Background: High dependence of cancer cells on glycolysis is a good target for cancer therapy.
Results: Tumor suppressor p53 represses the expression of key regulators of metabolic genes HIF1a and c-Myc and glucose transporters GLUT1 and GLUT12.
Conclusion: Blocking ATP production network by pharmacologically activated p53 contributes to cancer cell death.
Significance: Tumor-selective killing by reconstituted p53 might be in part due to inhibition of glycolysis.
Keywords: Cancer Therapy, Cell Death, Glucose Metabolism, Glycolysis, Oncogene, p53, Tumor Metabolism, Hypoxia-inducible Factor 1, c-Myc
Abstract
Unique sensitivity of tumor cells to the inhibition of glycolysis is a good target for anticancer therapy. Here, we demonstrate that the pharmacologically activated tumor suppressor p53 mediates the inhibition of glycolytic enzymes in cancer cells in vitro and in vivo. We showed that p53 binds to the promoters of metabolic genes and represses their expression, including glucose transporters SLC2A12 (GLUT12) and SLC2A1 (GLUT1). Furthermore, p53-mediated repression of transcription factors c-Myc and HIF1α, key drivers of ATP-generating pathways in tumors, contributed to ATP production block. Inhibition of c-Myc by p53 mediated the ablation of several glycolytic genes in normoxia, whereas in hypoxia down-regulation of HIF1α contributed to this effect. We identified Sp1 as a transcription cofactor cooperating with p53 in the ablation of metabolic genes. Using different approaches, we demonstrated that glycolysis block contributes to the robust induction of apoptosis by p53 in cancer cells. Taken together, our data suggest that tumor-specific reinstatement of p53 function targets the “Achilles heel” of cancer cells (i.e. their dependence on glycolysis), which could contribute to the tumor-selective killing of cancer cells by pharmacologically activated p53.
Introduction
The metabolism of most solid tumors is significantly different from that of surrounding normal tissues, which derive their energy from the oxidative phosphorylation. In contrast, increased aerobic glycolysis occurs in a wide spectrum of human cancers and is considered as one of the most fundamental alterations during malignant transformation (1). High dependence of cancer cells on glycolysis for ATP production in the presence of oxygen, known as the Warburg effect (2), is recognized as the seventh hallmark of cancer (3).
High glucose uptake exploited in cancer diagnosis and monitoring of treatment using the glucose analog tracer 18-fluorodeoxyglucose and positron emission tomography (4, 5) occurs due to the overexpression of glucose transporters, especially the glucose transporter isoform 1 (GLUT1) (6). Upon uptake, glucose molecules are irreversibly phosphorylated by hexokinases 1 and 2 (HK1 and 2), also overexpressed in cancers (7). Key glycolytic enzymes acting downstream of hexokinase include PFKFB3, PFK1, pyruvate dehydrogenase (PDH), and pyruvate dehydrogenase kinase (PDK) (Fig. 2C).
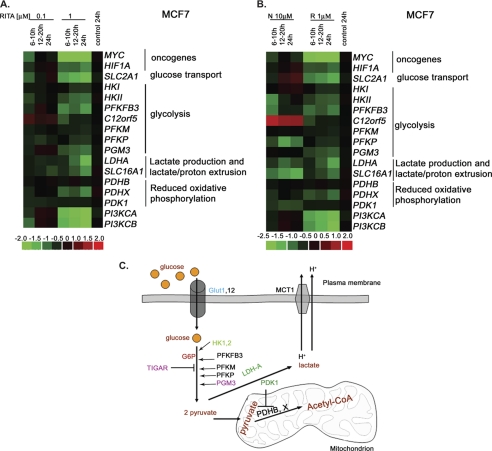
FIGURE 2.
Microarray analysis revealed dose- and time-dependent repression of metabolic genes upon RITA treatment. The microarray data are presented as heat maps made using dChip (DNA chip analyzer). The rows were standardized by subtracting -fold change of control and dividing by the S.D. value. Vertical columns indicate separate arrays, and horizontal rows indicate genes. A, heat map depicting the relative mRNA levels of genes involved in regulation of metabolism in MCF7 cells treated with two concentrations of RITA over the indicated periods of time. B, comparison of changes of metabolic gene expression in MCF7 cells treated with RITA or nutlin3a over the indicated periods of time. C, schematic representation of the ATP-generating pathways, indicating the set of metabolic genes altered upon RITA treatment and their regulators. GLUT, glucose transporter; HK, hexokinase; PFKFB, phosphofructokinase fructose biphosphate; PFKM, phosphofructokinase muscles; PFKP, phosphofructokinase platelets; PGM, phosphoglycerate mutase, TIGAR, TP53-induced glycolysis and apoptosis regulator; LDH, lactate dehydrogenase; PDK, pyruvate dehydrogenase kinase; PDH, pyruvate dehydrogenase; MCT, monocarboxylate transporter; G6P, glucose-6-phosphate. Adapted from Kroemer and Pouyssegur (56). Orange, glucose; dark orange, main metabolites produced during glycolysis and oxidative phosphorylation; dark green, targets of Myc, p53, and HIF1α; purple, p53 targets; green, Myc and HIF1α targets; blue, p53 and HIF1α targets.
The oncogenic networks, such as PI3K/Akt, c-Myc, and HIF1 influence the metabolic shift during cancerogenesis and support growth and proliferation of cancer cells under metabolic stress and hypoxia. HIF1a, a transcription factor stabilized under hypoxia (8) triggers the up-regulation of genes critical for the switch to glycolysis, including SLC2A1 (GLUT1), HKII, PDK1, PFK1, and lactate dehydrogenase LDHA (9). Transcription factor c-Myc, one of the major oncogenes, cooperates with HIF1 in promoting glycolysis by activating PDK1, HKII (3), and LDHA genes (10). Aberrations in the PI3K/Akt pathway constitute one of the most common sets of mutations in tumors (11). Enhanced PI3K/Akt signaling results in metabolic transformation via multiple pathways, including increased expression of genes involved in glycolysis and stimulation of hexokinase and PFK activities (10).
Targeting aerobic glycolysis for anticancer treatment is a very promising approach. Several glycolysis inhibitors are in preclinical and clinical development, such as lactate dehydrogenase A inhibitor FX11 (12) or hexokinase inhibitor 2-deoxyglucose (13).
p53 is a transcription factor that suppresses tumor development by regulating the expression of genes inducing cell cycle arrest, apoptosis, and senescence upon stress conditions (14). In order to survive, cancer cells render p53 inactive, either by point mutations (∼50% of human cancers) (15) or by increased degradation of wild type p53 due to the deregulation of E3 ubiquitin ligase MDM2 (16). Recently, p53 has been implicated in metabolic control by influencing the balance between glycolysis and oxidative phosphorylation via, for example, induction of TIGAR (17) and regulation of SCO2 (synthesis of cytochrome c oxidase 2) (18), which promote the switch from glycolysis to oxidative phosphorylation.5 Moreover, p53 inhibits the expression of glucose transporters GLUT1 and GLUT4 (19), indicating that p53 can impede metabolism by reducing glucose import. Additionally, wild-type p53 was shown to down-regulate oncogenic phosphoglycerate mutase (20). However, p53 involvement in metabolic regulation is rather complex; it may both inhibit and promote tumor growth (10, 21). Determining the stimuli that trigger different p53 responses affecting cell metabolism is very important, especially in light of the recent development of small molecules reactivating p53 function in cancer cells.
A number of strategies reactivating p53 (22) have been developed over the years. Our group has identified p53-reactivating compound RITA (reactivation of p53 and induction of tumor cell apoptosis) (23). RITA binds the p53 N-terminal domain and disrupts the interaction with its negative regulator MDM2, which results in p53 activation and induction of apoptosis (23, 24).5 Notably, we showed that RITA activates p53 in cells expressing oncogenes, whereas the effect in non-transformed cells is almost negligible (23, 25). In addition, we found that the response of tumor cells to different doses of RITA (0.1 and 1 μm) was similar in terms of induction of p53 and transcriptional activation of its apoptotic targets, but transcriptional repression of oncogenes c-Myc, PI3K, IGFR, Mcl-1, survivin, and others was triggered only by a higher dose (25). Oncogene repression correlated with apoptosis induction, indicating that it contributes to cancer cell killing by p53.
In the present study, we investigated whether pharmacological reconstitution of p53 can inhibit aerobic glycolysis in cancer cells in vitro and in vivo, using small molecule RITA as a research tool. We report a potent p53-dependent inhibition of the glucose transport and the first steps of glycolysis via the transcriptional repression of key players of these processes.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Tumor cells HCT116 (wtp53), MCF7 (wtp53), U2OS (wtp53), HCT116TP53−/− (p53-null), H1299 (p53-null), and Saos2 (p53-null) were maintained in Iscove's modified Dulbecco's medium supplemented with 10% fetal calf serum, penicillin/streptomycin (10 units/ml), and l-glutamine (2 mm) (all purchased from Sigma-Aldrich). The isogenic colon carcinoma cell lines HCT116 (p53-positive) and HCT116 TP53−/− (p53-null) were provided by B. Vogelstein. Small interfering RNA (siRNA) for HIF1α (sc-35561), c-Myc (sc-29226), and HK2 (sc-35621) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and green fluorescent protein (GFP) siRNA, which was used as a control, was purchased from Thermo Scientific Dharmacon. Transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions using 20 pmol of siRNA. Drug treatment was performed 32 h after transfection.
MCF7 cells stably depleted for Sp1 using an Sp1 shRNA lentivirus construct (Sigma) were treated with 1 μm RITA for 8 h to detect mRNA levels by quantitative RT-PCR (qRT-PCR)6 and microarray analysis or for 48 h to assess survival.
