarded as equilibration. Preliminary analyses of secondary structure propensities or contact patterns revealed that, given the reduced amount of sampling and increased friction, statistical convergence for the majority of readouts could not be obtained. Therefore, data from explicit solvent simulations are only included in Fig. 2.
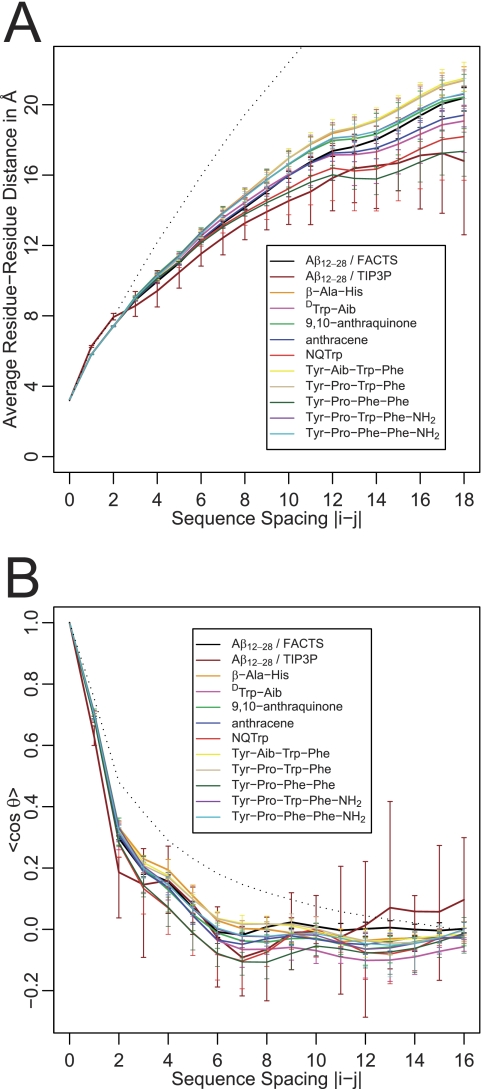
FIGURE 2.
Average polymeric properties for Aβ(12–28) alone and in the presence of various inhibitors. The indicated errors (error bars) are minimum and maximum values obtained from partitioning the data into six blocks. A shows the average atom-to-atom distance for pairs of residues as a function of sequence separation. Capping groups are included as separate residues in this analysis. Collapsed globules would be indicated by the internal distance reaching a plateau value for sequence spacings beyond the length scale of local rigidity. Random coil-like (good solvent) chains are significantly more extended, as indicated by the black, dotted line (data from simulation of Aβ(12–28) in the excluded volume limit (103)). B shows the angular correlation function of N → C vectors as a function of sequence separation. Negative values indicate the chain turning on itself (i.e. (partial) collapse). A good solvent chain leads to simple, monotonous decay of the correlation function (black dotted line).
cFEP Analysis
The 750,000 snapshots of each system were clustered by the Leader algorithm as implemented in Wordom (73) using the distance root mean square deviation of Cα atoms of residues 14–24 and a threshold of 1.0 Å. The cFEP (63) technique was used to identify metastable states of monomeric Aβ(12–28) and the change of their relative stability upon inhibitor binding. The input for the cFEP calculation is the network of conformational transitions, which is derived from the direct transitions between clusterized snapshots (nodes of the network) sampled at a given time interval (20 ps here) along the MD simulations. For each node, nodes are partitioned into two groups using the values of the mean first passage times to the reference node to define a cut. The free energy is related to the maximum flow across the cut and approximated as ΔG = −kTln(ZAB), where ZAB is the partition function of the mean first passage time-based cutting surface (for further details, see Ref. 63). The result is a one-dimensional profile along a reaction coordinate (the relative partition function, termed ZA/Z) that preserves the barrier height between well-separated free energy basins.
RESULTS AND DISCUSSION
The following analysis concerns the FACTS implicit solvent MD simulations at 300 K. The reference system is monomeric Aβ(12–28), whereas the simulations with inhibitors contained a single inhibitor molecule along with Aβ(12–28) at a final concentration of ∼2.5 mm. Each of these 11 systems was simulated for a total of 15 μs. If not stated otherwise, the statistical significance (i.e. convergence) of the simulation results was assessed by computing minimum/maximum errors from block averages over three or six blocks.
Monomeric Aβ(12–28) Is Partially Collapsed and Disordered
Visual inspection of the trajectories showed that Aβ(12–28) does not attain any specific, long lived structure akin to a folded ensemble. Conformational transitions are rapid and yield a disordered ensemble. To quantitatively assess the overall polymeric state of the Aβ peptide, we computed the scaling of internal distances with sequence spacing as well as the angular correlation function, as described in previous work (74). The data in Fig. 2 indicate that, independently of the presence of an inhibitor, the peptide populates a partially collapsed ensemble. Inhibitors appear to be able to cause both compaction and swelling of Aβ(12–28), but many of the differences are insignificant. For fully globular species, one would expect the internal scaling plot to yield a plateau (75) similar to what we observed for the full-length alloforms (21), whereas the theoretical prediction for a chain in a good solvent is significantly more expanded. As described under “Experimental Procedures,” we performed calculations in explicit solvent of the free system. These data are shown as well in Fig. 2. They crudely illustrate similarity between implicit and explicit solvent ensembles (Fig. 2A) and simultaneously demonstrate the difficulty to obtain converged explicit solvent results even for low dimensional readouts, such as the angular correlation function (Fig. 2B).
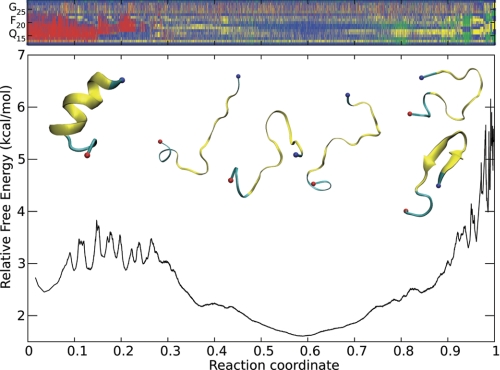
To demonstrate dominant disorder in Aβ(12–28), we show in Fig. 3 the cFEP of the free system. The profile is devoid of significant free energy barriers and dominated by a large entropic basin composed of conformations that lack canonical secondary structure. The remaining 30–40% of the ensemble is made up by various enthalpic basins, and schematic representations of representative snapshots have been added for illustrative purposes. Taken together, these results indicate that Aβ(12–28) is unstructured at physiological conditions in the absence of small molecule inhibitors of aggregation. Similar observations hold for Aβ(12–28) in the presence of any of the aggregation inhibitors (see supplemental Figs. S2–S11). The lack of a predominant structure and the small height of barriers for the ensemble of monomeric Aβ(12–28) are consistent with data from NMR spectroscopy experiments (76).4
FIGURE 3.
