Background: TP0453 is the only lipoprotein in T. pallidum known to be associated with the outer membrane (OM).
Results: TP0453 adopts closed and open conformations; opening is associated with membrane sensing/insertion.
Conclusion: Two amphipathic helices are responsible for amphiphilic behavior of the molecule.
Significance: TP0453 is a novel bacterial lipoprotein whose amphiphilic behavior may contribute to T. pallidum OM biogenesis.
Keywords: Crystal Structure, Lipids, Lipoprotein, Membrane Bilayer, Structural Biology, Spirochete, Amphipathic Helices, Closed and Open Conformations, Syphilis
Abstract
The molecular architecture and composition of the outer membrane (OM) of Treponema pallidum (Tp), the noncultivable agent of venereal syphilis, differ considerably from those of typical Gram-negative bacteria. Several years ago we described TP0453, the only lipoprotein associated with the inner leaflet of the Tp OM. Whereas polypeptides of other treponemal lipoproteins are hydrophilic, non-lipidated TP0453 can integrate into membranes, a property attributed to its multiple amphipathic helices (AHs). Furthermore, membrane integration of the TP0453 polypeptide was found to increase membrane permeability, suggesting the molecule functions in a porin-like manner. To better understand the mechanism of membrane integration of TP0453 and its physiological role in Tp OM biogenesis, we solved its crystal structure and used mutagenesis to identify membrane insertion elements. The crystal structure of TP0453 consists of an α/β/α-fold and includes five stably folded AHs. In high concentrations of detergent, TP0453 transitions from a closed to open conformation by lateral movement of two groups of AHs, exposing a large hydrophobic cavity. Triton X-114 phase partitioning, liposome floatation assay, and bis-1-anilino-8-naphthalenesulfonate binding revealed that two adjacent AHs are critical for membrane sensing/integration. Using terbium-dipicolinic acid complex-loaded large unilamellar vesicles, we found that TP0453 increased efflux of fluorophore only at acidic pH. Gel filtration and cross-linking experiments demonstrated that one AH critical for membrane sensing/insertion also forms a dimeric interface. Based on structural dynamics and comparison with Mycobacterium tuberculosis lipoproteins LprG and LppX, we propose that TP0453 functions as a carrier of lipids, glycolipids, and/or derivatives during OM biogenesis.
Introduction
Syphilis is a multistage, sexually transmitted illness caused by the spirochetal pathogen Treponema pallidum, an extracellular, highly motile bacterium (1). Although syphilis is one of the oldest recognized sexually transmitted diseases, the mechanisms underlying its pathogenesis are poorly understood in large part because T. pallidum cannot be cultivated in vitro (2). The molecular architecture of the T. pallidum cell envelope is believed to be a primary determinant of the capacity of syphilis spirochete to evade the robust immune responses it elicits in its obligate human host, a property that has led to its designation as “the stealth pathogen” (1, 3). Like Gram-negative bacteria, T. pallidum has both outer and cytoplasmic membranes; however, the composition and physical properties of its cell envelope differ markedly from those of prototypical diderms (4, 5). The OM4 of T. pallidum is devoid of lipopolysaccharide, contains phosphatidylcholine, phosphatidylserine, and uncharacterized glycolipids as its major lipid constituents, and is much more permeable to lipophilic compounds (6–9). In contrast to the OM of Escherichia coli, which contains a high density of integral β-barrel forming proteins, the OM of T. pallidum has a paucity of membrane-spanning proteins, often referred to collectively as rare OM proteins (4, 5, 10, 11). A major difference between the E. coli and T. pallidum cell envelopes relates to their lipoprotein components. In T. pallidum, with one known exception (described below), all lipoproteins that have been characterized to date have been localized to the periplasmic face of the inner membrane (5, 12, 13), whereas lipoproteins in E. coli are located predominantly at the periplasmic face of the OM (14).
Several years ago we devised a strategy for identifying OM-associated proteins in T. pallidum based upon the use of 3-(trifluoromethyl)-3-(m-[125I]iodophenyl-diazarene) (15, 16). This highly lipophilic molecule stably partitions into the hydrocarbon core of a membrane where upon UV photoactivation it promiscuously binds to intramembranous components (15). Using this reagent in conjunction with Triton X-114 phase-partitioning (17), we detected a single strongly radiolabeled polypeptide of ∼30 kDa (16), subsequently identified by peptide sequencing as TP0453 (p30.5) (18). Although this approach was intended to identify OM-spanning proteins, surprisingly, TP0453 was found to be a lipoprotein. Detailed investigation revealed, however, that TP0453 possesses unique physical properties in addition to being the only lipoprotein known to be localized to the T. pallidum OM. Whereas the polypeptide moieties of spirochetal lipoproteins typically are hydrophilic (19), the polypeptide portion of TP0453 is amphiphilic and was predicted to contain five AHs as membrane-insertion elements (18). Importantly, liposome incorporation assays revealed that non-lipidated p30.5 enhances the permeability of small unilamellar vesicles. Based on these results, we hypothesized that the polypeptide portion of native TP0453 exists in a dynamic equilibrium between OM-inserted and -uninserted forms and that transient perturbations in the OM caused by insertion of AHs facilitates ingress of water-soluble nutrients (18).
In the present study we solved the crystal structure of TP0453 and performed detailed physicochemical characterization of wild type (WT) and mutated forms of the protein to elucidate its contribution to the membrane biology of T. pallidum. We found that TP0453 is structurally dynamic, converting from a closed to an open form in the presence of high concentrations of nonionic detergent with concomitant exposure of a large hydrophobic cavity. As predicted, TP0453 possesses multiple AHs as part of its overall α/β/α-fold; however, only two adjacent AHs were found to be critical for membrane integration. In the closed form these helices contribute to a surface-exposed hydrophobic patch, which we hypothesize “senses” the membrane interior when the polypeptide is in proximity to the lipid bilayer. Assays using unilamellar vesicles (LUVs) confirmed the ability of the protein to enhance efflux of fluorophore but only under acidic conditions. Although a monomer as a major form in solution, TP0453 forms stable a dimer upon membrane insertion. The similarity between the structure of TP0453 and solved structures for two Mycobacterium tuberculosis lipoproteins, LprG and LppX (20, 21), suggests that TP0453 may function as a carrier of lipids, glycolipids, and/or derivatives during OM biogenesis in T. pallidum.
EXPERIMENTAL PROCEDURES
Propagation and Harvesting of T. pallidum
The animal protocol described in this work strictly follows the recommendations of the Guide for Care and Use of Laboratory Animals of the National Institutes of Health and was approved by the University of Connecticut Health Center Animal Care Committee under the auspices of Animal Welfare Assurance A347-01. The Nichols-Farmington strain of T. pallidum subspecies pallidum was propagated by intratesticular inoculation of adult New Zealand White rabbits with 1 × 108 treponemes per testis and harvested aseptically ∼10 days later (13). To remove rabbit testicular debris, the treponemal suspension was transferred to a sterile tube and centrifuged for 10 min at 500 × g. T. pallidum was enumerated using a Petroff-Hausser counting chamber (Hausser Scientific Company, Horsham, PA). The treponemes were sedimented by centrifugation for 20 min at 10,000 × g at 4 °C, and the pellet was washed with ice-cold phosphate-buffered saline (PBS) before processing for blue native polyacrylamide gel (BN-PAGE) (see below).
Cloning and Mutagenesis
The cloning of tp0453 into the expression vector pET4341 has been described elsewhere (18). pET4341::TP0453 encodes a glutathione S-transferase-hexahistidine chimera with a thrombin cleavage site fused to the N terminus of TP0453 lacking a signal peptide (i.e. residues 27–287). The TP0453 coding sequence in pET4341::TP0453 was mutated with a site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) using the primers listed in supplemental Table 1. The ompF gene (without the signal sequence-encoding portion) was PCR-amplified from E. coli DNA using the primers listed in supplemental Table 1 and cloned into the NheI (5′-end) and HindIII (3′-end) restriction sites of pET23b (Novagen, San Diego, CA). All constructs were verified by nucleotide sequencing.