Metabolic Chip Assay
HCT116 and its negative counterpart HCT116 p53-null cells were grown on the metabolic chips in DMEM supplemented with 10% fetal calf serum, penicillin/streptomycin (10 units/ml), and l-glutamine (2 mm) under standard conditions for 24 h. Before analysis, the cells were treated with 0.1 or 1 μm RITA reconstituted from the stock (0.1 m in 100% DMSO) for 12 h in a standard cell incubator. Afterward, the chips were transferred to the Bionas® 2500 analyzing system for 4 days with running medium supplemented only with 2% FCS. Programming of the Bionas® 2500 analyzing system was performed according to the user manual. Just before the measurements started, cell morphology was controlled microscopically and photographed for documentation (not shown). After the measurement, cells were killed with 0.2% Triton X-100 diluted in running medium. The general measurement schedule for the Bionas® 2500 analyzing system was as follows. In the first phase, base lines for acidification, respiration, and cell impedance were determined. After the stabilization phase of about 3 h, the values were standardized to 100%. The cells were measured in running medium (without compound) for 4 days. At the end of the experiment, the cells were killed by the addition of 0.2% Triton X-100 to the running medium. The values from the cells killed after Triton X-100 addition were set to 0%.
p53 ChIP-seq
Chromatin immunoprecipitation (ChIP) library preparation, massive parallel sequencing, and ChIP-seq primary analysis were performed as previously published (26). Data have been archived at the NCBI Sequence Read Archive (SRA) under accession number SRP007261.
Anti-p53 mouse monoclonal antibody DO1 (Genespin and Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)) and a nonspecific sc-2025 mouse IgG (Santa Cruz Biotechnology, Inc.) antibody were used to immunoprecipitate p53 from MCF7 cells treated with p53-activating compounds for 8 h. More detailed information on the ChIP-seq experiment and data processing will be provided in a separate paper.7
Microarrays
Total RNA was isolated with the RNeasy Mini kit (Qiagen). cDNA was synthesized with the One-Cycle cDNA synthesis kit from Affymetrix. cRNA was synthesized from cDNA by following the IVT labeling kit (Affymetrix) and purified with the GeneChip sample cleanup module from Affymetrix. Labeled cRNA was fragmented in fragmentation buffer (5× buffer: 200 mm Tris acetate (pH 8.1), 500 mm KOAc, 150 mm MgOAc) and hybridized to the microarrays in 200 μl of hybridization solution containing 10 μg of labeled target in 1× Mes buffer (0.1 m Mes, 1.0 m NaCl, 20 mm EDTA 0.01%, Tween 20) and 0.1 mg/ml herring sperm DNA, 0.5 mg/ml BSA, 50 pm control oligonucleotide B2, and 1× eukaryotic hybridization controls (bioB, bioC, bioD, and cre). Both control oligonucleotide B2 and eukaryotic hybridization controls were purchased from Affymetrix. Samples were then hybridized on Human Genome U133A 2.0 Arrays or U219 (Affymetrix). The arrays were then stained with a streptavidin-phycoerythrin conjugate (Molecular Probes), followed by 10 washing cycles of 4 mixes/cycle with 6× SSPE-T. To enhance the signals, the arrays were further stained with anti-streptavidin antibody solution for 10 min at 25 °C followed by a 10-min staining with a streptavidin-phycoerythrin conjugate. After 15 washing cycles of 4 mixes/cycle, the arrays were scanned using a confocal scanner (Affymetrix). The image data were analyzed by GCOS 1.4 (GeneChip Operating Software, Affymetrix).
F-Match Analysis of Selected Metabolic Genes
We performed F-Match analysis (27) using Explain 2.4.1 and geneXplain platform 1.0 software packages (available on the World Wide Web) (28) to search for overrepresented transcription factor binding sites in the promoter region. We used the TRANSFAC® data base (29) version 2010.4. As a background set, we chose a set of 1000 genes that did not show expression changes after nutlin3a or RITA treatment. The profile that was used for analysis contains a collection of vertebrate non-redundant transcription factor matrices. The promoter window was selected from −1000 to +100 from the transcription start site, and only best supported promoters of analyzed genes were used. To obtain only binding sites with high score, we chose cut-offs with a p value threshold of 0.01. Among matrices that were found, we selected those with high overrepresentation of the site frequency in promoters under study versus the background promoters (ratio > 1.3).
Key Node Analysis
We performed identification of potential master regulators in the signal transduction network using Explain 2.4.1 and geneXplain platform 1.0 software packages. The signal transduction network was provided by the manually curated data base, TRANSPATH® (30). The software applies an upstream analysis approach, which is based on implementation of machine learning and graph topological analysis algorithms in order to identify causality key nodes in the network of signal transduction (31). The algorithm starts from a set of transcription factors (found overrepresented in the promoters under study using the F-Match tool; see above) and performs a graph-topological search in the signal transduction network upstream of the transcription factors in order to identify “key nodes,” nodes in the network that can play a crucial role in transducing intracellular signaling from various receptors to the considered set of transcription factors. Such key nodes may be considered as master regulators of the process under study.
Animal Experiments
All animal studies were approved by the Northern Stockholm Animal Ethical Committee. The animal care was in accordance with Karolinska Institutet guidelines. Male SCID mice, 4–6 weeks old, were implanted subcutaneously with 1 × 106 HCT116 or HCT116 TP53−/− cells in 90% Matrigel (BD Biosciences). Palpable tumors were established 7 days after cell injection; at this point, we injected 1 mg/kg RITA into tumors in a total volume of 100 μl of phosphate-buffered saline.
Hypoxia and Drug Treatments
RITA (2,5-bis(5-hydroxymethyl-2-thienyl)-furan) was obtained from NCI, National Institutes of Health. RITA was dissolved to a concentration of 0.1 m in 100% dimethyl sulfoxide (DMSO). Afterwards, RITA was diluted in phosphate-buffered saline (PBS) and used at a final concentration of either 0.1 or 1 μm for different time points (indicated in the figures). Physiological hypoxia was achieved by incubating cells in 1% O2, 5% CO2, and 94% nitrogen in an In Vivo2 hypoxic work station 400 (Ruskinn Technology). Cells were put under hypoxic conditions for ∼18 h prior to treatment. The hypoxic mimetic agent CoCl2 (Sigma-Aldrich) was used at a final concentration of 100 μm for 24 h in combination with 1 μm RITA (8 h). The glycolysis inhibitor 2-deoxyglucose (Sigma-Aldrich) was used at a final concentration of 10 mm. Cells were pretreated with deoxyglucose 24 h prior to RITA treatment (8 h) in Iscove's modified Dulbecco's medium 2× diluted in PBS.
qRT-PCR
Total RNA was extracted and purified with a PerfectPure RNA kit (5 PRIME) using the manufacturer's protocol. RNA (1 μg) was reverse transcribed using the SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen). Real-time PCR was performed with SYBR Green reagent (Bio-Rad) using 20 ng of template in a 15-μl reaction mixture according to the manufacturer's protocol. Results were normalized to the GAPDH gene.
qRT-PCRs using tumor tissue were performed in xenograft samples obtained from previously conducted experiments (25). RNA from xenografts was purified using the PerfectPure RNA tissue kit from 5 PRIME GmBH according to the manufacturer's protocol.
All primers were designed using the PerlPrimer program (32) with one primer overlapping an exon boundary. Primer sequences are listed in supplemental Table 1.
Protein Extraction and Western Blot
Cells were washed with ice-cold PBS, and soluble proteins were extracted with cell lysis buffer (100 mm Tris-HCl, pH 8, 150 mm NaCl, 1% Nonidet P-40, phosphatase, and protease inhibitor mixture tablets (Roche Applied Science) according to the manufacturer's protocol). Insoluble material was removed by centrifugation (13,000 rpm, 30 min). The protein concentration was determined using the Bio-Rad Bradford assay and bovine serum albumin (BSA) standards. An equal amount of protein was separated by SDS-PAGE using Ready Made Gradient (4–20%) gels (Bio-Rad). Transfer was performed at 4 °C (180 mA, 80 V), and the following antibodies were used for Western blotting: p53 (DO-1), HK2 (sc-6521), TIGAR (sc-74577), and c-Myc (sc-764) (all from Santa Cruz Biotechnology, Inc.); ACL (4332; Cell Signaling); PUMA (PC686) and Noxa (OP180) (from Calbiochem); HIF1α (610959; BD Biosciences); and β-actin (Sigma). The latter was used to verify equal loading. Primary antibody incubation (1:500 dilution in 5% nonfat dry milk in PBS) was overnight (4 °C). Secondary HRP-conjugated antibodies (1:3000 dilution in 5% nonfat dry milk in PBS; 1 h incubation at room temperature) and SuperSignal West Dura extended duration substrate were from Pierce.
Cell Viability Assays
For colony formation assays, cells were seeded at a density of 30% and treated with RITA for 48 h. Afterward, cells on the plate were fixed with 70% ethanol and stained with crystal violet. After staining, the absorbance of the different wells was measured in a microplate reader (Victor X3, PerkinElmer Life Sciences) at a wavelength of 562 nm. The expected additive effect was calculated, adding the percentage of dead cells after siGFP combined with 0.1 μm RITA to the percentage of dead cells after siHK2 alone.
Statistical Analysis
The statistical significance of results was calculated by a parametric Student's t test (for variances, Fisher-Snedecor's test was applied, and the normality was assessed with Shapiro-Wilk's test). p < 0.05 values were considered statistically significant.
The statistical significance of qRT-PCR results for tumor samples was calculated by a one-way analysis of variance. p < 0.05 values were considered statistically significant. Statistical analysis was done using SAS version 9.2 (Raleigh, NC).