Monomeric Aβ(12–28) is mainly unstructured. Top, DSSP analysis of the sampling arranged according to the reaction coordinate of the cFEP (the color code is as follows: helix (DSSP assignment letters “H,” “G,” and “I”) in red; β-extended (letters “E” and “B”) in green; loop and turn (letter “T” or unassigned) in blue; DSSP assignment “S” corresponds to backbone conformations with high curvature and is plotted in yellow for clarity). Bottom, the cFEP of monomeric Aβ(12–28), using its most populated conformer as the reference state (the value of the reaction coordinate is zero for the reference state). Representative conformations of individual free energy basins are reported as insets (residues 14–24 are highlighted in yellow in the schematics), because only they were used in the determination of the cFEP.
Main Interactions between Monomeric Aβ(12–28) and Aggregation Inhibitors
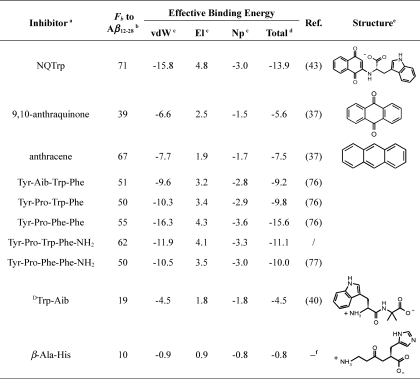
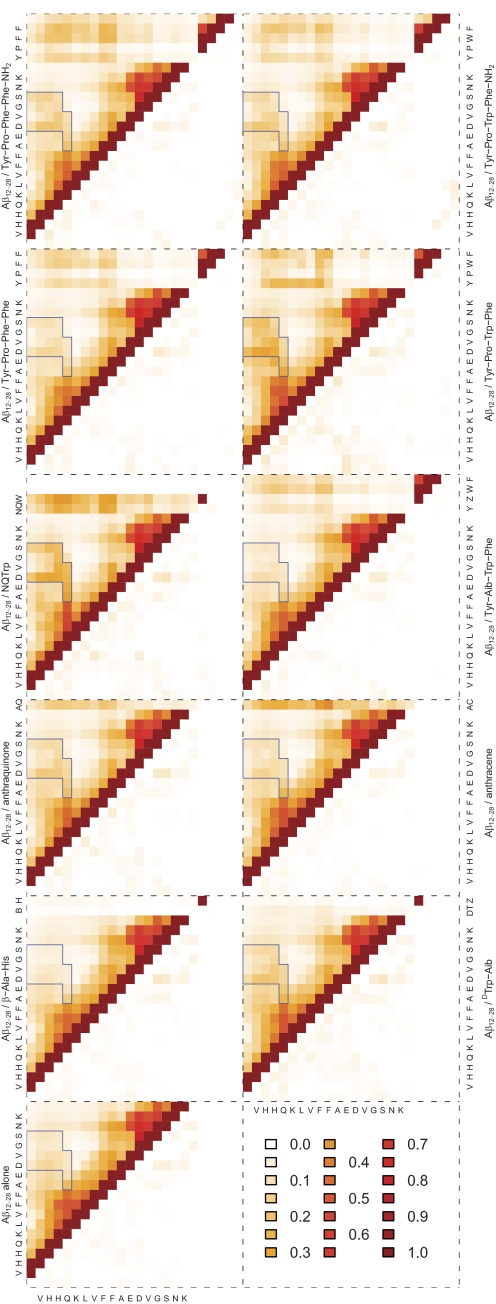
Table 1 shows estimates for how frequently each inhibitor is “bound” to the peptide in the simulations. To resolve which parts of the peptide the various inhibitors bind to, Fig. 4 shows intra- and intermolecular contact maps for the free system as well as for simulations in the presence of inhibitors. The most frequently observed intermolecular contacts are shared by all inhibitors and involve mainly the N-terminal segment (residues 13–20). With the exception of β-Ala-His, which largely remains dissociated, the large hydrophobic side chains of Phe-19 and Phe-20 constitute the site of highest interaction probability in all cases. It is interesting to note that the larger peptidic inhibitors appear to show more specific contact patterns than, for example, anthracene or 9,10-anthraquinone. This is consistent with the complexity of the interfaces presented by each molecule; anthracene is a completely homogeneous tricyclic molecule, whereas the tetrapeptides have three different aromatic moieties (two of which have polar sites), one or two formal charges, and a mainly rigid spacer. The segment 24–28 appears to be largely inert to inhibitor binding, consistent with the fact that residues in this region do not feature prominently in sequence analyses (62, 78, 79), mutation (80), or fragment binding studies (61) as being responsible for amyloid formation. Although this is also true for the N-terminal residues we truncated, the same cannot be said for C-terminal residues 29–40/42. A priori, we have no reason to assume that inhibitors would not bind to this portion of Aβ (see Fig. 1). However, the questions in this work center around the interaction of aromatic inhibitors with the central hydrophobic core as the suspected site of highest affinity (81). Given that several of the inhibitors have significant impact on oligomerization and fibrillization (36, 43) even at subequimolar concentration ratios, this may represent a reasonable simplification.
TABLE 1.