Purification and Selenomethionine Labeling of TP0453
For crystallographic studies, a 4-liter culture of Luria-Bertani (LB) was inoculated with 25 ml of an overnight culture of pET4341:TP0453 in Rosetta (DE3) cells (EMD Chemicals, Gibbstown, NJ) grown at 37 °C. At an A600 nm of 0.6, isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 0.5 mm; the cells were grown for an additional 3 h and then harvested by centrifugation at 6000 × g for 30 min at 4 °C. The pellets were suspended with 20 ml of 50 mm Tris (pH 7.5), 10% glycerol, 100 μg of lysozyme (Sigma), and 100 μl of protease inhibitor mixture (Sigma) and lysed by sonication. The sonicated lysate was centrifuged at 20,000 × g for 30 min at 4 °C followed by filtration through a 0.22 μm filter before loading onto a 5-ml nickel-nitrilotriacetic acid column equilibrated with 50 mm Tris (pH 7.5), 100 mm NaCl, 10 mm imidazole (Buffer A). The column initially was washed with Buffer A alone and subsequently twice with Buffer A containing 20 mm and then 40 mm imidazole. The protein was eluted with 50 ml of 50 mm Tris (pH 7.5), 100 mm NaCl, 400 mm imidazole (Buffer B) in 10-ml fractions, and the fractions containing eluted proteins were pooled. Pooled fractions from nickel-nitrilotriacetic acid column were mixed with pre-swollen glutathione-agarose beads and incubated overnight at 4 °C. The following day the glutathione-agarose with bound protein was loaded onto an empty column and washed extensively with 50 mm Tris (pH 7.5), 100 mm NaCl, and 10% glycerol. To excise the N-terminal GST tag, 100 units of thrombin were added to the resin followed by incubation for 12–14 h at 22 °C on a shaker. Cleaved TP0453 in the flow-through was loaded onto a cation exchange column packed with SP-Sepharose (Bio-Rad) and washed extensively with 25 mm Tris (pH 7.5), 10% glycerol, and eluted with a gradient of 0–1.0 m NaCl. Fractions containing TP0453 were pooled and subjected to size exclusion chromatography using Superdex 75 HR 10/300 column (GE Healthcare) equilibrated with 50 mm Tris (pH 7.5), 100 mm NaCl. All mutants (supplemental Table 1) were expressed and purified using the same protocol. The ProtParam tool provided by the ExPASy proteomics server (22) was used to calculate the molar extinction coefficient (m−1 cm−1) of the proteins. To generate selenomethionine-substituted TP0453, pET4341:TP0453 was transformed into E. coli B834 (Emd Chemicals), and a single colony was inoculated into 100 ml of M9 minimal medium supplemented with 50 μg/ml ampicillin. 25-ml aliquots of overnight culture were inoculated into 1 liter of M9 minimal medium, which then was incubated at 37 °C to an A600 nm of 0.6. Each 1-liter culture was then directly supplemented with a mixture of the following amino acids: 100 mg of lysine, phenylalanine, and threonine, 50 mg of isoleucine, leucine, and valine, and 60 mg of selenomethionine. The purification procedure for selenomethionine-incorporated TP0453 was the same as that used for the native protein.
Crystallization and Data Collection
Initial crystallization screening was carried out in the High Throughput Crystallization Laboratory at the Hauptman-Woodward Medical Research Institute utilizing a sparse-matrix screen designed for soluble proteins and a tailored grid screen optimized for membrane proteins that is based on detergent phase partitioning (23, 24). A 1536 mixture screen utilizing the microbatch-under-oil method was set up with purified TP0453 in 25 mm Tris (pH 7.2), 150 mm NaCl containing 0.15% (w/v) n-octyl-β-d-glucopyranoside (β-OG) at a protein concentration of 8.2 mg/ml. One crystallization lead generated from the tailored membrane protein screen was further optimized to generate the crystals utilized in this study. Specifically, crystals were grown at 20 °C in sitting drops by combining 2 μl of protein solution with 2 μl of 10–15% polyethylene glycol 3350 (PEG 3350), 100 mm MgCl2, 100 mm MES (pH 5.5), and 0–1.0% (w/v) β-OG. Different crystal morphologies were observed depending on the concentration of β-OG utilized in the crystallization setup. Two different crystal morphologies were observed when concentrations of β-OG between 0–0.5% (w/v) were utilized; two additional morphologies were observed when concentrations of β-OG were used between 0.5 and 0.75% (w/v) and between 0.75 and 1.0% (w/v), respectively. We designated the different crystal morphologies as Forms I-IV with Forms I and II generated from low concentrations of β-OG (0–0.5%) and Forms III and IV generated from high concentrations of β-OG (0.75–1.0%). Forms I and II were cryopreserved by initial transfer to a solution consisting of 15% PEG 3350, 100 mm MgCl2, 100 mm MES (pH 5.5) followed by subsequent transfers to solutions containing increasing concentrations of 2-methyl-2, 4-pentanediol up to a final concentration of 15% (w/v). Forms III and IV were cryopreserved via direct transfer to a solution consisting of 15% PEG 3350, 100 mm MgCl2, 100 mm MES (pH 5.5), 5% (w/v) glycerol, and 5% (w/v) ethylene glycol. After incubation in cryoprotectant, the crystals were looped and flash-cooled directly into the N2 gas stream before diffraction analysis. Crystals of the selenomethionine derivative of TP0453 were produced using the same techniques, generating a crystal morphology corresponding to Form III. Data were collected for Forms I, II, and IV on beamline A1 at the Cornell High Energy Synchrotron Source (Ithaca, NY) using an Area Detector Systems CCD Quantum-210 detector. For Form III, both native and selenomethionine multiwavelength anomalous diffraction data sets were collected on beamline 9-2 at the Stanford Synchrotron Research Laboratory (Menlo Park, CA) using remote data collection techniques (25). Data sets were measured at the selenium K-edge peak (0.97915 Å), inflection point (0.97932 Å), and remote (0.96396 Å) energy wavelengths using a MarMosaic-325 CCD detector. All data were processed and scaled using the HKL2000 program suite (26). Details of the data collection statistics are summarized in Table 1.
TABLE 1.
Crystallographic data collection and refinement statistics
r.m.s.d., root mean square deviations; MPD, 2-methyl-2,4-pentanediol.