RESULTS
RITA Inhibits Metabolism of Cancer Cell in a p53-dependent Manner
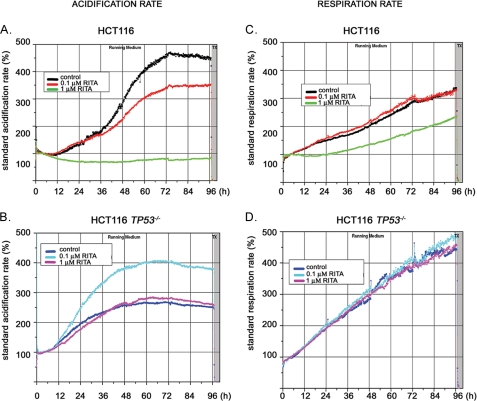
We addressed the question of whether reactivated p53 can mediate the inhibition of aerobic glycolysis, the key ATP-generating pathway in cancer cells. Using a pair of isogenic human colon cancer cell lines, HCT116 and HCT116 TP53−/−, varying only in p53 status, we analyzed their metabolic state upon treatment with p53-activating molecule RITA. As we found previously, p53 is differentially activated by 0.1 and 1 μm of RITA (25). Although p53 induction and transcriptional activation of proapoptotic targets was similar, transcriptional repression of oncogenes as well as cell death were achieved only in the presence of 1 μm RITA. Therefore, we treated cells with 0.1 and 1 μm RITA and continuously monitored their metabolism by assessing the acidification and respiration activity over a period of 96 h using metabolic chip (Bionas® metabolic chip SC 1000) (33). Upon treatment of HCT116 cells with 1 μm RITA, the acidification rate, which reflects the export of the end product of glycolysis, lactate, was reduced to ∼80% of the initial value (Fig. 1A, green line). In contrast, non-treated cells increased acidification to nearly 500% after 3 days (Fig. 1A, black line). Treatment with 0.1 μm RITA only slightly decreased acidification (Fig. 1A, red line). Thus, although treatment with 0.1 μm RITA induces p53 (25) (data not shown), a higher dose of RITA is required for the inhibition of cancer cell metabolism. These results are in line with our previous study, showing a higher threshold for the inhibition of survival genes by p53 compared with the induction of proapoptotic genes (1 μm RITA versus 0.1 μm RITA, respectively) (25).
FIGURE 1.
RITA inhibits respiration and induces acidosis in a p53-dependent manner. HCT116 and HCT116 TP53−/− cells treated with two different concentrations of RITA were analyzed on Bionas® metabolic chip SC 2500 in running medium supplemented with 2% FCS. A, in HCT116 cells, the acidification rate for a 1 μm concentration of RITA was reduced to ∼80%, whereas in control, it increased to nearly 500% after 3 days. Treatment with 0.1 μm resulted in a 350% increase of acidification rate. B, in HCT116 TP53−/− cells, the acidification rate increased to 280% and remained on this level for the last 2 days both in the control and in 1 μm RITA-treated cells, whereas upon treatment with 0.1 μm RITA, the acidification rate increased to 400%. Measurements were performed in running medium for 4 days. n = 2. C, the respiration rates were measured using the same experimental setup. In HCT116 cells treated with 1 μm RITA, the respiration increased up to 220%. In control and in cells treated with 0.1 μm RITA, the respiration rate increased to 320%. D, in treated and non-treated HCT116 TP53−/− cells, the respiration rate increased to 450–500%. Shown are the representative data of two independent experiments.
1 μm RITA did not change the metabolism of p53-null cells (Fig. 1B, control (dark blue line) and 1 μm RITA (pink line)). Thus, our data confirm that inhibition of cell metabolism as manifested by the levels of the end product of glycolysis, namely lactate, was p53-dependent.
Interestingly, 0.1 μm RITA increased the acidification rate of treated p53-null cells (Fig. 1B, light blue line). We speculate that this phenomenon could be due to RITA-mediated induction of ROS levels (34), which might lead to activation of glycolytic enzymes, resulting in higher acidification (see “Discussion”).
More detailed analysis at early time points allowed us to detect that the effect of 1 μm RITA on cell metabolism was manifested within a few h of treatment (data not shown), which indicates that the observed inhibition of cancer cell metabolism is not a consequence of apoptosis induction by RITA. Interestingly, p53-null cells seem to display a different metabolic rate compared with p53-positive cells, suggesting that p53 can modulate the metabolism of cells at basal level, even in the absence of external stimuli, which is in line with published data (21).
The respiration rate, analyzed on the same chip (Fig. 1C), was significantly reduced by 1 μm RITA compared with non-treated or 0.1 μm RITA-treated HCT116 cells (220% versus 370%), in line with results shown above. However, it was not reduced in p53-null cells (Fig. 1D). Taken together, data obtained using the metabolic chip analysis demonstrate the p53-dependent inhibition of cell metabolism upon reactivation by 1 μm RITA.
Genome-wide Gene Expression Analysis Reveals Transcriptional Repression of Key Metabolic Genes upon p53 Activation by 1 μm RITA
Next we assessed whether the changes in cell metabolism upon p53 activation occur at the transcriptional level. To estimate the expression levels of key enzymes driving glycolysis and oxidative phosphorylation (illustrated in Fig. 2C), we performed a microarray analysis of gene expression profiles in human breast cancer cell line MCF7 treated or non-treated with RITA. Our analysis revealed a substantial repression of a group of metabolic genes in MCF7 cells upon 1 μm RITA; among these, the set of glycolytic genes was clearly distinguishable (Fig. 2A). Time- and dose-dependent repression was identified for the following metabolic genes: SLC2A1 (glucose transporter 1, GLUT1), HKII (hexokinase 2), PFKFB3 (phosphofructokinase fructose biphosphate izosyme 3), PFK (phosphofructokinase isoforms P and M), and PGM3 (phosphoglycerate mutase). Further, LDHA (lactate dehydrogenase A) and SLC16A1 (monocarboxylate transporter 1) were found to be down-regulated.
mRNA levels of several factors known to be involved in the activation of some of the genes mentioned above were substantially repressed. These include oncogenic transcriptional factors c-Myc and HIF1α, as well as both catalytic subunits of PI3K, PIK3CA and PIK3CB (p110α and p110β, respectively) (Fig. 2A). Notably, mRNA of pyruvate dehydrogenase complex, component X, involved in oxidative phosphorylation, and its negative regulator, PDK1, were also down-regulated upon RITA treatment. Genes encoding oncogenic phosphoglycerate mutase (PGM3) and hexokinase 2 (HKII), whose expression has been shown to be ablated by wild-type p53 (20, 35), were inhibited in cancer cells upon p53 activation (Fig. 2, A and B). The functional role in glycolysis of differentially expressed factors mentioned above is indicated in Fig. 2C.
Further, induction of p53 with yet another p53 activator, nutlin3a (36), also resulted in inhibition of HKII, PFKBF3, LDHA, and SLC16A1 (Fig. 2B). However, there were differences in the pattern of gene expression upon RITA and nutlin3a treatment; some genes, such as MYC, HIF1A, PGM3, and SLC2A1, were not substantially inhibited by nutlin3a or were even up-regulated (SLC2A1; Fig. 2B). In contrast, we observed a more pronounced repression of PDK1 and PFKP and robust activation of the p53 target gene C12orf5 encoding the inhibitor of glycolysis TIGAR (17) upon nutlin3a treatment.
Notably, our microarray data suggest that the induction of p53 target genes involved in regulation of oxidative phosphorylation, C12orf5 (encoding TIGAR) and SCO2, depended on the type of p53-activating stimuli (supplemental Table 2). C12orf5 was induced by nulin3a, 5-FU, and 0.1 μm RITA but not by 1 μm RITA (Fig. 2, B and A, respectively, and supplemental Table 2). Induction of TIGAR correlated with the inefficient killing of MCF7 cells by nutlin3a and 0.1 μm RITA (24, 25) (data not shown), which is in line with the previously published data indicating that TIGAR can promote survival of cells in the absence of glycolysis (21). SCO2, on the other hand, was only marginally affected by RITA or 5-FU but was suppressed by nutlin3a (supplemental Table 2). These data suggest that distinct p53-activating agents differently affect transcriptional regulation of metabolic genes by p53. This is in line with previously published studies on p53-induced gene expression profiles, which show that the array of genes activated or repressed by p53 varies depending on the stimulus, the dose of the agent, and the cell type (37).
We compared the microarray data on expression profiles of metabolic genes upon RITA treatment of p53-positive and p53-null HCT116 cells and found that MYC, HIF1A, HKI, HKII, PGM1, PGM3, LDHA, SLC16A1, PDHX, and PDK1 were down-regulated in a p53-dependent manner (data not shown). Thus, pharmacological activation of p53 affects the expression of a number of genes driving ATP-generating pathways; depending on the type of p53 activator, different regulators of cancer cell metabolism could be affected.
p53-mediated Repression of Metabolic Genes Confirmed by qRT-PCR and Analysis of Protein Levels
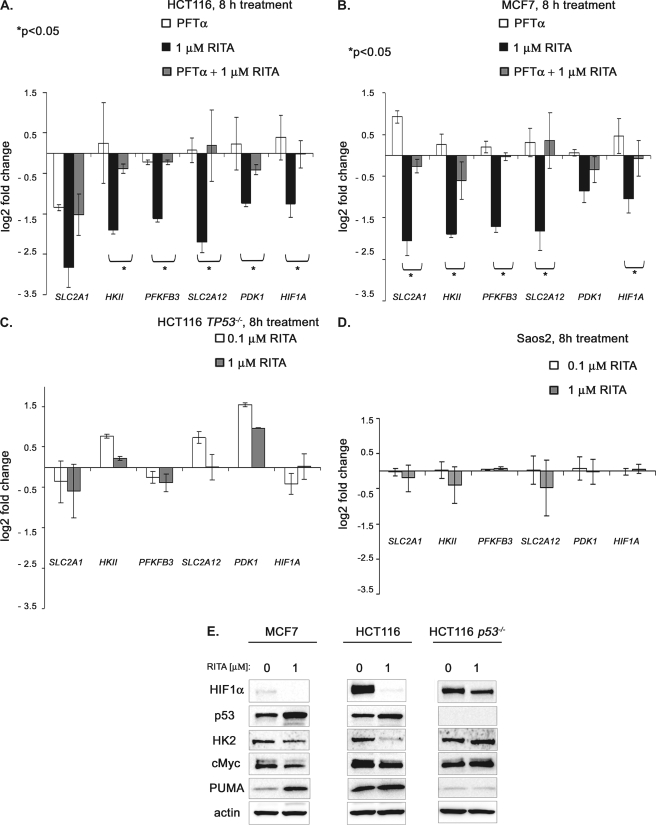
To further assess the p53-dependent inhibition of the metabolic genes upon RITA treatment and to validate the results of the microarray experiment, we selected a group of six genes that were significantly repressed in wild type p53-carrying cancer cell lines HCT116 and MCF7 and tested their expression by qRT-PCR. As shown in Fig. 3, A and B, a potent transcriptional inhibition of SLC2A1, HKII, PFKFB3, SLC2A12, PDK1, and HIF1A was detected by qRT-PCR. In addition, we confirmed transcriptional repression of metabolic genes in wild type p53-expressing osteosarcoma cell line U2OS (supplemental Fig. 1A). Moreover, these genes were not repressed upon RITA treatment of the p53-null cells HCT116 TP53−/−, Saos-2 (Fig. 3, C and D), and H1299 (supplemental Fig. 1B), demonstrating that the observed effect was strictly p53-dependent.