Small molecule inhibitors and their affinity to Aβ(12–28)
a Aib, α-aminoisobutyric acid; NQTrp, 1,4-napthoquinon-2-yl-l-tryptophan; Tyr-Aib-Trp-Phe, designed derivative of endomorphin-1; Tyr-Pro-Trp-Phe, endomorphin-1 with charged carboxyl group at the C terminus; Tyr-Pro-Phe-Phe, endomorphin-2 with charged carboxyl group at the C terminus; Tyr-Pro-Trp-Phe-NH2, endomorphin-1; Tyr-Pro-Phe-Phe-NH2, endomorphin-2; β-Ala-His, carnosine. Note that the endogenous endomorphins have the -NH2 group at the C terminus.
b Fb is the fraction of snapshots in which any atom of the inhibitor is within 7.5 Å of any atom of Aβ(12–28). Values are given as percentages.
c Each of the these three terms (van der Waals terms (vdW), electrostatic interactions (El), and nonpolar solvation terms (Np)) was calculated by subtracting the energy of Aβ(12–28) and inhibitor in the isolated state from the energy in the ensemble of bound conformations.
d The effective binding energy is the sum of the changes in the van der Waals energy, electrostatic energy, and non-polar solvation energy upon binding. All values are reported in kcal/mol. They include all contributions to the binding free energy except for changes in entropies of the solutes.
e Structure diagrams are omitted for tetrapeptides.
f —, F. Attanasio, M. Convertino, A. Caflisch, A. Corazza, G. Esposito, S. Cataldo, B. Pignataro, D. Milardi, and E. Rizzarelli, submitted for publication.
FIGURE 4.
Intra- and intermolecular contact maps of Aβ(12–28) alone and in the presence of various inhibitors. The color legend applies to all panels. Axes are labeled with single-letter amino acid codes. B, β-alanine; DT, DTrp; NQ, naphtoquinone; Z, α-aminoisobutyric acid; AC, anthracene; AQ, for anthraquinone. Capping groups for peptides are considered as separate residues but are not labeled on the axes. A contact is defined as any two atoms of the corresponding residues being separated by less than 5.5 Å. The diagonal is excluded. The top left half matrices contain average contact probabilities, and the bottom right half matrices show the corresponding S.E. values estimated as half the difference between the minimum and maximum values measured over six blocks. The region denoted by blue lines shows a specific contact pattern corresponding to the loop structure discussed under “Results and Discussion.”
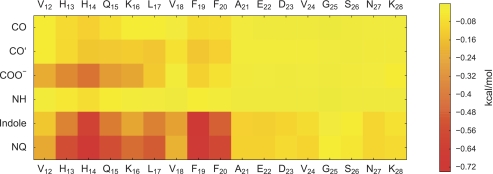
We can attempt to explain the interaction patterns within the confines of the employed computational model by decomposing system energetics. As an example, we focus on NQTrp, which has the highest affinity for Aβ(12–28) in the MD simulations (see Table 1) and is also one of the most potent inhibitors of Aβ peptide aggregation in experiments (43). In Fig. 5, the decomposition of the dominant term of the intermolecular energy into contributions from individual pairs of functional groups shows that the two aromatic moieties of NQTrp make favorable van der Waals interactions with the entire N-terminal stretch of the peptide (residues 13–20). In addition, binding appears to feature favorable electrostatic interactions involving the (charged) carboxyl group of NQTrp and peptide residues 12–16, which possess a wealth of polar hydrogens (His-13, His-14, Gln-15, and Lys-16). This can be inferred indirectly from the favorable van der Waals interactions between the carboxyl group and the peptide. We did not consider electrostatic interactions here because of effective multibody terms preventing pairwise decomposition. In general, good correspondence is seen between the interaction energy matrix and contact maps indicating that binding is largely enthalpic. This is expected in particular given that entropic contributions to ligand binding stemming from the solvent are accounted for implicitly in the continuum model.
FIGURE 5.
Matrix of van der Waals interaction energy between NQTrp and Aβ(12–28). The interaction energy between individual functional groups of NQTrp and Aβ(12–28) residues (backbone and side chain atoms) was computed by CHARMM using every 100th snapshot. NQTrp was decomposed into single functional groups with net integer charges (CO and CO′, COO−, NH, indole, and NQ, quinonic carbonyls, carboxyl, amide, indole, and naphtoquinone moieties, respectively (see Table 1 for the chemical structure of NQTrp)). The sum of all pairwise averages reported in the matrix is −13.3 kcal/mol.
It is possible to rationalize the relative affinities for Aβ(12–28) to the 10 inhibitors by focusing on the three main contributions (i.e. unfavorable desolvation of hydrophilic moieties, favorable burial of hydrophobic moieties, and favorable van der Waals interactions) (see Table 1 and supplemental Table S2). The total polar desolvation penalty ranges from 1 to 5 kcal/mol and scales roughly with the size of the inhibitor. No persistent salt bridges are formed, and the contribution is generally unfavorable. Burial of hydrophobic surface contributes favorably. This term correlates with the number of aromatic moieties present on the inhibitor. Last, dispersive interactions between inhibitor and Aβ(12–28) contribute the bulk of the favorable effective binding energy. It appears as if this contribution becomes less predictable with increasing numbers of hydrophobic moieties present on the inhibitor (e.g. NQTrp has a significantly more favorable van der Waals contribution than either Tyr-Aib-Trp-Phe or Tyr-Pro-Trp-Phe despite possessing fewer aromatic moieties). The computed effective binding energies appear to correlate by rank order with the fractions bound (Fb in Table 1) reasonably well. Two exceptions are as follows. The first is Tyr-Pro-Phe-Phe, which appears to incur a significant entropic penalty upon binding that is consistent with a significantly more structured free energy surface of Aβ(12–28) in the presence of Tyr-Pro-Phe-Phe (see supplemental Fig. S7). On the contrary, anthracene exhibits high affinity despite its modest effective binding energy. Here, the entropic penalty at the inhibitor level is essentially zero (rigid molecule), and the contact map indicates the most degenerate binding (see Fig. 4), suggesting a minimal entropic penalty at the peptide level as well.
Returning to Fig. 4, we note that inhibitor binding has an impact on intramolecular contacts of Aβ(12–28) as well. Although the contact maps continue to indicate degenerate long range interactions within the peptide, the presence of inhibitors seems to enhance contacts between residues 12–15 and 21–26 (marked in the plots). This is a relatively weak effect that is observed for all inhibitors but β-Ala-His and most prominently for NQTrp. It points to the ability of molecules composed of similar building blocks to exert a generic effect on the conformational properties of Aβ(12–28), and this is discussed next.