| Crystallographic Parameter | Form I | Form II | Form III | Form IV | Form III peak | Form III inflection | Form III high remote |
|---|---|---|---|---|---|---|---|
| PDB ID | 3K8G | 3K8H | 3K8J | 3K8I | |||
| Space group | P21212 | C2 | P21212 | P21212 | P21212 | P21212 | P21212 |
| No. in ASU | 2 | 2 | 1 | 1 | 1 | 1 | 1 |
| Unit cell (Å) | |||||||
| a | 87.74 | 145.30 | 45.68 | 45.42 | 45.78 | 45.79 | 45.80 |
| b | 159.87 | 44.76 | 157.51 | 143.73 | 157.66 | 157.78 | 157.77 |
| c | 44.53 | 96.97 | 43.03 | 42.92 | 43.03 | 43.04 | 43.04 |
| α = β = γ (°) | 90.00 | β = 99.75 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| Wavelength (Å) | 0.9789 | 0.9789 | 1.000 | 0.9789 | 0.97915 | 0.97932 | 0.96396 |
| Resolution (Å) | 50-1.95 | 50-2.40 | 50-2.20 | 40-2.20 | 50-2.50 | 50-2.50 | 50-2.50 |
| Res. last shell (Å) | 2.02-1.95 | 2.44-2.40 | 2.28-2.20 | 2.24-2.20 | 2.59-2.50 | 2.59-2.50 | 2.59-2.50 |
| Rmerge | 6.2 (45.5) | 6.2 (30.3) | 4.7 (27.0) | 5.6 (19.1) | 7.4 (32.5) | 6.7 (30.3) | 6.6 (30.5) |
| Total observations | 224,026 | 93,295 | 114,578 | 83,000 | 108,167 | 53,671 | 53,397 |
| Total unique | 46,264 | 24,202 | 16,362 | 14,662 | 11,484 | 11,464 | 11,444 |
| I/σ(I) | 23.3 (3.4) | 19.6 (3.1) | 43.9 (6.8) | 29.6 (4.5) | 42.9 (3.9) | 31.8 (2.9) | 30.9 (3.1) |
| Completeness (%) | 99.9 (100) | 98.1 (82.7) | 98.6 (88.4) | 98.8 (89.9) | 85.4 (48.8) | 82.8 (37.8) | 82.0 (39.1) |
| Multiplicity | 4.8 | 3.9 | 7.0 | 5.7 | 11.0 | 5.7 | 5.7 |
| Wilson B factor (Å2) | 27.6 | 46.8 | 36.3 | 38.6 | |||
| Atoms in refinement | 4,116 | 4,112 | 2,118 | 2,041 | |||
| Rwork | 21.5 | 20.6 | 21.2 | 22.8 | |||
| Rfree | 25.4 | 25.8 | 26.0 | 27.1 | |||
| Waters and ligand | 213 | 129 | 122 | 77 | |||
| MPD | 4 | 2 | 0 | 0 | |||
| Average B factor, protein (Å2) | 41.3 | 48.0 | 45.2 | 56.2 | |||
| Average B factor, solvent (Å2) | 34.6 | 37.0 | 38.6 | 39.3 | |||
| Mean positional error (Å) | 0.166 | 0.401 | 0.261 | 0.307 | |||
| r.m.s.d., bond length (Å) | 0.014 | 0.010 | 0.010 | 0.009 | |||
| r.m.s.d., bond angle (°) | 1.45 | 1.24 | 1.19 | 1.21 |
Structure Solution and Refinement
The structure of Form III was solved using multiwavelength anomalous diffraction phasing techniques. All three expected selenium sites were located using the program SHELX (27). The program SHARP (28) was used to further refine the heavy atom substructure and for density modification. The figure of merit was 0.48 after substructure refinement, with a subsequent increase to 0.75 after density modification. The program ARP/wARP (29) was utilized to generate an initial model for the TP0453 structure, comprising 141 of the 260 residues (54% total). At this stage 10 β-strands and 6 α-helices were identifiable. The remaining residues were built manually by performing iterative cycles of refinement and model building using the programs REFMAC (30) and COOT (31), respectively. The structures of Forms I, II, and IV were solved by molecular replacement using the program PHASER (32) and the structure of Form III as the search model. Iterative cycles of model building using COOT and refinement using REFMAC5 were performed to generate the final models for Forms I, II, and IV. Water molecules were added, and TLS refinement (33) was carried out on each crystal form in the final rounds of REFMAC5 refinement. Final refinement statistics are summarized in Table 1. All figures were made using PyMOL including hydrophobic (VASCo plugin) (34). The PISA program (35) was used to calculate the possible oligomeric assembly of all four crystal forms.
Expression and Purification of OmpF
OmpF was expressed in the BL21 (DE3) Rosetta-gami strain (Agilent Technologies). For OmpF purification, a 1-liter culture of LB was inoculated with 25 ml of overnight culture grown of (pET23b:ompF) at 37 °C. At an A600 nm of 0.6, isopropyl β-d-1-thiogalactopyranoside was added to the culture to a final concentration of 0.3 mm. Cells were grown for an additional 3 h and then harvested by centrifugation at 6000 × g for 30 min at 4 °C. The pellets were resuspended with 20 ml of 50 mm Tris (pH 7.5), 100 mg of lysozyme, and 100 ml of protease inhibitor mixture, and the bacterial suspension was lysed by sonication. The pellet was recovered by centrifugation at 20,000 × g for 30 min at 4 °C and then incubated in solubilization buffer (50 mm Tris (pH 7.0), 8 m urea) for 30 min at 4 °C; the remaining insoluble material was removed by centrifugation at 20,000 × g for 30 min at 4 °C. The supernatant was added to a Q-Sepharose column equilibrated with solubilization buffer and was washed with wash buffer (50 mm Tris (pH 7.0), 8 m urea). Bound protein was eluted off the column using a linear gradient of NaCl from 0 to 0.5 m. Purified protein was incubated in folding buffer (0.5% n-dodecyl-β-d-glucopyranoside, 0.5% n-dodecyl-β-d-maltoside, 100 mm NaCl, 50 mm Tris) for 24 h at 4 °C to ensure complete folding of the protein. The samples were centrifuged at 20,000 × g for 30 min at 4 °C to remove misfolded aggregates.
Preparation of Liposomes
All phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL). Large LUVs were generated using an Avanti Mini-Extruder. A mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-l-serine] (sodium salt), 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-(racemic)-glycerol] (sodium salt), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (69.3:17:13:0.7 mol %, respectively) in chloroform was dried at room temperature under argon and kept in vacuum for at least 4 h. This ratio simulates the phospholipid composition of the T. pallidum OM (7). To hydrate the lipid mixture, 0.5 ml of 50 mm HEPES (pH 7.5), 100 mm NaCl was added to the dried phospholipids, and the sample was incubated for 30 min at 37 °C. The lipids were resuspended by vortexing and then passed 21 times at 23 °C through the extruder equipped with a 100-nm pore size polycarbonate filter. The filter dictates the final size of the vesicles (mean diameter ∼110 nm) and reduces the chances of contamination with larger particles or foreign material. The resulting liposomes were stored at 4 °C for 2 weeks. Liposomes containing Tb(DPA)33− were prepared as above except that buffer A, including 3 mm terbium(III) chloride (TbCl3) and 9 mm 2,6-pyridinedicarboxylic acid (Sigma) neutralized to pH 7 was added to the lipid film. The suspended phospholipid mixtures were frozen in liquid N2 and thawed at 37 °C a total of 6 times to reduce the number of multilamellar liposomes and to enhance the trapped volumes of the vesicles. Loaded liposomes were separated from non-encapsulated Tb(DPA)33− by gel filtration using Superdex TM 75 (GE Healthcare) in 50 mm HEPES (pH 7.5), 100 mm NaCl.
Circular Dichroism Spectroscopy
CD measurements were carried out using a Jasco J-715 spectropolarimeter (Jasco, Easton, MD), and the results were expressed as relative ellipticity. The CD spectra were obtained at protein concentrations of 10 μm in 50 mm Tris (pH 7.5), 100 mm NaCl in a 1-mm cell at 25 °C. The values obtained were normalized by subtracting the base line recorded for the buffer having the same concentration of NaCl under similar conditions. Deconvolution of the CD data was performed using the DICHROWEB server (36).
Immunologic Reagents
Rat polyclonal antiserum directed against TP0453 was described previously (18). A mouse monoclonal antibody (hybridoma clone HIS-1) specific for polyhistidine tags was purchased from Sigma.
SDS-PAGE and Immunoblot Analysis
Samples were resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes (0.45 μm pore size, GE Healthcare) at 25 V for 30 min using a semidry apparatus (Bio-Rad). Membranes were blocked for 1 h with PBS, 5% nonfat dry milk, 5% fetal bovine serum, and 0.1% Tween 20 and probed overnight at 4 °C with primary antibodies at dilutions of 1:3000 (anti-TP0453) or 1:1000 (anti-His tag). After washing with PBS and 0.05% Tween 20 (PBST), the membranes were incubated for 1 h at 4 °C with horseradish peroxidase-conjugated goat anti-rat or anti-mouse antibodies (Southern Biotech, Birmingham, AL) at a dilution of 1:30,000. After additional washes with PBST, the immunoblots were developed using the SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific).
Triton X-114 Phase Partitioning
10 μg of each protein were incubated in 250 μl of PBS containing 2% Triton X-114 (Sigma) and 0.005% protease inhibitor mixture (Sigma) and diluted with 990 μl of PBS. After incubation for 1 h at 4 °C, each sample was dialyzed overnight against PBS at 4 °C. After dialysis, the samples were incubated at 37 °C for 10 min followed by centrifugation at 20,000 × g for 20 min at 4 °C. After centrifugation, the aqueous and detergent phases were separated, and both phases were washed 5 times. All washed samples were precipitated with 10 volumes of acetone overnight at −80 °C for subsequent SDS-PAGE and immunoblot analysis.