FIGURE 3.
p53-dependent ablation of selected metabolic factors upon RITA treatment. A and B, down-regulation of SLC2A1 (GLUT1), HKII (HK2), PFKFB3, SLC2A12 (GLUT12), PDK1, and HIF1A 8 h after treatment with 1 μm RITA in p53-positive cancer cell lines as assessed by qRT-PCR (mean ± S.E. (error bars), n = 3). A, down-regulation of selected metabolic genes in colon cancer cell line HCT116 (black bars) and upon inhibition of p53 by small molecule pifithrin-α (gray bars). B, down-regulation of selected metabolic genes in wild type p53 human breast cancer cells MCF7 (black bars) and upon inhibition of p53 by small molecule pifithrin-α (gray bars). C and D, comparison of changes in mRNA levels of metabolic genes in p53-null cancer cell lines HCT116 TP53−/− and Saos2 (mean ± S.E., n = 3). E, p53-dependent down-regulation of selected metabolic factors on protein level as assessed by immunoblotting.
To further assess p53 dependence, we evaluated the changes in mRNA levels of selected metabolic genes upon specific block of p53 activity by p53 inhibitor pifithrin-α (38). Pifithrin-α (PFTα) is a superior p53 inhibitor compared with p53 shRNA because it completely blocks p53 induction by RITA, whereas p53 shRNA, although depleting 70–80% of basal p53 levels, could not prevent its induction by RITA (25). Indeed, repression of the metabolic genes was successfully prevented by PFT-α pretreatment (Fig. 3, A and B, grey bars).
Importantly, protein analysis of HK2 and HIF1α revealed a strong depletion of these factors upon RITA treatment (Fig. 3E). In accordance with the data obtained using qRT-PCR, down-regulation of these proteins was observed only in p53-positive HCT116, MCF7, and U2OS and not in p53-negative HCT116TP53−/−, Saos2, and H1299 cells (Fig. 3E), (data not shown).
RITA-activated p53 Mediates the Inhibition of Selected Metabolic Genes in Human Tumor Xenografts
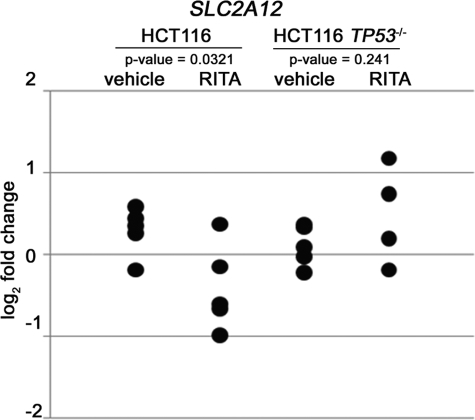
We previously reported that RITA efficiently inhibits the growth of HCT116 and HeLa human tumor xenografts in mice (23, 39). Therefore, we examined p53-induced inhibition of metabolic genes in a more physiological environment resembling a clinical setting using HCT116 and HCT116 TP53−/− xenografts grown in immunodeficient mice. Animals were treated with a 1 mg/kg intratumor injection of RITA for 18 h, and the status of metabolic genes was analyzed using qRT-PCR (supplemental Table 1). The expression of selected genes was repressed upon RITA treatment in at least some tumors. SLC2A12 (encoding glucose transporter type 12) was the most statistically significantly repressed gene upon p53 activation in tumors. Notably, this effect was p53-dependent because it was not observed in p53-null xenografts (Fig. 4). Thus, our data demonstrate that RITA-activated p53 inhibited SLC2A12 (GLUT12), one of the key metabolic factors crucial for providing ATP to cancer cells in vivo.
FIGURE 4.
RITA represses the expression of SLC2A12 (GLUT12) in tumor xenografts in vivo in a p53-dependent manner. qRT-PCR analysis of mRNA levels of SLC2A12 in HCT116 and HCT116 TP53−/− tumor xenografts treated with RITA for 18 h. The figure shows the data obtained from five tumor samples treated with vehicle and five RITA-treated tumors (1 mg/kg). Primers were designed to be specific for human SLC2A12.
Assessment of p53 Binding to Metabolic Genes Using ChIP-seq
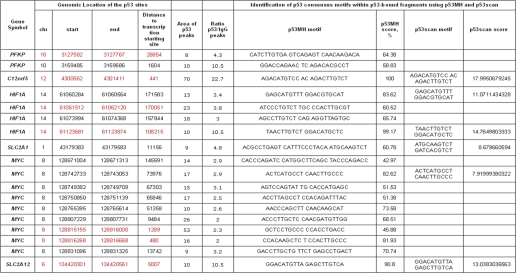
In order to assess whether p53 might play a direct role in regulation of expression of a set of metabolic genes, we investigated whether p53 binds the promoter regions of these genes in cellulo by analyzing p53 genome-wide chromatin occupancy upon 1 μm RITA treatment in MCF7 cells using chromatin immunoprecipitation coupled to deep sequencing (ChIP-seq). 6 million sequencing reads were mapped to the human genome (NCBI36) and used to calculate the height of the peaks. As a negative control, IgG-precipitated sample was used. Prefiltering of the obtained data according to the height of the p53 peak allowed us to identify 21,000 high quality p53 peaks. Detailed information on this experiment will be published elsewhere.7
Among the top score 10,957 peaks with height >8 and p values <0.05, significantly enriched over IgG samples (ratio p53/IgG > 2) that were present upon p53 activation with RITA, several peaks were located within metabolic genes. We found that upon RITA treatment, p53 bound DNA in vicinity to a number of genes regulating cell metabolism: PFKP, C12orf5, HIF1A, SLC2A1, SLC2A12, and MYC (Table 1). Binding of p53 to C12orf5 and MYC promoters is in line with previously reported data identifying these as direct p53 target genes (17, 40). In most of the genes, we identified p53-bound DNA fragments in proximity to the transcriptional starting site ±10 kbp (e.g. SLC2A12). HIF1A, on the other hand, had several sites where p53 was bound that were located much farther (100 kbp) from the transcription start site (Table 1). This observation suggests that HIF1A might be regulated by p53 through distal interactions via a so-called “looping” mechanism (41).
TABLE 1.
In vivo p53 binding to metabolic genes, determined by ChIP-seq
Chromosomal coordinates of p53-occupied sites in vicinity of metabolic genes upon 1 μm RITA treatment are shown. Red color indicates the p53-bound fragments occupied also upon 100 μm 5-FU and 10 μm nutlin3a treatments. Higher score indicates better fit to the consensus binding site.
We identified p53 consensus motifs within p53-bound DNA fragments using p53MH and p53Scan programs (42, 43). The presence of p53 consensus sites in HIF1A, SLC2A1, and SLC2A12 was confirmed by both approaches, along with consensus sites in MYC and C12orf5 (Table 1). Taken together with expression profile data in cells and in tumor xenografts, these results suggest that SLC2A12, SLC2A1, and HIF1A are directly repressed by p53. Because HIF1α and c-Myc are transcriptional factors that can regulate the expression of several genes involved in metabolism (Fig. 2C), we addressed their contribution to inhibition of metabolic genes by activated p53.
Involvement of c-Myc in p53-mediated Ablation of Metabolic Genes
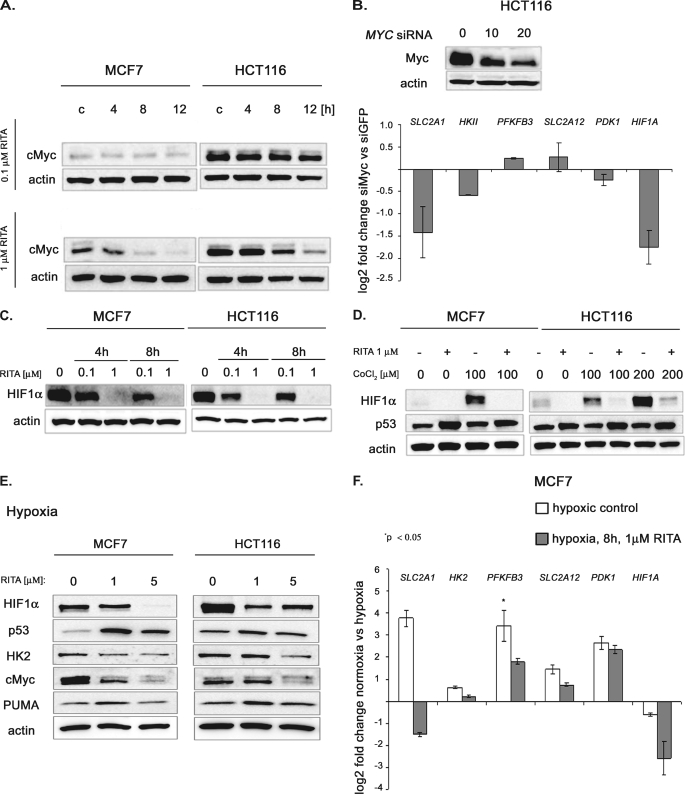
Several lines of evidence suggest that c-Myc can up-regulate the expression of SCL2A1 (44), LDHA (45, 46), PDK1, HKII (46, 47), and HIF1α (48). c-Myc was down-regulated by p53 upon treatment with 1 μm but not 0.1 μm RITA, as we showed previously (25) and in Fig. 5A.