Changes in the Free Energy Surface of Aβ(12–28) upon Inhibitor Binding
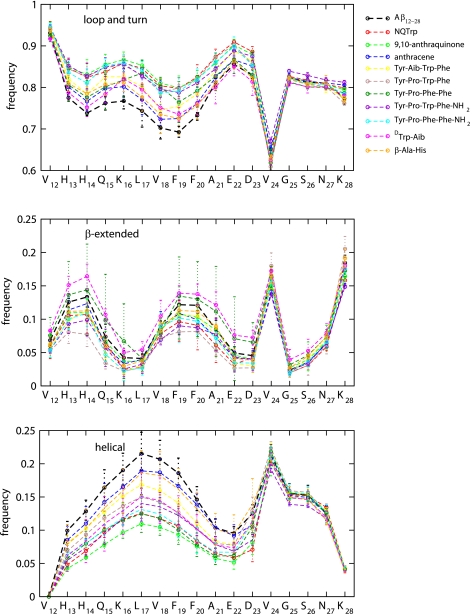
The DSSP strings in supplemental Figs. S1–S11 suggest that the overall secondary structure content of Aβ(12–28) is not strongly altered in the presence of inhibitors compared with the free system (Fig. 3). Fig. 6 shows that the inhibitors generally increase the content of loop, bend, and turn conformations in the segment 13–23 at the expense of regular secondary structure, particularly of helix. These changes are qualitatively similar for all inhibitors. As expected, the low affinity compounds β-Ala-His and DTrp-Aib exhibit only weak effects. The same is true for anthracene despite its affinity being the second highest (Table 1). This is consistent with the entropic binding mode described above. Over the same sequence, β-secondary structure is significantly enhanced only in the presence of DTrp-Aib and Tyr-Pro-Phe-Phe. In contrast to residues 13–23, there are negligible differences for residues 24–28, which is consistent with the lack of interactions between the inhibitors and the C-terminal segment of Aβ(12–28) (see Fig. 4). The effects are overall subtle and confirm the prevalence of disorder in binding.
FIGURE 6.
Influence of different inhibitors of aggregation on secondary structure propensities of monomeric Aβ(12–28). The secondary structure content was calculated by the DSSPcont algorithm (104) as implemented in Wordom (73). Top, loop and turn, DSSP letters L, S, and T. Middle, β-extended consists of DSSP letters E and B. Bottom, helical, DSSP letters G, H, and I. The error bars indicate the range between the maximum and minimum value over three independent 5-μs simulations. Please refer to Fig. 3 regarding details of DSSP assignments.
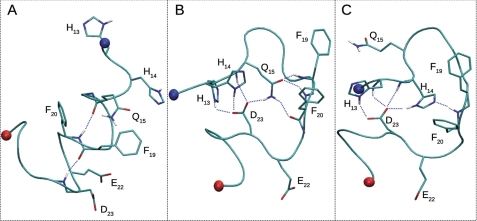
As noted above, the presence of inhibitors does seem to enhance intramolecular Aβ(12–28) contacts between residues 12–15 and 21–26 (see Fig. 4). This points to the potential of inhibitors to alter the free energy surface of Aβ(12–28) in a manner that cannot be captured clearly by evaluating ensemble-averaged readouts like secondary structure propensities. Cut-based FEPs represent an appropriate way to condense trajectory information and to show approximately the distributions of barriers and basins. In Fig. 3, free Aβ(12–28) exhibited no predominantly populated conformation. When clustering data using the Cα atoms of residues 14–24, the most populated conformer corresponds to a straight helical structure (see Fig. 7) with a disordered C terminus. The entire helical basin encompasses partially helical as well as completely helical structures that constitute a total statistical weight of about 10%. It is separated by a wide barrier region composed of minor basins with partially helical states (population of about 10%) from a broad entropic basin (70%). The latter consists of fluctuating conformers devoid of regular secondary structure content. Supplemental Figs. S2–S11 show the same type of data for simulations in the presence of inhibitors. In all cases, a large entropic basin has the largest statistical weight, indicating that inhibitors are not able to lock Aβ(12–28) into a specific conformation. There are changes, however, to the relative weights of ordered conformers. In particular, a basin is increasingly populated that corresponds to a compact structure characterized by a specific loop conformation spanning residues 14–24. It is stabilized (see Fig. 7) by the formation of a hydrogen-bonded network of side chains including Asp-23, His-13, and either Gln-15 or His-14. Its statistical weight for each inhibitor is summarized in supplemental Table S1 and ranges from ∼2 to 20%. Given that the population of the loop is lowest for the free system and in the presence of largely inert β-Ala-His and that it is only transiently populated in either case (see supplemental Figs. S12 and S13), one may ask how a specific Aβ(12–28) conformation can be universally enhanced by other inhibitors in the absence of a specific binding mode.
FIGURE 7.
Helical and loop conformations identified with cFEP. The helical conformation is shown in A. The loop conformation is stabilized by the hydrogen bonds between the side chain of Asp-23 (D23) and backbone NH groups as well as the side chains of either Gln-15 (Q15) (B) or His-14 (H14) (C).
The answer is that the inhibitors provide an additional non-aqueous interface (i.e. they allow patterns of sequestering residues from solvent to change). Normalized by their propensity to bind Aβ(12–28), the indole-containing inhibitors NQTrp (see supplemental Table S1) and DTrp-Aib exert the largest effect in this regard. Indole groups have been heavily implicated in Aβ aggregation inhibitor design (39) and were also analyzed in systematic fashion as inhibitors of hen egg white lysozyme aggregation (41). Interestingly, the inhibitor is not required to interact directly with the polar residues mentioned but can act indirectly or allosterically. Such a mechanism of action is entropically favored on account of the peptide's high intrinsic flexibility (this is true, for example, for DTrp-Aib, as seen in supplemental Fig. S14). However, supplemental Fig. S14 also shows the following counterexample. Direct binding of anthracene to the side chains of Asp-23 and Gln-15 occurs with greater frequency than binding in general, and population of the loop conformer in the absence of binding is essentially identical to that seen for the free system. These data resemble a conformational selection mechanism described in binding equilibria of intrinsically disordered proteins (82).
An interfacial effect as described is intriguing because it allows for an explanation as to why so many different inhibitors, which share some structural hallmarks, have been characterized (83, 84). If one compares the requirements for small molecule design in enzyme inhibition with that of inhibition of oligomerization and fibrillization, one would be forced to conclude that the intrinsic disorder of the target translates to the inhibitor level. This could imply that “polyfunctional” moieties combining largely hydrophobic parts with polar sites are well suited to bind disordered Aβ. Disordered interactions of such poly-functional units with peptide moieties may also explain why mutation studies have failed to establish the necessity for aromatic residues Phe-19 and Phe-20 in Aβ to be present for aromatic inhibitors of the general flavor tested here to be effective (85).