Liposome Floatation Assay
WT and mutant proteins (300 nm final concentration) were incubated with ∼100 μg of LUVs for 1 h at 22 °C in low (50 mm Tris (pH 7.5), 100 mm NaCl) and high salt (50 mm Tris (pH 7.5), 1.0 m NaCl) buffers. Incorporation was terminated by adding sucrose to a final concentration of 50%, and the mixtures were transferred to an ultracentrifuge tube. Discontinuous sucrose gradients were made by gently adding layers of 40 and 6% sucrose to the ultracentrifuge tubes. After centrifugation at 90,000 × g for 1 h at 4 °C, the liposome (top layer)- and non-liposome (middle and bottom layers)-containing fractions were carefully removed.
Pore Formation Assay
For end-point measurements, liposomes loaded with Tb(DPA)33− were diluted to a final lipid concentration of 100 μm at the different pH values using buffer 50 mm Tris (pH 7.5), 100 mm NaCl supplemented with 5 mm EDTA. The net initial emission intensity (F0) was determined after equilibration of the sample at 25 °C for 5 min. Aliquots of TP0453 or OmpF were added to the liposome suspension at 100 nm final concentration, and samples were incubated for 30 min at 37 °C. After re-equilibration to 25 °C, the final net emission intensity (Ff) of the sample was determined (i.e. after blank subtraction and dilution correction) and the fraction of Tb(DPA)33− quenched was estimated using Ff/F0. For kinetic measurements, the same procedure was used with the difference that Ff was monitored continuously right after TP0453 or OmpF addition at 25 °C, and Ff/F0 was plotted as a function of time.
Bis-ANS Binding Kinetics
1 μm concentrations of each sample in 50 mm Tris (pH 7.5), 50 mm NaCl was stirred in a 1-cm path length cuvette. Subsequently, graded aliquots (1–25 μm final concentration) of bis-ANS (Sigma) were added to the cuvette, and the excitation wavelengths were set to 390 nm. The emission spectra were collected from 400 to 600 nm in a Hitachi F-2500 fluorescence spectrophotometer. For each measurement, the fluorescence intensity was corrected by subtracting the fluorescence of the sample containing only bis-ANS. The data were plotted against the total concentration of bis-ANS. The apparent Kd was estimated using the equation,
For binding analyses, TP0453 (1 or 5 μm) was incubated in 50 mm Tris (pH 7.5), 50 mm NaCl in the presence of 5 μm bis-ANS for 20 min at 22 °C. Emission spectra were collected from 400 to 600 nm; net fluorescence intensities were corrected by subtracting the fluorescence of the sample containing only bis-ANS.
Analytical Gel Filtration
Gel filtration experiments were carried out using a SuperdexTM 75 column on a Bio-Rad FPLC system. The column was calibrated using molecular weight standard markers (GE Healthcare). All experiments were carried out using 50 mm Tris (pH 7.5), 50 mm NaCl. Typically, 500 μl of sample was loaded on the column and run at 25 °C at a flow rate of 0.3 ml/min, with detection at 280 nm. The relative elution volume (KAV) was calculated based on the equation,
where Ve is the elution volume, Vo is the void volume determined by elution of blue dextran 2000 kDa, and Vg is the geometric column volume.
Glutaraldehyde Cross-linking
The cross-linking of protein samples was carried out in the presence of 1% glutaraldehyde in low (50 mm Tris (pH 7.5), 100 mm NaCl) and high salt (50 mm Tris (pH7.5), 1 m NaCl) conditions. WT, α2I62E, and α8V249E were used at a concentration of 1 μm. The molecular masses of the cross-linked products were determined by 10% SDS-PAGE and subsequent immunoblot analysis.
BN-PAGE of Native TP0453
Freshly harvested treponemes were solubilized overnight at 4 °C with 50 mm Tris (pH 7.0), 2% n-dodecyl β-d-maltoside (Calbiochem), and 5% protease inhibitor mixture. Before electrophoresis, lysates were cleared of detergent-insoluble material by centrifugation at 20,000 × g for 20 min at 4 °C. Native lysates consisting of 2.0 × 108 spirochetes were resolved in a 4–12% Bis-Tris acrylamide gel (Bio-Rad) at 4 °C using the BN-PAGE method (37, 38). The cathode buffer (50 mm Tricine (pH 7.0) and 15 mm Bis-Tris) contained 0.02% Coomassie Brilliant Blue G-250 for the first one-third of the run, after which the gel was run with fresh cathode buffer without Coomassie Brilliant Blue G-250. For the duration of the run the anode buffer consisted of 50 mm Bis-Tris (pH 7.0). Resolved lysates were transferred to a nitrocellulose membrane in 50 mm Tricine (pH 7.0) followed by immunoblotting using TP0453 antiserum.
RESULTS
TP0453 Exists in Closed and Open Conformations
As noted earlier, we previously proposed that the polypeptide moiety of TP0453 exists in membrane-integrated and -unintegrated forms (18). To obtain structures representing these hypothesized different states, we performed crystallization experiments using concentrations of β-OG ranging from 0 to 1.0%. No crystals were obtained in the absence of detergent. Two forms (I and II) were obtained using 0.1–0.5% β-OG; two additional forms (III and IV, respectively) were obtained in 0.5–0.75% and 0.75–1.0% detergent. The structure of Form III was solved by multiwavelength anomalous diffraction and refined to a resolution of 2.2 Å with R and Rfree values of 21.2 and 26%, respectively. The structures of Forms I, II, and IV were solved by molecular replacement using Form III as a search model (see “Experimental Procedures”). The most relevant crystal parameters and refinement statistics for all crystal forms are summarized in Table 1. The final refined structures of Forms I and II contain two molecules in an asymmetric unit, whereas Forms III and IV have only one.
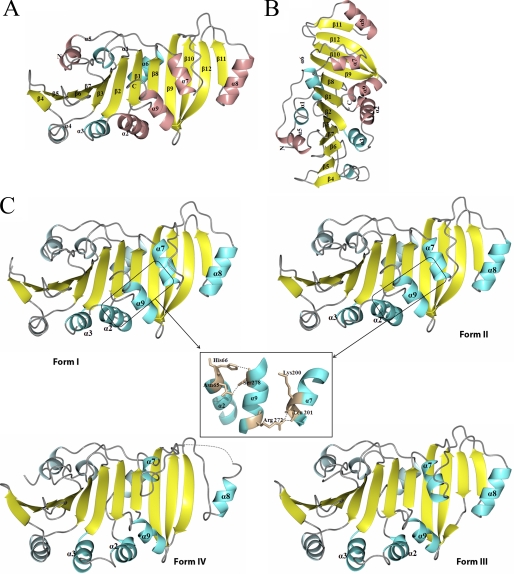
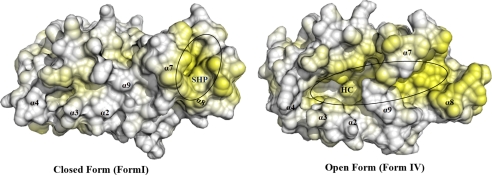
As shown in Fig. 1, A and B, the TP0453 structure has a globular shape with approximate dimensions of 64 × 44 × 44 Å. The overall structure of TP0453 consists of a typical α/β/α-fold consisting of a 12-stranded anti-parallel β-sheet (β1–12) sandwiched between a total of 9 α helices (α-1 to α-9). The β-sheet (strand order 11–12-10–9-8–1-2–3-7–6-5–4) forms a U-shaped β-half barrel in which strands 9, 10, and 12 are twisted in relation to the other 9 strands. Six of the nine α-helices (α2–4 and α7–9) form a lid-like structure overlying the concave face of the β-sheet, whereas three α helices (α1, α5, and α6) overlie the convex face. α2, α5, and α7–9 correspond to the AHs predicted in our previous report (18). The root mean square deviations between all Cα atoms in the four forms, depicted in Fig. 1 C, were calculated by the LSQKAB program (39) (supplemental Table 2). For comparison purposes, only molecules 1 of Forms I and II were utilized. Forms I and II are highly similar with only small deviations in the positions of α2 and α3. In contrast, a marked structural change occurs between Forms I/II and III; two groups of helices (α2/α3/α9 and α7/α8) move laterally away from each other, exposing the β-sheet, which forms the floor of a large hydrophobic cavity (calculated volume of ∼3549 Å3, Figs. 1C and 2). The transition from the closed (I/II) to open (III) form primarily involves breakage of hydrogen bonds between α7 (Lys200, Leu201) and α9 (Arg272) and between α2 (Asn65, His66) and α9 (Ser278) (Fig. 1C, inset). The principal change between Forms III and IV involves α7 and α8, which move outwards and away from each other, widening the mouth of the cavity and increasing the exposure of β strands 9, 10, and 12 (Fig. 1C). Analysis of lipophilic potential by VASCo (34) reveals that the surfaces of the closed forms contain a horseshoe-shaped hydrophobic patch (designated SHP (surface hydrophobic patch), Fig. 2) comprised of a small portion of α7 and much of α8, which is disrupted by the opening of the molecule. A DALI search (40) failed to reveal any structural homologs in the databases.