FIGURE 5.
Contribution of c-Myc and HIF1α inhibition to the repression of metabolic genes upon p53 reactivation under normoxia and hypoxia, respectively. A, c-Myc protein level is down-regulated upon 1 μm but not upon 0.1 μm RITA in wild type p53 MCF7 and HCT116 cancer cells, as assessed by Western blot. B, top, the extent of c-Myc depletion by siRNA was assessed by immunoblotting. Bottom, inhibition of c-Myc levels by siRNA led to the down-regulation of the SLC2A1, HKII, and HIF1A mRNA in HCT116 cells, as detected by qRT-PCR (mean ± S.E. (error bars), n = 3). C, RITA down-regulates HIF1α protein levels in a dose- and time-dependent manner under normoxia, as detected by Western blot (short exposure). D, efficient down-regulation of HIF1α by RITA treatment upon its induction by hypoxia mimetic CoCl2 as assessed by immunoblotting (long exposure). E, Western blot analysis revealed the induction of p53 and its target PUMA by 1 and 5 μm of RITA in hypoxic conditions. Down-regulation of c-Myc and HK2 correlated with p53 induction. HIF1α, which is induced by hypoxia, was down-regulated by RITA as well. F, qRT-PCR analysis revealed transcriptional repression of SLC2A1, HK2, PFKFB3, SLC2A12, and HIF1α in hypoxia upon treatment with 1 μm RITA. Shown is the -fold change in hypoxia compared with normoxia (mean ± S.E., n = 3).
Thus, we addressed the question of whether down-regulation of these genes upon RITA treatment is due to c-Myc inhibition. We determined the mRNA levels of metabolic genes upon MYC depletion by siRNA in HCT116 cells. c-Myc knockdown resulted in down-regulation of SLC2A1, HKII, and HIF1A, suggesting that transcriptional repression of these genes upon RITA treatment could be, at least partially, due to c-Myc ablation (Fig. 5B).
Inhibition of HIF1α by Reactivated p53 Contributes to the Ablation of Metabolic Genes in Hypoxic but Not Normoxic Conditions
Many tumors encounter a hypoxic environment during their development, which results in the overexpression of HIF1α (49). It has been shown previously that p53 acts as a potent negative regulator of HIF1α at the protein level (50) and that upon hypoxic conditions, HIF1α translation is inhibited by RITA-reactivated p53 (51). In the present study, we show that p53 down-regulated HIF1α mRNA (2–4) as well as HIF1α protein (Fig. 5C) in MCF7 and HCT116 cells. To test whether the induction of HIF1α can rescue it from p53-dependent inhibition, we induced the levels of HIF1α by cobalt chloride (CoCl2), a widely used experimental hypoxia mimetic, which up-regulates the expression of HIF1α (52).
As expected, HIF1α-expression was increased in both HCT116 and MCF7 cell lines after treatment with CoCl2 (Fig. 5D). Due to the potent induction of HIF1a upon CoCl2 treatment, the membranes were exposed for a shorter time; hence, the basal level seems lower than in the previous figure. However, CoCl2 did not rescue HIF1α from down-regulation by activated p53. Upon treatment with RITA, even robustly accumulated HIF1α was almost completely ablated in MCF7 and HCT116 cells (Fig. 5D).
Cell viability assays revealed that cells were killed by RITA to the same extent in the presence or absence of CoCl2 (data not shown). This suggests that HIF1α cannot confer survival to tumor cells upon p53 reactivation, even upon its massive induction by CoCl2. However, the inability of CoCl2 to rescue HIF1α levels did not allow us to assess a possible contribution of HIF1α inhibition to the growth suppression effect of RITA. Next, we studied whether the induction of HIF1α under hypoxic conditions might rescue cancer cells from p53-dependent inhibition of ATP-generating pathways.
The viability assay based on cell morphology analysis confirmed previous data (51) indicating that RITA induces cell death in MCF7 and HCT116 cells to the same extent under hypoxia as under normoxia (supplemental Fig. 2, A and B). Further, protein analysis demonstrated that upon RITA administration c-Myc and HK2 were decreased to comparable levels under hypoxia and normoxia (Figs. 3E and 5E). HIF1α levels were down-regulated somewhat differently in HCT116 and MCF7 cells; HIF1α was substantially ablated already by 1 μm RITA in HCT116 cells, whereas for MCF7 cells, 5 μm RITA was required to achieve complete inhibition of HIF1α (Fig. 5E). Thus, p53 activated by RITA is capable of inhibiting HIF1α overexpressed under hypoxic conditions. Next, we evaluated the efficiency of RITA to ablate the expression of metabolic genes under hypoxia and performed qRT-PCR analysis in MCF7 cells.
SLC2A1, PFKFB3, SLC2A12, and PDK1 mRNAs were up-regulated in hypoxia (Fig. 5F), along with HIF1α protein level, in line with being known HIF1 targets. Reactivation of p53 by RITA in hypoxia led to a significant repression of HIF1α mRNA (Fig. 5F). Moreover, we observed a dramatic repression of SLC2A1, more than 5-fold, whereas in normoxia it was inhibited 2-fold. These data suggest the contribution of HIF1α inhibition to repression of SLC2A1 upon RITA treatment in hypoxia. Despite the induction of PFKFB3 and SLC2A12 in hypoxia, they were still down-regulated by RITA ∼2-fold, similarly to normoxia (Fig. 5F). Thus, our data indicate that down-regulation of HIF1α contributes to the transcriptional repression of at least some metabolic genes observed upon p53 reactivation in hypoxia.
Identification of Sp1 as a Transcriptional Cofactor Involved in p53-mediated Repression of Metabolic Genes
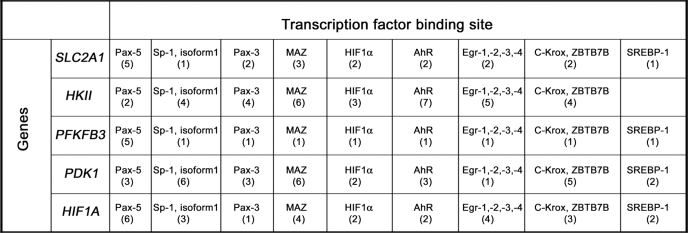
In search of other candidate transcription factors involved in the regulation of the identified set of genes, we analyzed the transcription factor binding sites in the promoters of these genes based on the microarray data obtained for MCF7 cells. Using the F-Match program and TRANSFAC® database, we selected those transcription factors whose potential binding sites (one or more) were significantly overrepresented (frequency ratio >1.3 and p value <0.01) in the promoters of metabolic genes differentially expressed upon RITA treatment (Fig. 2A). Our analysis revealed several transcription factor candidates for a common co-regulator(s) of the metabolic genes (supplemental Table 3), including the paired box family of transcription factors (Pax-5 and Pax-3), c-Myc-associated zinc finger protein (Maz), Sp1, Ahr, HIF1α, ZNF148, Egr1, and Ap2. Data for two factors, ZNF148 and Sp1, demonstrated the most significant p values exceeding the multiple testing correction threshold (10−6) (Bonferroni correction). Interestingly, many of the factors co-occur in several promoters of the metabolic genes under study (Table 2).
TABLE 2.
Transcription factors whose binding sites were found in the promoters of selected metabolic genes
Transcriptional factors were identified whose specific binding sites were overrepresented in the promoters of the selected set of metabolic genes in comparison with 1000 genes that did not change their expression upon RITA treatment.
In order to identify the most important regulatory molecules crucial for the p53 transcriptional response, we applied another bioinformatics method. We performed the key node analysis of our microarray data (as described under “Experimental Procedures”), which revealed p53 as one of the top ranking key nodes (by the key node score, p53 was ranked number 12 among more than 2400 other factors). Importantly, the Sp1 transcription factor was found to be tightly linked to p53 in our key node diagram (see supplemental Fig. 3). Sp1 was the most significantly overrepresented transcriptional factor that belongs to the constructed p53 network (supplemental Fig. 3). Thus, two bioinformatics approaches suggest that Sp1 might be involved in p53-dependent transcriptional regulation upon RITA treatment. This is in line with previously published studies suggesting that Sp1 is the p53 co-repressor for several genes, including Cdc25B and DNMT1 (53, 54). Therefore, we selected Sp1 as a candidate p53 cofactor for regulation of metabolic genes and performed a series of experiments to address the question of whether Sp1 plays a role in p53-mediated repression of glycolytic genes.
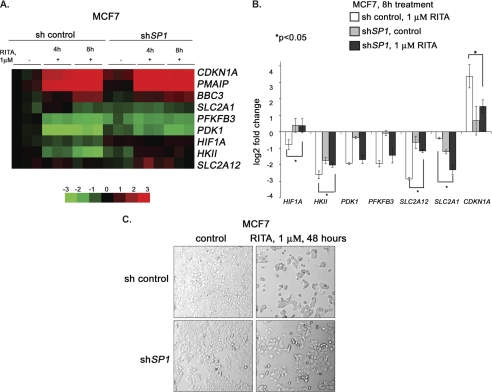
To study the impact of Sp1, we stably depleted it in MCF7 cells and performed microarray analysis. Expression profiling showed that depletion of Sp1 in MCF7 cells significantly protected them from the repression of metabolic genes PDK1, HIF1A, HKII, SLC2A1, and SLC2A12 upon RITA treatment (Fig. 6A). Assessment of mRNA levels by qRT-PCR revealed that the repression of HIF1A, HKII, and SLC2A12 was significantly attenuated in the absence of Sp1 (Fig. 6B), suggesting that Sp1 contributes to the p53-mediated inhibition of these genes. Interestingly, the repression of the SLC2A1 gene was even more pronounced in Sp1-depleted cells, which suggests a different mechanism of its transcriptional regulation. Furthermore, Sp1 depletion partially prevented RITA-mediated growth suppression (Fig. 6C), indicating that the contribution of Sp1 to the p53-mediated transcriptional repression of HIF1A and other glycolytic genes plays a role in the biological outcome.