It should be noted that the partial desolvation of a charged moiety (Asp-23) may raise questions about the accuracy of the computational model in use. We wish to emphasize that we assign little importance to the structural details of the aforementioned loop structure on account of these concerns, although desolvation of Asp-23 is seen experimentally in the NMR structures (86) and in explicit solvent simulations (87) of similar fragments. It is worthwhile to point out that in our simulations, the statistical weight of the loop never exceeds 20%, meaning that most computed ensemble averages would only be moderately affected.
Comparison with NMR NOE Data
Aside from keeping the project computationally tractable, an added motivation for studying the specific fragment Aβ(12–28) was the availability of NMR spectroscopic data in the presence of different inhibitors. For the inhibitor Tyr-Aib-Trp-Phe, Frydman-Marom et al. (76) report a few intermolecular NOEs indicating preferential binding of Aib to residues 20–22. Undoubtedly, this is qualitatively compatible with our data as presented in Fig. 4. For Aβ(12–28) in the presence of NQTrp (4:1 excess of Aβ(12–28)), Scherzer-Attali et al. (43) report eight long range NOEs. Four of them cover a sequence spacing of only i, i + 4, and five of them involve Val-18. However, with the exception of an NOE between the α-proton of Asp-23 and the backbone amide proton of Gln-15, all of them are weak. When computing predicted NOEs from simulation data, we found a significant number of long range NOEs that should have been detectable by NMR irrespective of whether we use third or sixth power averaging (88). This presence of “false positives” indicates either incompatible ensembles, NMR-intrinsic issues when studying disordered systems (e.g. issues pertaining to the lifetime of conformational states) (89), or inapplicability of the model used for prediction, considering that most hydrogen positions had to be modeled a posteriori. Coupled to the fact that the source of NOE signals at a 4:1 excess concentration of Aβ(12–28) can hardly be expected to delineate Aβ-intrinsic and inhibitor-induced signals, we conclude that a quantitative comparison between measured NMR parameters and simulation data carries more caveats than potential. This is particularly true given the lack of reliable long range NOEs observed for Aβ(12–28) alone.4
Structural ensembles derived from NMR data suggest that Aβ(12–28) adopts looplike or hairpin-like structures devoid of canonical backbone hydrogen bonds (40, 43), and this trend is indirectly supported by independent studies on different fragments (86, 90, 91). Interestingly, the aforementioned loop state that is stabilized in the presence of inhibitors features a prominent contact between residues Gln-15 and Asp-23 (see Fig. 4). In our simulations, it is, however, the amide hydrogens of the glutamine side chain that mediate this contact (see Fig. 7B). Last, due to the very weak impact of inhibitors on average secondary structure populations (see Fig. 6), we do not attempt to correlate those changes to chemical shift differences observed in the presence of inhibitors.
CONCLUSIONS
The FACTS implicit solvation model allowed us to obtain data with statistical errors that permit establishing subtle, quantitative effects of inhibitor binding (e.g. see Fig. 6). Therefore, five main results emerge from the comparative analysis of MD simulations of Aβ(12–28) alone and in the presence of small molecules that have been shown experimentally to interfere with Aβ peptide self-assembly. 1) The free energy surface of Aβ(12–28) shows a broad entropic basin devoid of a predominant conformation (Fig. 3 and supplemental Figs. S1–S11). Upon inhibitor binding, significant but subtle changes to the free energy landscape occur, as exemplified by changes in average secondary structure propensities and intramolecular contacts (Figs. 4 and 6). 2) Different inhibitors share common intermolecular interactions, but none of them exhibits a specific binding mode irrespective of binding affinity (Table 1 and Fig. 4). 3) Differences in affinity can be partially rationalized by the relative number of aromatic groups and charged groups in the inhibitors (Table 1). The former provide a favorable contribution through hydrophobic interactions mainly with Phe-19 and Phe-20 and through mixed polar and nonpolar interactions with residues 13–18. The desolvation of charged groups is generally unfavorable (i.e. inhibitor binding does not rely on salt bridge formation) (Figs. 4 and 5). 4) The simulation data demonstrate that detailed predictions of interactions at the molecular level cannot just rely on simple heuristics (92). As an example, the relative binding properties of the different endomorphin variants appear nearly impossible to rationalize with purely sequence-based approaches (Table 1). 5) The data show how small molecules composed of similar building blocks can incur a generic interfacial effect. Such effect is the cause of the enhancement of a specific loop conformation of Aβ(12–28) (supplemental Table S1 and Fig. S13).
Taken together, the results show how aggregation inhibitors can have subtle but significant effects on the behavior of a fragment of Aβ encompassing the central hydrophobic cluster that is most commonly implied in the peptide's amyloidogenicity. It is important to ask whether our results could apply to other aggregation-prone IDPs as well. We use α-synuclein, the disease protein of Parkinson disease, as an example, which is significantly larger than Aβ and adopts more extended conformations in solution (93). Despite large differences in sequence properties, monomeric ensembles, and oligomer distributions, there exists a comparable range of compounds possessing one or more aromatic moieties that have been shown to bind monomeric α-synuclein (94) and to interfere with α-synuclein aggregation (53, 95–98) and oligomerization (99, 100). It is not necessary and perhaps unlikely that all of these inhibitors act in equivalent fashion for diverse systems. Returning to Fig. 1, this means, for example, that the multiple equilibria involving inhibitor binding may not all contribute significantly. As an example, our data for β-Ala-His suggest that this compound is unlikely to interfere with Aβ dimerization at the low concentrations typically in use in vitro. Similarly, DTrp-Aib exhibited no significant effects on early oligomer formation of Aβ(1–42) at concentration ratios of up to 40:1 excess of DTrp-Aib (40) and is also largely unbound in our simulations. This suggests a specific role in step VI for those two compounds. Conversely, evidence for generic effects at the oligomer level (in particular step V in Fig. 1) is not only given in the wider context of enzyme activity assays (101) but has also been proposed specifically in the context of aggregation experiments (102).