FIGURE 1.
Crystal structures of TP0453. A, shown is the overall structure of TP0453 depicted as a ribbon model in which α helices are shown in cyan or pink, β strands are shown in yellow, and loops are shown in gray. Previously predicted AHs (18) are shown in pink. B, TP0453, shown in A, is rotated by 90° along the z axis. C, the different crystal forms of TP0453 obtained with increasing concentrations of β-OG are shown: Form I (0.1–0.25%), Form II (0.25–0.5%), Form III (0.5–0.75%), Form IV (0.75–1.0%). The helices that move are labeled and depicted in dark cyan. Dotted lines represent disordered regions in the crystal structures. The inset shows the hydrogen bonds which maintain the closed state but are disrupted in the open forms. The following residues are missing in the final refined structure: Form I (68 in molecule-1 and residues 63–69,104–108, and 119–120 in molecule-2), Form II (122 in molecule 1 and 119–120 in molecule 2); Form III (122), and Form IV (231–239). Otherwise, the electron densities throughout all of the polypeptide chains were high quality.
FIGURE 2.
Surface hydrophobicity of closed (I) and open (IV) forms of TP0453. The positions of the SHP and hydrophobic cavity (HC) are indicated; hydrophobic residues are highlighted in yellow.
Membrane Insertion of TP0453 Occurs via α7 and α8 and Involves Surface-accessible Residues
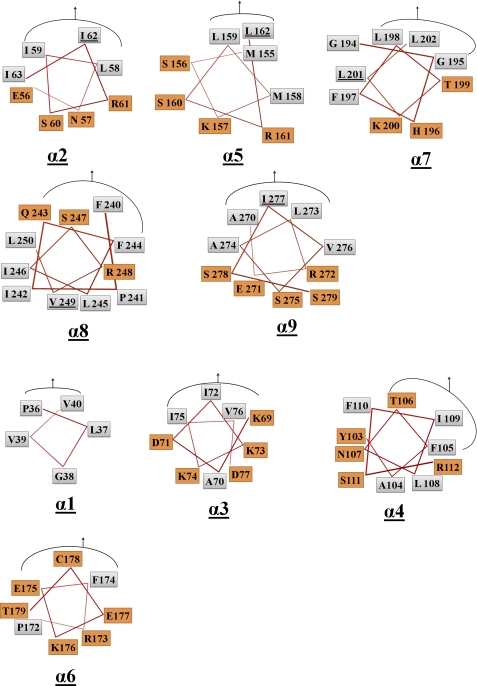
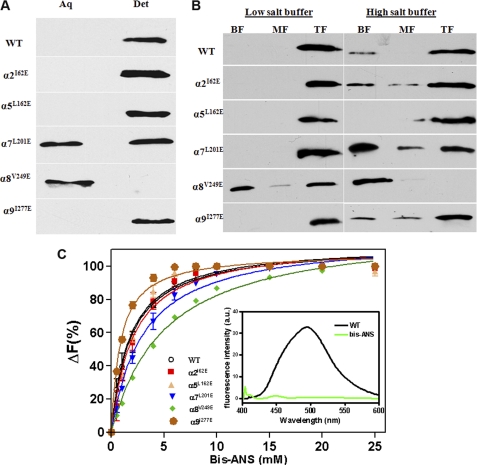
We next conducted experiments to identify the regions of TP0453 responsible for membrane integration, turning our attention to its AHs as potential membrane insertion elements. A defining feature of AHs is that their hydrophobic and polar residues segregate to opposite faces of the helix; upon insertion into a lipid bilayer, the hydrophobic face adopts an orientation parallel to the membrane (41, 42). A helical wheel analysis using PyMOL revealed that α2, α5, and α7–9 are the only AHs in TP0453 (Fig. 3). Inspection of the TP0453 closed structures reveals that the hydrophobic faces of α2, α5, α9 are oriented toward the β sheet, whereas as noted earlier, the hydrophobic faces of both α7 and α8 contribute to varying extents to the SHP (Figs. 2 and 3). α7 and α8, therefore, appear to be the AHs most likely to be involved in membrane insertion. To examine this prediction, we generated mutant proteins in which a residue on the hydrophobic face of each AH was replaced by a glutamic acid (Fig. 3 and supplemental Table 1). No significant differences in secondary structure were observed between WT and mutants as determined by far-UV CD spectroscopy (supplemental Fig. 1). We then used Triton X-114 phase partitioning to compare the amphiphilic properties of WT and mutants. As shown in Fig. 4A, WT and three of the mutants (α2I62E, α5L162E, and α9I277E) partitioned exclusively into the detergent-enriched phase; two of the mutants (α8V249E and α7L201E), however, showed marked deviations in phase partitioning behavior. α7L201E partitioned into both aqueous and detergent phases, whereas α8V249E partitioned exclusively into the aqueous phase.
FIGURE 3.
Identification of AHs in TP0453. PyMOL was used to construct helical wheels for all nine AHs based on the closed structures in Fig. 1. For each helical wheel, the hydrophobic and polar amino acids are shown in gray and orange boxes, respectively; the curved line plus arrow denotes the residues facing toward the β sheet. The five AHs (α2, α5, α7–9) indentified by this and our previous analysis are grouped together and correspond to the pink helices in figure 1. Residues selected for mutagenesis are underlined.
FIGURE 4.
α7 and α8 are required for membrane insertion of TP0453. A, Triton X-114 phase partitioning of WT and mutants is shown. Samples were subjected to SDS-PAGE followed by immunoblotting with monospecific antiserum directed against TP0453. aq, aqueous; det, detergent. B, incorporation of WT and mutant proteins into LUVs is shown. Lanes, bottom (BF), middle (MF) and top fractions (TF) from sucrose gradients were analyzed by SDS-PAGE and immunoblotting. C, binding curves of WT and mutants with bis-ANS show the relative changes in fluorescence intensity at 490 nm plotted against bis-ANS concentration. The inset represents a fluorescence emission spectrum of WT (1 μm) or buffer alone in the presence of 1 μm bis-ANS.
We extended these results using a liposome floatation assay. WT and mutant proteins were incubated with LUVs followed by discontinuous sucrose gradient centrifugation to separate membrane-associated (top fraction) from unassociated material (middle and bottom fractions); incubation under low and high salt conditions was performed to distinguish electrostatic interactions from those involving membrane integration (43). As shown in Fig. 4B, WT was predominantly liposome-associated under high salt, as were the same three mutants (α2I62E, α5L162E, and α9I277E) that partitioned into the Triton X-114-enriched phase. The floatation results for α7L201E and α8V249E in high salt mirrored their aberrant phase-partitioning behaviors; a substantial amount of α7L201E mutant was unassociated with vesicles, whereas H8V249E was recovered exclusively in the bottom fraction.