FIGURE 6.
Sp1 cooperates with p53 to repress metabolic genes. A, expression of metabolic genes was compared between cells with depleted Sp1, untreated or treated with RITA for 4 or 8 h, and control transfected cells, untreated or treated with RITA, using microarray analysis. Data are presented as a heat map. Vertical columns indicate separate arrays, and horizontal rows indicate genes. The rows were standardized by subtracting the mean of the first column. B, Sp1 depletion partially rescued p53-mediated repression of HIF1A, SLC2A12, and HKII as assessed by qRT-PCR. *, statistically significant differences, calculated using Student's t test. Error bars, S.E. C, Sp1 knockdown rescued MCF7 cells from growth suppression mediated by RITA as assessed by the microscopy analysis of cell morphology.
Ablation of Glycolysis Contributes to the Full Scale Induction of Apoptosis by RITA-activated p53
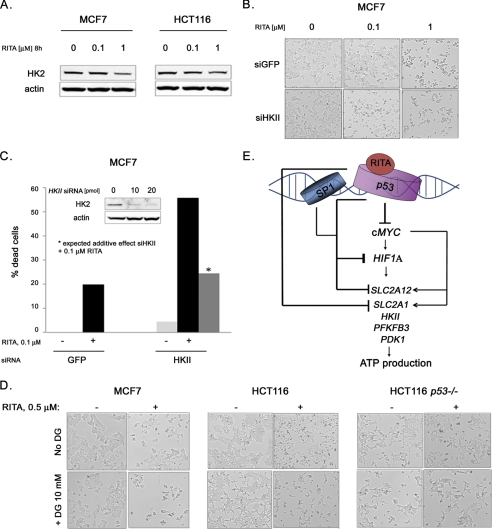
There is currently great interest in the development of inhibitors of glycolysis for treating cancer. Therefore, we addressed the question of whether inhibition of glycolysis plays a role in growth suppression by RITA. In line with the absence of robust effects of 0.1 μm RITA on cell metabolism and viability, we did not detect down-regulation of HK2 protein at 0.1 μm RITA in both MCF7 and HCT116 cells (Fig. 7A) despite p53 induction. Thus, we investigated whether the depletion of HK2 can facilitate the growth suppression effect of p53 upon 0.1 μm RITA. HKII was depleted by corresponding siRNA (see Fig. 7C), and cells were treated with 0.1 μm RITA. Although down-regulation of HKII by siRNA on its own had a barely detectable effect on cell viability, it synergized with 0.1 μm RITA in apoptosis induction in MCF7 cells (Fig. 7, B and C). These data indicate that HK2 ablation is important for apoptosis induction by RITA-reactivated p53.
FIGURE 7.
Ablation of HK2 contributes to RITA-induced apoptosis. A, HK2 was down-regulated by RITA in a dose-dependent manner, as detected by Western blot analysis in MCF7 and HCT116 cells. B, inhibition of HK2 synergized with 0.1 μm RITA. Microscopy analysis shows the extent of cell death induction by 0.1 and 1 μm RITA in the presence or absence of HK2 depletion by siRNA. C, down-regulation of HK2 by siRNA synergized with 0.1 μm RITA treatment in apoptosis induction in MCF7 cells. Quantification of cell death induction by RITA treatment in the presence or absence of HK2 depletion by siRNA was performed using trypan blue staining. D, combination of 0.5 μm RITA with 2-deoxyglucose with 0.5 μm RITA potentiated growth inhibition of p53-positive HCT116 and MCF7 cells but not of the HCT116 TP53−/− cells. E, model depicting the regulatory pathways governing cancer cell metabolism that are affected upon pharmacological activation of p53 by small molecule RITA. For more details, see “Discussion.”
Besides gene silencing, hexokinase could be inhibited by treatment with the inactive analog of glucose, 2-deoxyglucose. Treatment with 2-deoxyglucose changed the morphology of MCF7, HCT116, and HCT116 TP53−/− cells but did not affect their viability (Fig. 7D). However, the combination of 2-deoxyglucose with 0.5 μm RITA resulted in stronger induction of apoptosis in MCF7 and HCT116 but not in HCT116 TP53−/− cells (Fig. 7D).
Taken together, our results suggest that inhibition of glycolysis contributes to the induction of apoptosis by RITA. These data show that the combination of small molecule RITA with glycolytic inhibitors analogous to 2-deoxyglucose could be a promising therapeutic approach to induce robust apoptosis in cancer cells.
DISCUSSION
Despite a high genetic diversity, cancer cells exhibit a common set of functional characteristics, one of them being the “Warburg effect” (i.e. continuously high glucose uptake and higher rate of glycolysis than in normal cells). Extensive studies have provided evidence that a number of genes that initiate tumorigenesis are linked to the metabolic regulation (i.e. several oncogenes are very prominent inducers of glycolytic phenotype (reviewed in Refs. 55–58)). Such altered tumor cells are uniquely sensitive to the inhibition of glycolysis, suggesting the potential to exploit changes in tumor cell metabolism for anti-cancer therapy. Thus, development of molecules targeting metabolic pathways can open a new vista for cancer treatment.
A “triad” of transcription factors, c-Myc, HIF1α, and p53, have been implicated in the control of transcription of genes involved in energy production and metabolism (3). Our previous work demonstrates that upon pharmacological activation of p53 in cancer cells by small molecule RITA, the c-Myc oncogene is potently inhibited at the transcriptional and translation levels as well as destabilized at the protein level (25). Well documented involvement of c-Myc in regulation of glycolysis prompted us to investigate whether the activation of p53 by small molecules can affect metabolism of cancer cells.
Indeed, metabolic chip data revealed p53-dependent inhibition of respiration and acidification rates in cells already a few hours after RITA treatment. To elucidate the molecular mechanism of the observed phenotype, we used microarray analysis to study gene expression profiles in wild type p53-expressing MCF7 and HCT116 cancer cells upon pharmacological p53 reactivation and found the transcriptional repression of a set of key factors involved in ATP-generating pathways. Interestingly, changes in expression profiles of metabolic genes induced by nutlin3a, another p53-activating molecule, differed from those induced by RITA. It has been shown previously that p53 transcriptional response can differ significantly upon induction by different stimuli (16). This might be due to different posttranslational modifications of p53 and/or different cofactors bound by p53.
Small molecules are most likely to have more than one target in cells because they are generally too small to have as high specificity as, for example, antibodies. It is thus plausible that their effect on additional targets contributes to the overall response. We have previously shown that RITA binds and inhibits TrxR1, an enzyme important for keeping ROS levels in check (34). Induction of ROS due to the effect of RITA on TrxR1 together with p53-dependent induction of DNA damage response (51, 59)8 might contribute to the differential p53 modifications upon RITA treatment, compared with nutlin3a. Indeed, our collaborators showed p53 phosphorylation at Ser-46 upon RITA, but not nutlin3a treatment, which can affect p53-mediated response (60). However, p53 induction by RITA is independent of DNA damage signaling and ATM, as we recently showed (61); therefore, the additional effects of RITA are not the cause of p53 activation, although they might contribute to it by amplifying the signal.
We validated our microarray data using qRT-PCR in p53-positive MCF7 and HCT116, U2OS, and p53-null cells HCT116TP53−/−, Saos-2, and H1299, which demonstrated that 1 μm RITA significantly down-regulated the expression of glucose transporters SLC2A1 and SLC2A12, enzymes HKII, PFKFB3, and PDK1, and transcriptional regulator of metabolic genes HIF1α in a p53-dependent manner. Thus, p53 activated by RITA ablates the first steps of glycolysis, namely glucose uptake, primary phosphorylation of glucose molecules, and the rate-limiting step of glycolysis: conversion of fructose-6-phosphate to fructose-1,6-bisphosphate (Fig. 2C). Additionally, microarray data pointed to the inhibition of lactate production and the lactate/proton removal pathway.
Analysis of ChIP-seq data provided valuable insights into the p53-mediated regulation of metabolic genes upon RITA treatment. We found that p53 binds the promoters of several metabolic genes in vivo and identified p53-binding elements in the promoter regions of SLC2A12, SLC2A1, HIF1A, and MYC, suggesting that they are directly repressed by p53.
Interestingly, down-regulation of HIF1α by RITA under hypoxia has been reported previously (51, 62), but it has been attributed to the decreased translation of HIF1α. p53 has been also shown to promote degradation of the HIF1α protein (50). In this study, we found yet another level of negative regulation of HIF1α by p53 (i.e. inhibition of its transcription). In contrast to the previous studies (51, 62), down-regulation of HIF1α upon RITA treatment was observed both in normoxia and hypoxia. We tested whether p53 might directly affect the transcription of HIF1α by binding to its promoter. Our ChIP-seq data indeed indicated that p53 binds to HIF1α promoter at two sites, within which we found p53 consensus binding sites (Table 1). Importantly, p53 binding to the same sites in HIF1α promoter was observed upon p53 activation with RITA, nutlin3a and 5-FU. Because p53 binding sites are located quite far from the transcriptional starting site, it is possible that a DNA looping mechanism is involved in p53-mediated regulation of HIF1A.
In addition, indirect transcriptional inhibition of HIF1α upon RITA treatment might be attributed to the repression of c-Myc because HIF1A was identified as the high affinity c-Myc target (48). This notion is supported by our data showing that c-Myc depletion by siRNA results in transcriptional down-regulation of HIF1α in MCF7 cells.
The HIF1α level is regulated through oncogenic PI3K acting upstream of Akt (63). We have shown previously potent p53-dependent ablation of the IGFR/PI3K/Akt pathway upon RITA treatment (27); thus, inhibition of both subunits PI3KCA and PI3KCB might contribute to the repression of HIF1α as well. It appears that the combined inhibition of several pathways regulating HIF1α results in its potent repression.