Finally, the consistency between simulation results and experimental data (NMR spectroscopy analysis of Aβ(12–28) and characterization of inhibition of Aβ40 aggregation) suggest that several of the conclusions are independent of the details of the simulation model. Encouraged by these results, we are currently using the same methodology to perform a medium throughput screening of a novel class of compounds meant to interfere with Aβ oligomerization and fibrillization.
Acknowledgments
We thank Dr. Riccardo Pellarin, Andrea Magno, and François Marchand for interesting discussions and suggestions. Most of the simulations were carried out on the Schrödinger cluster at the Informatikdienste of the University of Zurich.
This work was supported by a grant of the Swiss National Science Foundation (to A. C.) and a grant of the UZH Forschungskredit (to A. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S14.
F. Attanasio, M. Convertino, A. Caflisch, A. Corazza, G. Esposito, S. Cataldo, B. Pignataro, D. Milardi, and E. Rizzarelli, submitted for publication.
- AD
- Alzheimer disease
- Aβ
- β-amyloid peptide
- Aib
- α-aminoisobutyric acid
- IDP
- intrinsically disordered protein
- MD
- molecular dynamics
- cFEP
- cut-based free energy profile
- NQTrp
- 1,4-napthoquinon-2-yl-l-tryptophan
- DTrp
- d-tryptophan.
REFERENCES
- 1. Bharadwaj P. R., Dubey A. K., Masters C. L., Martins R. N., Macreadie I. G. (2009) J. Cell. Mol. Med. 13, 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Selkoe D. J. (1991) Neuron 6, 487–498 [DOI] [PubMed] [Google Scholar]
- 3. Terry R. D. (1994) Prog. Brain Res. 101, 383–390 [DOI] [PubMed] [Google Scholar]
- 4. Lomakin A., Chung D. S., Benedek G. B., Kirschner D. A., Teplow D. B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1125–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fawzi N. L., Okabe Y., Yap E. H., Head-Gordon T. (2007) J. Mol. Biol. 365, 535–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pellarin R., Caflisch A. (2006) J. Mol. Biol. 360, 882–892 [DOI] [PubMed] [Google Scholar]
- 7. Rochet J. C., Lansbury P. T., Jr. (2000) Curr. Opin. Struct. Biol. 10, 60–68 [DOI] [PubMed] [Google Scholar]
- 8. Serio T. R., Cashikar A. G., Kowal A. S., Sawicki G. J., Moslehi J. J., Serpell L., Arnsdorf M. F., Lindquist S. L. (2000) Science 289, 1317–1321 [DOI] [PubMed] [Google Scholar]
- 9. Wu C., Shea J. E. (2011) Curr. Opin. Struct. Biol. 21, 209–220 [DOI] [PubMed] [Google Scholar]
- 10. Kim S., Takeda T., Klimov D. K. (2010) Biophys. J. 99, 1949–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tarus B., Straub J. E., Thirumalai D. (2005) J. Mol. Biol. 345, 1141–1156 [DOI] [PubMed] [Google Scholar]
- 12. Tarus B., Straub J. E., Thirumalai D. (2008) J. Mol. Biol. 379, 815–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lansbury P. T., Lashuel H. A. (2006) Nature 443, 774–779 [DOI] [PubMed] [Google Scholar]
- 14. Haass C., Selkoe D. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 101–112 [DOI] [PubMed] [Google Scholar]
- 15. Uversky V. N., Oldfield C. J., Midic U., Xie H., Xue B., Vucetic S., Iakoucheva L. M., Obradovic Z., Dunker A. K. (2009) BMC Genomics 10, S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Uversky V. N., Oldfield C. J., Dunker A. K. (2008) Annu. Rev. Biophys. 37, 215–246 [DOI] [PubMed] [Google Scholar]
- 17. Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004) J. Mol. Biol. 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 18. Mittag T., Forman-Kay J. D. (2007) Curr. Opin. Struct. Biol. 17, 3–14 [DOI] [PubMed] [Google Scholar]
- 19. Uversky V. N., Gillespie J. R., Fink A. L. (2000) Proteins Struct. Funct. Genet. 41, 415–427 [DOI] [PubMed] [Google Scholar]
- 20. Dunker A. K., Obradovic Z. (2001) Nat. Biotechnol. 19, 805–806 [DOI] [PubMed] [Google Scholar]
- 21. Vitalis A., Caflisch A. (2010) J. Mol. Biol. 403, 148–165 [DOI] [PubMed] [Google Scholar]
- 22. Chen Y. R., Glabe C. G. (2006) J. Biol. Chem. 281, 24414–24422 [DOI] [PubMed] [Google Scholar]
- 23. Wright P. E., Dyson H. J. (2009) Curr. Opin. Struct. Biol. 19, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pappu R. V., Wang X., Vitalis A., Crick S. L. (2008) Arch. Biochem. Biophys. 469, 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Linding R., Schymkowitz J., Rousseau F., Diella F., Serrano L. (2004) J. Mol. Biol. 342, 345–353 [DOI] [PubMed] [Google Scholar]
- 26. Fields G. B., Alonso D. O. V., Stigter D., Dill K. A. (1992) J. Phys. Chem. 96, 3974–3981 [Google Scholar]
- 27. Cohen F. E., Kelly J. W. (2003) Nature 426, 905–909 [DOI] [PubMed] [Google Scholar]
- 28. Necula M., Kayed R., Milton S., Glabe C. G. (2007) J. Biol. Chem. 282, 10311–10324 [DOI] [PubMed] [Google Scholar]
- 29. Kokkoni N., Stott K., Amijee H., Mason J. M., Doig A. J. (2006) Biochemistry 45, 9906–9918 [DOI] [PubMed] [Google Scholar]
- 30. Yan L. M., Velkova A., Tatarek-Nossol M., Andreetto E., Kapurniotu A. (2007) Angew. Chem. Int. Ed. Engl. 46, 1246–1252 [DOI] [PubMed] [Google Scholar]
- 31. Ban T., Hoshino M., Takahashi S., Hamada D., Hasegawa K., Naiki H., Goto Y. (2004) J. Mol. Biol. 344, 757–767 [DOI] [PubMed] [Google Scholar]