Binding of bis-ANS has been used extensively to probe proteins for surface-accessible hydrophobic regions (44, 45). In aqueous buffer this compound is minimally fluorescent with an emission maximum at 525 nm but upon binding to a hydrophobic surface undergoes a marked increase in fluorescence intensity with a blue shift to 490 nm. Fig. 4C shows the binding curves for WT and mutants with increasing concentrations of bis-ANS; corresponding dissociation constants are presented in Table 2. The collective results show that bis-ANS bound well to TP0453, α2I62E, α5L162E, α9I277E, and α7L201E but only weakly to α8V249E.
TABLE 2.
Bis-ANS dissociation constants (Kd) values of TP0453 and mutants
| Constructs | Kd value |
|---|---|
| μm | |
| WT | 1.792 ± 0.1939 |
| α2I62E | 2.043 ± 0.2489 |
| α5L162E | 1.723 ± 0.1746 |
| α7L201E | 2.985 ±.0.2526 |
| α8V249E | 5.047 ± 0.2486 |
| α9I277E | 0.8581 ± 0.04994 |
TP0453 Increases Membrane Permeability at Acidic pH
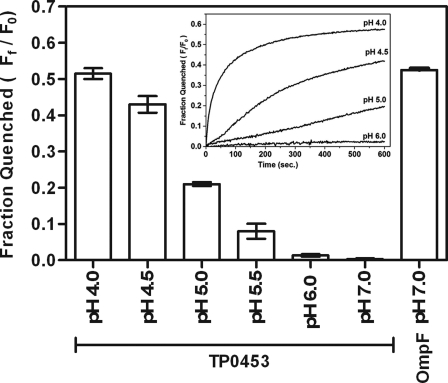
We previously demonstrated that the displacement of phospholipids after the incorporation of TP0453 into small unilamellar vesicles increases membrane permeability as measured by calcein efflux assay (18). The findings herein that only two of the protein five AHs are involved in membrane insertion prompted us to re-examine the polypeptide capacity to alter membrane permeability using a more stringent methodology involving LUVs and the fluorophore Tb(DPA)33− (46). As shown in Fig. 5, TP0453 did increase efflux of fluorophore but only at acidic pH, whereas the conventional porin, E. coli OmpF, increased efflux at pH 7.0.
FIGURE 5.
TP0453 increases efflux of fluorophore in LUVs at acidic pH. The graphs show pH-dependent quenching of Tb(DPA)33− from the encapsulated LUVs, after incubation with TP0453. Pore formation at pH 7.0 by E. coli OmpF was used as a positive control. The fractions quenched were determined as described under “Experimental Procedures.” The inset represents initial kinetic profiles for quenching of Tb(DPA)33− from the encapsulated LUVs at different pH values.
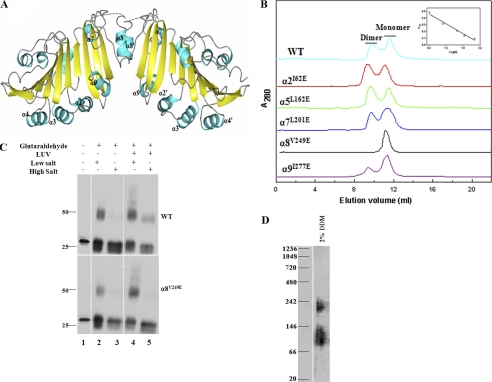
α8 Contributes to Formation of a Membrane-inserted Dimer
Examination of the crystal lattice using the PISA program (35) indicated that Forms II, III, and IV weakly dimerize via α8 (Fig. 6A and supplemental Table 3). Initial experiments were conducted using size exclusion chromatography to determine whether TP0453 can form a dimer in solution. As shown in Fig. 6B, WT eluted in low salt buffer as a mixture of monomer and dimer. α2I62E, α5L162E, α9I277E, and αL201E exhibited virtually identical elution profiles, whereas α8V249E eluted exclusively as a monomer. These findings led us to assess whether membrane-integrated TP0453 forms dimers and whether α8 contributes to dimer formation as well as membrane insertion. WT, α2I62E, and α8V249E were incubated in the absence and presence of LUVs under low and high salt conditions, as described for the experiments in Fig. 4C, followed by cross-linking with glutaraldehyde; the samples examined by SDS-PAGE and immunoblotting. The upper and lower panels of Fig. 6C show the results obtained, respectively, with WT and α8V249E (not shown are the results obtained with α2I62E, which were identical to WT). In the absence of both liposomes and glutaraldehyde, WT and α8V249E migrated as monomers (lanes 1). Cross-linking of both WT and α8V249E without liposomes yielded dimers (lanes 2) that were dissociated by high salt (lanes 3). However, when incubated with liposomes, cross-linked WT, but not α8V249E, formed a dimer that was stably liposome-associated in high salt (lanes 4 and 5).
FIGURE 6.
α8 is required for dimerization as well as membrane insertion of TP0453. A, shown is the dimerization mode of TP0453 predicted by the PISA server. Monomers in TP0453 assemble side-by-side along an interface established by α8. B, shown are elution profiles of WT and mutant proteins on a SuperdexTM 75 HR column in 50 mm Tris (pH 7.5), 100 mm NaCl at 25 °C. The inset represents the calibration curve of the column. C, WT, α2I62E, and α8V249E were incubated in the absence and presence of LUVs under low and high salt conditions followed by glutaraldehyde cross-linking and SDS-PAGE/immunoblot analysis. Not shown are the results for α2I62E, which were identical to WT. D, shown is BN-PAGE and immunoblot analysis of T. pallidum lysates (2 × 108 organisms) solubilized in 2% n-dodecyl β-d-maltoside with 25 mm Tris (pH 7.0).
Last, we used BN-PAGE and immunoblot analysis to assess the relevance of these results for native TP0453. As shown in Fig. 6D, TP0453 in T. pallidum lysate migrated predominantly with a calculated molecular mass ranging from 80 to 130 kDa, a result consistent with monomer-dimer equilibrium after correcting for the approximate 50-kDa size of the n-dodecyl β-d-maltoside micelles (47).
DISCUSSION
In recent years an increasing number of proteins have been reported that utilize AHs as membrane anchors (48–51). The term amphitropic was coined to describe proteins whose AHs are unstructured in an aqueous milieu but adopt a helical structure upon integration into a membrane (42, 52). Previously, we postulated that the polypeptide moiety of the T. pallidum OM-associated lipoprotein TP0453 is amphitropic and contains five AHs that mediate reversible membrane insertion (18). Furthermore, based on our finding that membrane integration of the TP0453 polypeptide increases bilayer permeability, we hypothesized that TP0453 represents a new class of pore-forming molecule. To examine these predictions, we solved the crystal structure of non-lipidated TP0453 using graded concentrations of detergent to simulate the putative membrane-uninserted and -inserted forms of the polypeptide. The structural analysis verified the predicted AHs (α2, α5 α7–9) but also revealed that these domains are stably folded. Rather than converting between non-structured and structured forms, four AHs (α2, α7–9) were found to move during the closed to open conformational change observed with increasing concentrations of surfactant. These structural data enabled us to devise an experimental strategy to relate the membrane-interactive properties of individual AHs to the conformational changes, a key step toward elucidating the functionality of this unusual lipoprotein.
The AHs in the closed state of TP0453 exhibit two basic orientations. The hydrophobic faces of α2, α5, and α9 are oriented entirely toward the β sheet (Fig. 3), whereas portions of the hydrophobic faces of α7 and α8 face away from the β-sheet and are exposed to the aqueous milieu. The distinctive orientation of α7 and α8 suggested them as the likely membrane insertion elements. We confirmed this supposition by examining the Triton X-114 phase-partitioning behaviors and membrane interactions of mutants in which the hydrophobic faces of individual AHs were disrupted. Results obtained by probing the surfaces of WT and mutant proteins with the planar fluorescent dye bis-ANS further revealed that α8 likely functions as part of a membrane sensor that initiates the conformational changes required for membrane integration. Interestingly, we found that the same residue of α8 (Val249) critical for membrane sensing/insertion is also essential for dimerization. An obvious question is whether this one helix can serve as a membrane insertion element and dimeric interface. Analysis of accessible surface area using Naccess (39) reveals that α8 does in fact contain sufficient hydrophobic surface to function in both capacities (supplemental Fig. 2). The demonstration of a putative native dimer in T. pallidum supports the biologic relevance of these results.