Our results demonstrate that the ablation of both c-Myc and HIF1α, transcriptional factors that cooperate in positive regulation of expression of glycolytic genes (reviewed in Refs. 3, 9, and 64) further promotes the inhibition of glycolytic genes, both in normoxia and hypoxia.
Bioinformatics analysis of transcription factor binding sites present in the promoters of the repressed genes and key node analysis allowed us to identify Sp1 as a p53 cofactor facilitating the repression of metabolic genes, including novel p53 target genes that we identified, HIF1A and SLC2A12.
We did not observe the induction of C12orf5 and SCO2 genes, shown to be activated by p53 (17, 18) upon 1 μm RITA treatment, which suggests the p53-dependent ablation of both energy production pathways, aerobic glycolysis and oxidative phosphorylation. We reported previously that RITA induces ROS in a p53-dependent manner due to the inhibition of TrxR, which contributes to cancer cell killing (34). We speculate that the absence of TIGAR induction might contribute to a ROS increase in RITA-treated cells, along with the inhibition of TrxR.
We observed transcriptional repression of C12orf5 (TIGAR) by 1 μm RITA, in contrast to its induction by 0.1 μm RITA, 5-FU, and nutlin3a, whereas p53 was bound to the C12orf5 promoter irrespective of the type of treatment (Table 1). These data suggest that under certain conditions, p53 might be converted from transcriptional activator to a transcriptional repressor, as has been shown, for example, in case of p53-mediated regulation of the survivin gene (65). An interesting example of another transcription factor that can both activate and repress the same genes upon binding to their promoters is MyoD. Depending on its interaction with LSD1 or HDAC1 at the promoter of its target genes, MyoD can repress or activate genes, respectively (66, 67).
Previous studies demonstrated that, depending on the dose of activating agent, p53 can promote either cell survival or cell death by differentially regulating anti-oxidant or pro-oxidant genes, respectively (68). Further, the small molecule nutlin3a also can act in a dose-dependent manner; a high dose of nutlin promotes p53-dependent inhibition of mTOR, whereas a lower dose does not (69). It is plausible that p53 posttranslational modifications and cofactors that associate with p53 can serve as the bars of the “bar code” that governs p53 transcriptional activity, thereby forming the underlying basis of the heterogeneity of p53 response, depending on the type and dose of stimuli as well as cell type (70). Because there are more than 50 known p53 partner proteins, which can modulate p53-mediated regulation of gene expression, high throughput approaches are required to identify the bars of the code that confer the differential gene regulation in response to different p53-activating molecules. In order to address this, we initiated proteomics and functional genomics studies aimed to identify cofactors differentially bound to p53 upon nutlin3a and RITA treatment.
Based on our results, we propose a model suggesting that pharmacologically activated p53 triggers a set of events that ablate the network of key regulators of ATP production. Upon activation by RITA, p53 inhibits c-Myc and HIF1α transcriptional factors regulating a number of genes involved in cell metabolism (Fig. 7E). Combined inhibition of c-Myc and HIF1α results in down-regulation of a whole set of metabolic genes. Further, p53 directly inhibits the expression of glucose transporters 1 and 12, facilitating ablation of glycolysis. Sp1 cooperates with p53 in transcriptional repression of HIF1A and SLC2A12 and probably other metabolic genes. Taken together, several pathways triggered by p53 result in robust inhibition of energy production in cells, leading to efficient elimination of cancer cells.
HIF1α was ablated by RITA-reactivated p53 under hypoxic and normoxic conditions in MCF7 and HCT116 cells. This can be of special interest for tumor therapy of hypoxic tumors because HIF1α can increase the tumor's resistance to chemotherapeutic agents and radiotherapy (71).
We showed that RITA can down-regulate at least some metabolic genes, such as SLC2A12, in HCT116 xenografts in vivo, suggesting that glycolytic gene inhibition by RITA-reactivated p53 is not restricted to the in vitro phenomenon. This indicates the potential of RITA as a glycolytic inhibitor in conditions resembling a clinical setting and is in line with our previous data on inhibition of specific oncogenes upon RITA treatment in HCT116 but not in HCT116 TP53−/− xenografts (25).
Hexokinases and glucose transporters are frequently overexpressed in tumors along with other gycolytic proteins (3, 72). Hexokinase plays an important role in immortalizing cancer cells (73). We found that depletion of HK2 by siRNA or inhibition by 2-deoxyglucose facilitates the induction of apoptosis, suggesting that inhibition of glycolytic factors contributes to the robust apoptosis triggered by RITA. Further, it suggests that combination of p53-reactivating drug with inhibitors of glycolysis might be a promising strategy for anti-cancer therapy. Although we cannot rule out the possibility that RITA and 2-deoxyglucose activate different pathways that collaborate to induce cell death, our data obtained in Sp1-depleted cells suggest that the repression of HIF1A and other metabolic genes plays a role in the growth suppression effect of RITA.
Drug combinations are currently regarded as an efficient way to solve the problem of de novo resistance to cancer therapies, a formidable barrier to the successful cure of cancer. Our data indicate that drugs targeting metabolic pathways can be efficiently combined with p53-reactivating compounds. In summary, our data suggest that reactivation of p53 by small molecules, such as RITA, has a high potential for cancer therapy because it simultaneously targets several key enzymes involved in glycolysis.
Acknowledgments
We are indebted to Dr. Maria C. Shoshan (Karolinska Institute) for useful discussion and suggestions, to Bartosz Ferens for technical assistance, and to Maria Timofeeva (IARC) for help with statistical analysis of tumor samples. We especially thank Prof. Lars Holmgren and his group (Department of Oncology-Pathology, Karolinska Institute) for providing help and devices for hypoxia studies.
This study was supported by grants from the Swedish Cancer Society, the Swedish Research Council, and the Torsten and Ragnar Söderberg Foundation (to G. S.). This work was also supported by EC FP6 projects “Mutant p53,” “Active p53,” and “Net2Drug.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. 1–3.
J. Zawacka-Pankau, V. V. Grinkevich, A. Vema, K. Ridderstråle, N. Issaeva, E. Hedström, C. Spinnler, V. Andreotti, A. Inga, L.-G. Larsson, A. Pramanik, A. Karlen, A. Okorokov, and G. Selivanova, submitted for publication.
F. Nikulenkov, C. Spinnler, T. Tonelli, M. Turunen, H. Li, T. Kivioja, A. Kel, J. Taipale, and G. Selivanova, submitted for publication.
Y. Shi, F. Nikulenkov, and G. Selivanova, manuscript in preparation.
- qRT-PCR
- quantitative RT-PCR
- ROS
- reactive oxygen species
- 5-FU
- 5-fluorouracil.
REFERENCES
- 1. Gatenby R. A., Gillies R. J. (2004) Nat. Rev. Cancer 4, 891–899 [DOI] [PubMed] [Google Scholar]
- 2. Warburg O. (1956) Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 3. Yeung S. J., Pan J., Lee M. H. (2008) Cell Mol. Life Sci. 65, 3981–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hawkins R. A., Phelps M. E. (1988) Cancer Metastasis Rev. 7, 119–142 [DOI] [PubMed] [Google Scholar]
- 5. Czernin J., Phelps M. E. (2002) Annu. Rev. Med. 53, 89–112 [DOI] [PubMed] [Google Scholar]
- 6. Younes M., Lechago L. V., Somoano J. R., Mosharaf M., Lechago J. (1996) Cancer Res. 56, 1164–1167 [PubMed] [Google Scholar]
- 7. Rempel A., Bannasch P., Mayer D. (1994) Biochim. Biophys. Acta 1219, 660–668 [DOI] [PubMed] [Google Scholar]
- 8. Wang G. L., Semenza G. L. (1995) J. Biol. Chem. 270, 1230–1237 [DOI] [PubMed] [Google Scholar]
- 9. Denko N. C. (2008) Nat. Rev. Cancer 8, 705–713 [DOI] [PubMed] [Google Scholar]
- 10. Jones R. G., Thompson C. B. (2009) Genes Dev. 23, 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shaw R. J., Cantley L. C. (2006) Nature 441, 424–430 [DOI] [PubMed] [Google Scholar]
- 12. Le A., Cooper C. R., Gouw A. M., Dinavahi R., Maitra A., Deck L. M., Royer R. E., Vander Jagt D. L., Semenza G. L., Dang C. V. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2037–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scatena R., Bottoni P., Pontoglio A., Mastrototaro L., Giardina B. (2008) Expert Opin. Investig. Drugs 17, 1533–1545 [DOI] [PubMed] [Google Scholar]
- 14. Levine A. J., Hu W., Feng Z. (2006) Cell Death Differ. 13, 1027–1036 [DOI] [PubMed] [Google Scholar]