- 32. Kanapathipillai M., Lentzen G., Sierks M., Park C. B. (2005) FEBS Lett. 579, 4775–4780 [DOI] [PubMed] [Google Scholar]
- 33. Gervais F., Paquette J., Morissette C., Krzywkowski P., Yu M., Azzi M., Lacombe D., Kong X., Aman A., Laurin J., Szarek W. A., Tremblay P. (2007) Neurobiol. Aging 28, 537–547 [DOI] [PubMed] [Google Scholar]
- 34. Mishra R., Bulic B., Sellin D., Jha S., Waldmann H., Winter R. (2008) Angew. Chem. Int. Ed. Engl. 47, 4679–4682 [DOI] [PubMed] [Google Scholar]
- 35. Porat Y., Mazor Y., Efrat S., Gazit E. (2004) Biochemistry 43, 14454–14462 [DOI] [PubMed] [Google Scholar]
- 36. Bastianetto S., Krantic S., Quirion R. (2008) Mini Rev. Med. Chem. 8, 429–435 [DOI] [PubMed] [Google Scholar]
- 37. Convertino M., Pellarin R., Catto M., Carotti A., Caflisch A. (2009) Protein Sci. 18, 792–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chebaro Y., Derreumaux P. (2009) Proteins Struct. Funct. Bioinform. 75, 442–452 [DOI] [PubMed] [Google Scholar]
- 39. Cohen T., Frydman-Marom A., Rechter M., Gazit E. (2006) Biochemistry 45, 4727–4735 [DOI] [PubMed] [Google Scholar]
- 40. Frydman-Marom A., Rechter M., Shefler I., Bram Y., Shalev D. E., Gazit E. (2009) Angew. Chem. Int. Ed. Engl. 48, 1981–1986 [DOI] [PubMed] [Google Scholar]
- 41. Morshedi D., Rezaei-Ghaleh N., Ebrahim-Habibi A., Ahmadian S., Nemat-Gorgani M. (2007) FEBS J. 274, 6415–6425 [DOI] [PubMed] [Google Scholar]
- 42. Pickhardt M., Gazova Z., von Bergen M., Khlistunova I., Wang Y., Hascher A., Mandelkow E. M., Biernat J., Mandelkow E. (2005) J. Biol. Chem. 280, 3628–3635 [DOI] [PubMed] [Google Scholar]
- 43. Scherzer-Attali R., Pellarin R., Convertino M., Frydman-Marom A., Egoz-Matia N., Peled S., Levy-Sakin M., Shalev D. E., Caflisch A., Gazit E., Segal D. (2010) PLoS One 5, e11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jiang P., Li W. F., Shea J. E., Mu Y. G. (2011) Biophys. J. 100, 1550–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Evers F., Jeworrek C., Tiemeyer S., Weise K., Sellin D., Paulus M., Struth B., Tolan M., Winter R. (2009) J. Am. Chem. Soc. 131, 9516–9521 [DOI] [PubMed] [Google Scholar]
- 46. Bieschke J., Russ J., Friedrich R. P., Ehrnhoefer D. E., Wobst H., Neugebauer K., Wanker E. E. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 7710–7715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takeda T., Kumar R., Raman E. P., Klimov D. K. (2010) J. Phys. Chem. B 114, 15394–15402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim S., Chang W. E., Kumar R., Klimov D. K. (2011) Biophys. J. 100, 2024–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Landau M., Sawaya M. R., Faull K. F., Laganowsky A., Jiang L., Sievers S. A., Liu J., Barrio J. R., Eisenberg D. (2011) PLoS Biol. 9, e1001080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sievers S. A., Karanicolas J., Chang H. W., Zhao A., Jiang L., Zirafi O., Stevens J. T., Münch J., Baker D., Eisenberg D. (2011) Nature 475, 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schütz A. K., Soragni A., Hornemann S., Aguzzi A., Ernst M., Böckmann A., Meier B. H. (2011) Angew. Chem. Int. Ed. Engl. 50, 5956–5960 [DOI] [PubMed] [Google Scholar]
- 52. Lorenzo A., Yankner B. A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 12243–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Braga C. A., Follmer C., Palhano F. L., Khattar E., Freitas M. S., Romão L., Di Giovanni S., Lashuel H. A., Silva J. L., Foguel D. (2011) J. Mol. Biol. 405, 254–273 [DOI] [PubMed] [Google Scholar]
- 54. De Felice F. G., Vieira M. N., Saraiva L. M., Figueroa-Villar J. D., Garcia-Abreu J., Liu R., Chang L., Klein W. L., Ferreira S. T. (2004) FASEB J. 18, 1366–1372 [DOI] [PubMed] [Google Scholar]
- 55. Riek R., Güntert P., Döbeli H., Wipf B., Wüthrich K. (2001) Eur. J. Biochem. 268, 5930–5936 [DOI] [PubMed] [Google Scholar]
- 56. Cohlberg J. A., Li J., Uversky V. N., Fink A. L. (2002) Biochemistry 41, 1502–1511 [DOI] [PubMed] [Google Scholar]
- 57. Feng Y., Yang S. G., Du X. T., Zhang X., Sun X. X., Zhao M., Sun G. Y., Liu R. T. (2009) Biochem. Biophys. Res. Commun. 390, 1250–1254 [DOI] [PubMed] [Google Scholar]
- 58. Straub J. E., Guevara J., Huo S., Lee J. P. (2002) Acc. Chem. Res. 35, 473–481 [DOI] [PubMed] [Google Scholar]
- 59. Zhang S., Casey N., Lee J. P. (1998) Fold Des. 3, 413–422 [DOI] [PubMed] [Google Scholar]
- 60. Porat Y., Abramowitz A., Gazit E. (2006) Chem. Biol. Drug Des. 67, 27–37 [DOI] [PubMed] [Google Scholar]
- 61. Tjernberg L. O., Näslund J., Lindqvist F., Johansson J., Karlström A. R., Thyberg J., Terenius L., Nordstedt C. (1996) J. Biol. Chem. 271, 8545–8548 [DOI] [PubMed] [Google Scholar]
- 62. Williams A. D., Portelius E., Kheterpal I., Guo J. T., Cook K. D., Xu Y., Wetzel R. (2004) J. Mol. Biol. 335, 833–842 [DOI] [PubMed] [Google Scholar]
- 63. Krivov S. V., Karplus M. (2006) J. Phys. Chem. B 110, 12689–12698 [DOI] [PubMed] [Google Scholar]
- 64. Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) J. Comput. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Haberthür U., Caflisch A. (2008) J. Comput. Chem. 29, 701–715 [DOI] [PubMed] [Google Scholar]
- 66. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 67. Bussi G., Donadio D., Parrinello M. (2007) J. Chem. Phys. 126, 014101 [DOI] [PubMed] [Google Scholar]