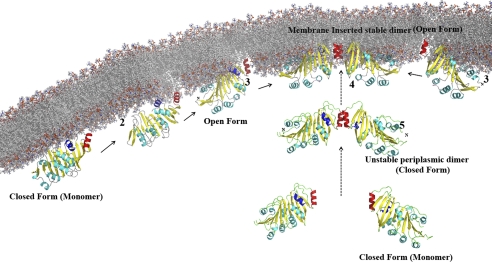
On the basis of data presented here and elsewhere (18), it is possible to propose a scenario for membrane insertion and dimerization by the TP0453 polypeptide (Fig. 7). Initial electrostatic interactions between the charged surface of the protein and phospholipids head groups of the periplasmic leaflet would position the SHP so that it can sense transient gaps that expose the hydrophobic interior of the bilayer. In this regard it is noteworthy that TP0453 integrates more efficiently into liposomes that contain the negatively charged phospholipid, phosphatidylserine (18), a constituent of the T. pallidum OM (7). The energy released from the resulting hydrophobic interactions between the SHP and acyl chains within the membrane would overcome the hydrogen bonds that maintain the closed conformation, freeing α7 and α8 for insertion. Dimerization would then occur between monomers free to diffuse laterally within the lipid bilayer. It is also possible that dimerization either proceeds or occurs in parallel with membrane insertion. Significantly, the above scheme can be accommodated by our current conception of the spirochete machinery for OM localization of lipoproteins. T. pallidum, like Gram-negative bacteria (53), contains a Lol system for chaperoning lipoproteins across the periplasm (8, 18). Unlike cytoplasmic membrane-associated lipoproteins (e.g. TpN47) (5, 13), which presumably have Lol avoidance signals (53), newly exported lipidated TP0453 would be engaged by the Lol transporter (LolC, -D, and -E) (54) and subsequently transferred to the periplasmic chaperone LolA (53). T. pallidum does not contain an ortholog for LolB, the OM-associated lipoprotein in Gram-negative bacteria that accepts nascent lipoproteins from LolA and inserts them into the OM inner leaflet (53). The ability of the TP0453 polypeptide to spontaneously insert into membranes would, however, obviate the need for the terminal component of the Lol pathway. Lipoproteins bind to the hydrophobic cavity of LolA via their N termini; the membrane-interactive elements of TP0453 bound to LolA would, therefore, be freely accessible for interaction with the periplasmic leaflet of the OM. Release of the N terminus from LolA after integration of the polypeptide would liberate the lipid moiety for insertion into the OM. Two aspects of the OM topology of TP0453 warrant emphasis (Fig. 7). First, the hydrophobic cavity of a monomer anchored to the inner leaflet by α7 and α8 would open toward the periplasmic space. Second, the positive (convex) curvature of the dimer would “match” the concavity of the OM periplasmic face.
FIGURE 7.
Proposed sequence for membrane insertion and dimerization of the TP0453 polypeptide. 1, shown are initial electrostatic interactions between the closed form and phospholipids head groups position α7 and α8 proximal to the membrane. 2, α7 and α8 sense transient gaps within the lipid bilayer. 3, α7 and α8 insert into the lipid bilayer, and the molecule adopts the open conformation. 4, adjacent membrane-inserted-monomers dimerize via α8. 5, alternatively, monomers could dimerize before membrane sensing/insertion. In each stage, α7 and α8 are shown in blue and red, respectively.
Our earlier proposal that TP0453 functions as a pore-forming molecule was supported by results from a calcein efflux assay using small unilamellar vesicles (18). Our refined model of membrane insertion and topology prompted us to reinvestigate the molecule pore-forming properties with an alternative methodology employing LUVs, which are much less sensitive to minor perturbations (55, 56). The data obtained here demonstrate that under the physiological conditions in which membrane insertion occurs, increase in membrane permeability is not observed. Given that T. pallidum is an extracellular organism that is rapidly degraded within phagolysosomes (57, 58), the enhanced efflux of fluorophore observed at acidic pH is unlikely to be physiologically relevant.
Our search for an alternative function of TP0453 called our attention to the recently solved crystal structures of LprG and LppX, lipoproteins that translocate complex lipids in M. tuberculosis (20, 21). Both LprG and LppX contain α helices that move to accommodate large, amphipathic ligands within hydrophobic cavities delimited by β sheet (supplemental Fig. 3). The DALI server failed to identify these two lipoproteins as structural homologs because TP0453 possesses a different β-strand order and additional α-helices. Nevertheless, the structural similarities between these three molecules, readily evident upon inspection (supplemental Fig. 3), suggest that TP0453 might be a carrier of lipids, glycolipids, and/or derivatives during OM biogenesis in T. pallidum. TP0453 in the closed form would be unable to bind ligand during export; binding could occur, however, once the lipoprotein localizes to the OM and assumes its open conformation.
Acknowledgments
This work was based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation under National Science Foundation Award DMR-0225180 using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by National Center for Research Resources, National Institutes of Health Award RR-01646. Portions of this research were also carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Laboratory Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the NIGMS. We acknowledge the excellent technical assistance of Morgan LeDoyt and thank Star Dunham-Ems and V. Kuruthihalli (University of Connecticut Health Center) for valuable discussions.
This work was supported by Public Health Service Grants AI-26756 (to J. D. R.) and RG-71111-N (to A. P. H.). This work was also supported, in whole or in part, by National Institutes of Health Grants GM074899, GM094611, and GM094597 (NIGMS Protein Structure Initiative to the High throughput screening laboratory at the Hauptman Woodward Institute).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–2 and Figs. 1–3.
The atomic coordinates and structure factors (codes 3K8G, 3K8H, 3K8J, and 3K8I) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- OM
- outer membrane
- TP
- T. pallidum
- AHs
- amphipathic helices
- LUV
- large unilamellar vesicle
- SHP
- surface hydrophobic patch
- ANS
- anilino-8-naphthalenesulfonate
- BN-PAGE
- blue native PAGE
- β-OG
- n-octyl-β-d-glucopyranoside
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Tb(DPA)33−
- terbium dipicolinic acid complex.