- 15. Hainaut P., Hollstein M. (2000) Adv. Cancer Res. 77, 81–137 [DOI] [PubMed] [Google Scholar]
- 16. Vousden K. H., Prives C. (2009) Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 17. Bensaad K., Tsuruta A., Selak M. A., Vidal M. N., Nakano K., Bartrons R., Gottlieb E., Vousden K. H. (2006) Cell 126, 107–120 [DOI] [PubMed] [Google Scholar]
- 18. Matoba S., Kang J. G., Patino W. D., Wragg A., Boehm M., Gavrilova O., Hurley P. J., Bunz F., Hwang P. M. (2006) Science 312, 1650–1653 [DOI] [PubMed] [Google Scholar]
- 19. Schwartzenberg-Bar-Yoseph F., Armoni M., Karnieli E. (2004) Cancer Res. 64, 2627–2633 [DOI] [PubMed] [Google Scholar]
- 20. Kondoh H., Lleonart M. E., Gil J., Wang J., Degan P., Peters G., Martinez D., Carnero A., Beach D. (2005) Cancer Res. 65, 177–185 [PubMed] [Google Scholar]
- 21. Vousden K. H., Ryan K. M. (2009) Nat. Rev. Cancer 9, 691–700 [DOI] [PubMed] [Google Scholar]
- 22. Selivanova G. (2010) Semin. Cancer Biol. 20, 46–56 [DOI] [PubMed] [Google Scholar]
- 23. Issaeva N., Bozko P., Enge M., Protopopova M., Verhoef L. G., Masucci M., Pramanik A., Selivanova G. (2004) Nat. Med. 10, 1321–1328 [DOI] [PubMed] [Google Scholar]
- 24. Enge M., Bao W., Hedström E., Jackson S. P., Moumen A., Selivanova G. (2009) Cancer Cell 15, 171–183 [DOI] [PubMed] [Google Scholar]
- 25. Grinkevich V. V., Nikulenkov F., Shi Y., Enge M., Bao W., Maljukova A., Gluch A., Kel A., Sangfelt O., Selivanova G. (2009) Cancer Cell 15, 441–453 [DOI] [PubMed] [Google Scholar]
- 26. Wei G. H., Badis G., Berger M. F., Kivioja T., Palin K., Enge M., Bonke M., Jolma A., Varjosalo M., Gehrke A. R., Yan J., Talukder S., Turunen M., Taipale M., Stunnenberg H. G., Ukkonen E., Hughes T. R., Bulyk M. L., Taipale J. (2010) EMBO J. 29, 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kel A., Voss N., Jauregui R., Kel-Margoulis O., Wingender E. (2006) BMC Bioinformatics 7, Suppl. 2, S13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kel A., Voss N., Valeev T., Stegmaier P., Kel-Margoulis O., Wingender E. (2008) SAR QSAR Environ. Res. 19, 481–494 [DOI] [PubMed] [Google Scholar]
- 29. Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Voss N., Stegmaier P., Lewicki-Potapov B., Saxel H., Kel A. E., Wingender E. (2006) Nucleic Acids Res. 34, D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choi C., Krull M., Kel A., Kel-Margoulis O., Pistor S., Potapov A., Voss N., Wingender E. (2004) Comp. Funct. Genomics 5, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stegmaier P., Voss N., Meier T., Kel A., Wingender E., Borlak J. (2011) PLoS One 6, e17738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marshall O. J. (2004) Bioinformatics 20, 2471–2472 [DOI] [PubMed] [Google Scholar]
- 33. Ceriotti L., Kob A., Drechsler S., Ponti J., Thedinga E., Colpo P., Ehret R., Rossi F. (2007) Anal. Biochem. 371, 92–104 [DOI] [PubMed] [Google Scholar]
- 34. Hedström E., Eriksson S., Zawacka-Pankau J., Arnér E. S., Selivanova G. (2009) Cell Cycle 8, 3576–3583 [DOI] [PubMed] [Google Scholar]
- 35. Smith T. A., Sharma R. I., Thompson A. M., Paulin F. E. (2006) J. Nucl. Med. 47, 1525–1530 [PubMed] [Google Scholar]
- 36. Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 37. Zhao R., Gish K., Murphy M., Yin Y., Notterman D., Hoffman W. H., Tom E., Mack D. H., Levine A. J. (2000) Genes Dev. 14, 981–993 [PMC free article] [PubMed] [Google Scholar]
- 38. Komarov P. G., Komarova E. A., Kondratov R. V., Christov-Tselkov K., Coon J. S., Chernov M. V., Gudkov A. V. (1999) Science 285, 1733–1737 [DOI] [PubMed] [Google Scholar]
- 39. Zhao C. Y., Szekely L., Bao W., Selivanova G. (2010) Cancer Res. 70, 3372–3381 [DOI] [PubMed] [Google Scholar]
- 40. Ho J. S., Ma W., Mao D. Y., Benchimol S. (2005) Mol. Cell. Biol. 25, 7423–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jackson P., Mastrangelo I., Reed M., Tegtmeyer P., Yardley G., Barrett J. (1998) Oncogene 16, 283–292 [DOI] [PubMed] [Google Scholar]
- 42. Hoh J., Jin S., Parrado T., Edington J., Levine A. J., Ott J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8467–8472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smeenk L., van Heeringen S. J., Koeppel M., van Driel M. A., Bartels S. J., Akkers R. C., Denissov S., Stunnenberg H. G., Lohrum M. (2008) Nucleic Acids Res. 36, 3639–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Osthus R. C., Shim H., Kim S., Li Q., Reddy R., Mukherjee M., Xu Y., Wonsey D., Lee L. A., Dang C. V. (2000) J. Biol. Chem. 275, 21797–21800 [DOI] [PubMed] [Google Scholar]
- 45. Shim H., Dolde C., Lewis B. C., Wu C. S., Dang G., Jungmann R. A., Dalla-Favera R., Dang C. V. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim J. W., Zeller K. I., Wang Y., Jegga A. G., Aronow B. J., O'Donnell K. A., Dang C. V. (2004) Mol. Cell Biol. 24, 5923–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim J. W., Gao P., Liu Y. C., Semenza G. L., Dang C. V. (2007) Mol. Cell Biol. 27, 7381–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fernandez P. C., Frank S. R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., Amati B. (2003) Genes Dev. 17, 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhong H., De Marzo A. M., Laughner E., Lim M., Hilton D. A., Zagzag D., Buechler P., Isaacs W. B., Semenza G. L., Simons J. W. (1999) Cancer Res. 59, 5830–5835 [PubMed] [Google Scholar]
- 50. Ravi R., Mookerjee B., Bhujwalla Z. M., Sutter C. H., Artemov D., Zeng Q., Dillehay L. E., Madan A., Semenza G. L., Bedi A. (2000) Genes Dev. 14, 34–44 [PMC free article] [PubMed] [Google Scholar]
- 51. Yang J., Ahmed A., Poon E., Perusinghe N., de Haven Brandon A., Box G., Valenti M., Eccles S., Rouschop K., Wouters B., Ashcroft M. (2009) Mol. Cell Biol. 29, 2243–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ardyanto T. D., Osaki M., Tokuyasu N., Nagahama Y., Ito H. (2006) Int. J. Oncol. 29, 549–555 [PubMed] [Google Scholar]
- 53. Dalvai M., Mondesert O., Bourdon J. C., Ducommun B., Dozier C. (2011) Oncogene 30, 2282–2288 [DOI] [PubMed] [Google Scholar]
- 54. Lin R. K., Wu C. Y., Chang J. W., Juan L. J., Hsu H. S., Chen C. Y., Lu Y. Y., Tang Y. A., Yang Y. C., Yang P. C., Wang Y. C. (2010) Cancer Res. 70, 5807–5817 [DOI] [PubMed] [Google Scholar]
- 55. Zhao Y., Liu H., Riker A. I., Fodstad O., Ledoux S. P., Wilson G. L., Tan M. (2011) Front. Biosci. 16, 1844–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kroemer G., Pouyssegur J. (2008) Cancer Cell 13, 472–482 [DOI] [PubMed] [Google Scholar]
- 57. Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009) Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ortega A. D., Sánchez-Aragó M., Giner-Sánchez D., Sánchez-Cenizo L., Willers I., Cuezva J. M. (2009) Cancer Lett. 276, 125–135 [DOI] [PubMed] [Google Scholar]
- 59. Ahmed A., Yang J., Maya-Mendoza A., Jackson D. A., Ashcroft M. (2011) Cell Death Dis. 2, e160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rinaldo C., Prodosmo A., Siepi F., Moncada A., Sacchi A., Selivanova G., Soddu S. (2009) Cancer Res. 69, 6241–6248 [DOI] [PubMed] [Google Scholar]
- 61. Spinnler C., Hedstrom E., Li H., de Lange J., Nikulenkov F., Teunisse A. F., Verlaan-de Vries M., Grinkevich V., Jochemsen A. G., Selivanova G. (2011) Cell Death Differ., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lou J. J., Chua Y. L., Chew E. H., Gao J., Bushell M., Hagen T. (2010) PLoS One 5, e10522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jiang B. H., Jiang G., Zheng J. Z., Lu Z., Hunter T., Vogt P. K. (2001) Cell Growth Differ. 12, 363–369 [PubMed] [Google Scholar]
- 64. Kondoh H. (2008) Exp. Cell Res. 314, 1923–1928 [DOI] [PubMed] [Google Scholar]
- 65. Riley T., Sontag E., Chen P., Levine A. (2008) Nat. Rev. Mol. Cell Biol. 9, 402–412 [DOI] [PubMed] [Google Scholar]
- 66. Choi J., Jang H., Kim H., Kim S. T., Cho E. J., Youn H. D. (2010) Biochem. Biophys. Res. Commun. 401, 327–332 [DOI] [PubMed] [Google Scholar]
- 67. Mal A., Sturniolo M., Schiltz R. L., Ghosh M. K., Harter M. L. (2001) EMBO J. 20, 1739–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sablina A. A., Budanov A. V., Ilyinskaya G. V., Agapova L. S., Kravchenko J. E., Chumakov P. M. (2005) Nat. Med. 11, 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Leontieva O. V., Gudkov A. V., Blagosklonny M. V. (2010) Cell Cycle 9, 4323–4327 [DOI] [PubMed] [Google Scholar]
- 70. Murray-Zmijewski F., Slee E. A., Lu X. (2008) Nat. Rev. Mol. Cell Biol. 9, 702–712 [DOI] [PubMed] [Google Scholar]
- 71. Williams K. J., Telfer B. A., Xenaki D., Sheridan M. R., Desbaillets I., Peters H. J., Honess D., Harris A. L., Dachs G. U., van der Kogel A., Stratford I. J. (2005) Radiother. Oncol. 75, 89–98 [DOI] [PubMed] [Google Scholar]
- 72. Altenberg B., Greulich K. O. (2004) Genomics 84, 1014–1020 [DOI] [PubMed] [Google Scholar]
- 73. Robey R. B., Hay N. (2006) Oncogene 25, 4683–4696 [DOI] [PubMed] [Google Scholar]