- 68. Parrinello M., Rahman A. (1981) J. Appl. Phys. 52, 7182–7190 [Google Scholar]
- 69. Buck M., Bouguet-Bonnet S., Pastor R. W., MacKerell A. D., Jr. (2006) Biophys. J. 90, L36–L38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) J. Chem. Phys. 79, 926–935 [Google Scholar]
- 71. Darden T., York D., Pedersen L. (1993) J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 72. Hess B., Bekker H., Berendsen H. J. C., Fraaije J. G. (1997) J. Comput. Chem. 18, 1463–1472 [Google Scholar]
- 73. Seeber M., Felline A., Raimondi F., Muff S., Friedman R., Rao F., Caflisch A., Fanelli F. (2011) J. Comput. Chem. 32, 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vitalis A., Wang X., Pappu R. V. (2007) Biophys. J. 93, 1923–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schäfer L. (1999) Excluded Volume Effects in Polymer Solutions: As Explained by the Renormalization Group, 1st Ed., Springer, Berlin, Germany [Google Scholar]
- 76. Frydman-Marom A., Convertino M., Pellarin R., Lampel A., Shaltiel-Karyo R., Segal D., Caflisch A., Shalev D. E., Gazit E. (2011) ACS Chem. Biol., in press [DOI] [PubMed] [Google Scholar]
- 77. Szegedi V., Juhász G., Rózsa E., Juhász-Vedres G., Datki Z., Fülöp L., Bozsó Z., Lakatos A., Laczkó I., Farkas T., Kis Z., Tóth G., Soós K., Zarándi M., Budai D., Toldi J., Penke B. (2006) FASEB J. 20, 1191–1193 [DOI] [PubMed] [Google Scholar]
- 78. Tartaglia G. G., Cavalli A., Pellarin R., Caflisch A. (2005) Protein Sci. 14, 2723–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Caflisch A. (2006) Curr. Opin. Chem. Biol. 10, 437–444 [DOI] [PubMed] [Google Scholar]
- 80. Wurth C., Guimard N. K., Hecht M. H. (2002) J. Mol. Biol. 319, 1279–1290 [DOI] [PubMed] [Google Scholar]
- 81. Azriel R., Gazit E. (2001) J. Biol. Chem. 276, 34156–34161 [DOI] [PubMed] [Google Scholar]
- 82. Csermely P., Palotai R., Nussinov R. (2010) Trends Biochem. Sci. 35, 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hawkes C. A., Ng V., McLaurin J. (2009) Drug Dev. Res. 70, 111–124 [Google Scholar]
- 84. Re F., Airoldi C., Zona C., Masserini M., La Ferla B., Quattrocchi N., Nicotra F. (2010) Curr. Med. Chem. 17, 2990–3006 [DOI] [PubMed] [Google Scholar]
- 85. Armstrong A. H., Chen J., McKoy A. F., Hecht M. H. (2011) Biochemistry 50, 4058–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhang S., Iwata K., Lachenmann M. J., Peng J. W., Li S., Stimson E. R., Lu Y., Felix A. M., Maggio J. E., Lee J. P. (2000) J. Struct. Biol. 130, 130–141 [DOI] [PubMed] [Google Scholar]
- 87. Baumketner A., Krone M. G., Shea J. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6027–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zagrovic B., van Gunsteren W. F. (2006) Proteins Struct. Funct. Bioinform. 63, 210–218 [DOI] [PubMed] [Google Scholar]
- 89. Eliezer D. (2009) Curr. Opin. Struct. Biol. 19, 23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lazo N. D., Grant M. A., Condron M. C., Rigby A. C., Teplow D. B. (2005) Protein Sci. 14, 1581–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sgourakis N. G., Yan Y., McCallum S. A., Wang C., Garcia A. E. (2007) J. Mol. Biol. 368, 1448–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Stempler S., Levy-Sakin M., Frydman-Marom A., Amir Y., Scherzer-Attali R., Buzhansky L., Gazit E., Senderowitz H. (2011) J. Comput. Aided Mol. Des. 25, 135–144 [DOI] [PubMed] [Google Scholar]
- 93. Uversky V. N. (2007) J. Neurochem. 103, 17–37 [DOI] [PubMed] [Google Scholar]
- 94. Latawiec D., Herrera F., Bek A., Losasso V., Candotti M., Benetti F., Carlino E., Kranjc A., Lazzarino M., Gustincich S., Carloni P., Legname G. (2010) PLoS One 5, e9234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hong D. P., Fink A. L., Uversky V. N. (2008) J. Mol. Biol. 383, 214–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhou W., Gallagher A., Hong D. P., Long C., Fink A. L., Uversky V. N. (2009) J. Mol. Biol. 388, 597–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kim J., Harada R., Kobayashi M., Kobayashi N., Sode K. (2010) Mol. Neurodegener. 5, 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pandey N., Strider J., Nolan W. C., Yan S. X., Galvin J. E. (2008) Acta Neuropathol. 115, 479–489 [DOI] [PubMed] [Google Scholar]
- 99. Caruana M., Högen T., Levin J., Hillmer A., Giese A., Vassallo N. (2011) FEBS Lett. 585, 1113–1120 [DOI] [PubMed] [Google Scholar]
- 100. Hillmer A. S., Putcha P., Levin J., Högen T., Hyman B. T., Kretzschmar H., McLean P. J., Giese A. (2010) Biochem. Biophys. Res. Commun. 391, 461–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Shoichet B. K. (2006) Drug Discov. Today 11, 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Feng B. Y., Toyama B. H., Wille H., Colby D. W., Collins S. R., May B. C., Prusiner S. B., Weissman J., Shoichet B. K. (2008) Nat. Chem. Biol. 4, 197–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tran H. T., Pappu R. V. (2006) Biophys. J. 91, 1868–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Carter P., Andersen C. A., Rost B. (2003) Nucleic Acids Res. 31, 3293–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]