REFERENCES
- 1. Lafond R. E., Lukehart S. A. (2006) Clin. Microbiol. Rev. 19, 29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cox D. L. (1994) Methods Enzymol. 236, 390–405 [DOI] [PubMed] [Google Scholar]
- 3. Radolf J. D., Hazlett K. R. O., Lukehart S. A. (2006) in Pathogenic Treponemes: Cellular and Molecular Biology (Radolf J. D., Lukehart S. A. eds) pp. 197–236, Caister Academic Press, Norfolk, UK [Google Scholar]
- 4. Radolf J. D. (1995) Mol. Microbiol. 16, 1067–1073 [DOI] [PubMed] [Google Scholar]
- 5. Cameron C. E. (2006) in Pathogenic Treponemmes: Cellular and Molecular Biology (Radolf J. D., Lukehart S. A. eds) pp. 237–266, Caister Academic Press, Norfolk, UK [Google Scholar]
- 6. Belisle J. T., Brandt M. E., Radolf J. D., Norgard M. V. (1994) J. Bacteriol. 176, 2151–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Radolf J. D., Robinson E. J., Bourell K. W., Akins D. R., Porcella S. F., Weigel L. M., Jones J. D., Norgard M. V. (1995) Infect. Immun. 63, 4244–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fraser C. M., Norris S. J., Weinstock G. M., White O., Sutton G. G., Dodson R., Gwinn M., Hickey E. K., Clayton R., Ketchum K. A., Sodergren E., Hardham J. M., McLeod M. P., Salzberg S., Peterson J., Khalak H., Richardson D., Howell J. K., Chidambaram M., Utterback T., McDonald L., Artiach P., Bowman C., Cotton M. D., Fujii C., Garland S., Hatch B., Horst K., Roberts K., Sandusky M., Weidman J., Smith H. O., Venter J. C. (1998) Science 281, 375–388 [DOI] [PubMed] [Google Scholar]
- 9. Cox D. L., Radolf J. D. (2001) Microbiology 147, 1161–1169 [DOI] [PubMed] [Google Scholar]
- 10. Walker E. M., Zampighi G. A., Blanco D. R., Miller J. N., Lovett M. A. (1989) J. Bacteriol. 171, 5005–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Desrosiers D. C., Anand A., Luthra A., Dunham-Ems S. M., LeDoyt M., Cummings M. A., Eshghi A., Cameron C. E., Cruz A. R., Salazar J. C., Caimano M. J., Radolf J. D. (2011) Mol. Microbiol. 80, 1496–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu J., Howell J. K., Bradley S. D., Zheng Y., Zhou Z. H., Norris S. J. (2010) J. Mol. Biol. 403, 546–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cox D. L., Luthra A., Dunham-Ems S., Desrosiers D. C., Salazar J. C., Caimano M. J., Radolf J. D. (2010) Infect. Immun. 78, 5178–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Narita S., Matsuyama S., Tokuda H. (2004) Arch. Microbiol. 182, 1–6 [DOI] [PubMed] [Google Scholar]
- 15. Brunner J. (1996) Trends Cell Biol. 6, 154–157 [DOI] [PubMed] [Google Scholar]
- 16. Shevchenko D. V., Sellati T. J., Cox D. L., Shevchenko O. V., Robinson E. J., Radolf J. D. (1999) Infect. Immun. 67, 2266–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brusca J. S., Radolf J. D. (1994) Methods Enzymol. 228, 182–193 [DOI] [PubMed] [Google Scholar]
- 18. Hazlett K. R., Cox D. L., Decaffmeyer M., Bennett M. P., Desrosiers D. C., La Vake C. J., La Vake M. E., Bourell K. W., Robinson E. J., Brasseur R., Radolf J. D. (2005) J. Bacteriol. 187, 6499–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones J. D., Bourell K. W., Norgard M. V., Radolf J. D. (1995) Infect. Immun. 63, 2424–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sulzenbacher G., Canaan S., Bordat Y., Neyrolles O., Stadthagen G., Roig-Zamboni V., Rauzier J., Maurin D., Laval F., Daffé M., Cambillau C., Gicquel B., Bourne Y., Jackson M. (2006) EMBO J. 25, 1436–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drage M. G., Tsai H. C., Pecora N. D., Cheng T. Y., Arida A. R., Shukla S., Rojas R. E., Seshadri C., Moody D. B., Boom W. H., Sacchettini J. C., Harding C. V. (2010) Nat. Struct. Mol. Biol. 17, 1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003) Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luft J. R., Collins R. J., Fehrman N. A., Lauricella A. M., Veatch C. K., DeTitta G. T. (2003) J. Struct. Biol. 142, 170–179 [DOI] [PubMed] [Google Scholar]
- 24. Koszelak-Rosenblum M., Krol A., Mozumdar N., Wunsch K., Ferin A., Cook E., Veatch C. K., Nagel R., Luft J. R., Detitta G. T., Malkowski M. G. (2009) Protein Sci. 18, 1828–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soltis S. M., Cohen A. E., Deacon A., Eriksson T., González A., McPhillips S., Chui H., Dunten P., Hollenbeck M., Mathews I., Miller M., Moorhead P., Phizackerley R. P., Smith C., Song J., van dem Bedem H., Ellis P., Kuhn P., McPhillips T., Sauter N., Sharp K., Tsyba I., Wolf G. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 1210–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Otwinowski Z., Minor W. (1997) in Methods in Enzymology (Carter C. W., Jr. ed) pp. 307–326, Academic Press, Inc., New York: [DOI] [PubMed] [Google Scholar]
- 27. Sheldrick G. M. (2010) Acta Crystallogr D. Biol. Crystallogr. 66, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de La Fortelle E., Bricogne G. (1997) in Methods in Enzymology (Carter C. W., Jr. ed) pp. 472–494, Academic Press, Inc., New York: [DOI] [PubMed] [Google Scholar]
- 29. Morris R. J., Perrakis A., Lamzin V. S. (2003) Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 30. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Acta Crystallogr D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr D. Biol. Crystallogr 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 34. Steinkellner G., Rader R., Thallinger G. G., Kratky C., Gruber K. (2009) BMC Bioinformatics 10, 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu Q., Canutescu A. A., Wang G., Shapovalov M., Obradovic Z., Dunbrack R. L., Jr. (2008) J. Mol. Biol. 381, 487–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Whitmore L., Wallace B. A. (2004) Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schägger H., von Jagow G. (1991) Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 38. Wittig I., Braun H. P., Schägger H. (2006) Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 39. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Holm L., Sander C. (1995) Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 41. Hristova K., Wimley W. C., Mishra V. K., Anantharamiah G. M., Segrest J. P., White S. H. (1999) J. Mol. Biol. 290, 99–117 [DOI] [PubMed] [Google Scholar]
- 42. Cornell R. B., Taneva S. G. (2006) Curr. Protein Pept. Sci. 7, 539–552 [DOI] [PubMed] [Google Scholar]
- 43. Romano F. B., Rossi K. C., Savva C. G., Holzenburg A., Clerico E. M., Heuck A. P. (2011) Biochemistry 50, 7117–7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Farris F. J., Weber G., Chiang C. C., Paul I. C. (1978) J. Am. Chem. Soc. 100, 4469–4474 [Google Scholar]
- 45. Smoot A. L., Panda M., Brazil B. T., Buckle A. M., Fersht A. R., Horowitz P. M. (2001) Biochemistry 40, 4484–4492 [DOI] [PubMed] [Google Scholar]
- 46. Heuck A. P., Tweten R. K., Johnson A. E. (2003) J. Biol. Chem. 278, 31218–31225 [DOI] [PubMed] [Google Scholar]
- 47. Rosevear P., VanAken T., Baxter J., Ferguson-Miller S. (1980) Biochemistry 19, 4108–4115 [DOI] [PubMed] [Google Scholar]
- 48. Szeto T. H., Rowland S. L., Rothfield L. I., King G. F. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15693–15698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lenarcic R., Halbedel S., Visser L., Shaw M., Wu L. J., Errington J., Marenduzzo D., Hamoen L. W. (2009) EMBO J. 28, 2272–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marcia M., Ermler U., Peng G., Michel H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9625–9630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dasanayake D., Richaud M., Cyr N., Caballero-Franco C., Pittroff S., Finn R. M., Ausió J., Luo W., Donnenberg M. S., Jardim A. (2011) Mol. Microbiol. 81, 734–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Seelig J. (2004) Biochim. Biophys. Acta 1666, 40–50 [DOI] [PubMed] [Google Scholar]
- 53. Okuda S., Tokuda H. (2011) Annu. Rev. Microbiol. 65, 239–259 [DOI] [PubMed] [Google Scholar]
- 54. Yakushi T., Masuda K., Narita S., Matsuyama S., Tokuda H. (2000) Nat. Cell Biol. 2, 212–218 [DOI] [PubMed] [Google Scholar]
- 55. Hope M. J., Bally M. B., Mayer L. D., Janoff A. S., Cullis P. R. (1986) Chem. Phys. Lipids 40, 89–107 [DOI] [PubMed] [Google Scholar]
- 56. Moreira J. N., Almeida L. M., Geraldes C. F., Costa M. L. (1996) J. Mater. Sci. Mater. Med. 7, 301–303 [Google Scholar]
- 57. Lukehart S. A., Miller J. N. (1978) J. Immunol. 121, 2014–2024 [PubMed] [Google Scholar]
- 58. Moore M. W., Cruz A. R., LaVake C. J., Marzo A. L., Eggers C. H., Salazar J. C., Radolf J. D. (2007) Infect. Immun. 75, 2046–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]