Background: Given the biological importance of glycosphingolipids (GSLs), a widespread need exists for sensitive, rapid, and quantitative GSL-glycome analysis.
Results: Rhodococcal endoglycosylceramidase (EGCase)-assisted glycan cleavage was optimized for the major GSL classes and combined with glycoblotting to unveil cellular glycomic profiles.
Conclusion: Cellular GSL-glycomes were quantitatively and qualitatively characterized by newly established technique.
Significance: GSL-glycomics provides a unique way to delineate/characterize cells.
Keywords: Ganglioside, Glycolipid Structure, Glycolipids, Glycomics, Mass Spectrometry (MS), Rhodococcal Endoglycosylceramidase, Embryonal Carcinoma, Glycoblotting, Glycosphingolipids
Abstract
Glycosphingolipids (GSLs) are crucially important components of the cellular membrane, where they comprise microdomains with many critical biological functions. Despite this fact, qualitative and quantitative techniques for the analysis of GSLs still lag behind the needs of researchers. In this study, a reliable procedure for the elucidation of cellular GSL-glycomes was established based on (a) enzymatic glycan cleavage by endoglycosylceramidases derived from Rhodococcus sp. in combination with (b) glycoblotting-assisted sample preparation. The mixture of endoglycosylceramidase I and II was employed to maximize the release of glycan moieties from the major classes of GSLs (i.e. ganglio-, (neo)lacto- and globo-series GSLs). The glycoblotting technique enabled the quantitative detection of GSL-glycans using as few as 2 × 105 cells. Thirty-seven different kinds of cellular GSL glycans were successfully observed in 11 kinds of cells, including Chinese hamster ovary cells and their lectin-resistant mutants as well as murine and human embryonic carcinoma cells. Furthermore, in-depth structural clarification in terms of discrimination of isomers was achieved by MALDI-TOF/TOF mass spectrometry analysis and/or linkage-specific glycosidase digestion. These novel analytical techniques were shown to be capable of delineating cell-specific GSL-glycomes. Thus, they are anticipated to have a broad range of applications for the characterization, description, and comparison of various cellular/tissue samples in the fields of drug discovery and regenerative medicine.
Introduction
Glycosphingolipids (GSLs),3 carbohydrate- containing derivatives of a sphingoid or a ceramide, are among the most ubiquitously distributed glycoconjugates. They are the major component of biologically functional microdomains on ectoplasmic membranes and play crucial roles in neuritogenesis, cell proliferation, and receptors for certain bacterial toxins and viruses (1, 2). GSLs display a huge diversity of structures as a result of the great heterogeneity in the glycan moiety as well as the length of the fatty acid in ceramide.
The glycosylation of GSLs is initiated by the incorporation of a glucose (Glc) or a galactose (Gal) residue onto the ceramide portion of the molecule. The glycans are further elongated to yield complex structures that allow the classification of GSLs into subtypes such as the ganglio-, globo-, and (neo)lacto-series GSLs. Better elucidation of the different types and structures of GSL-derived glycans would be desirable to improve our understanding of the physiological and biological roles of GSLs and to aid in the discovery of novel carbohydrate-related biomarkers suitable for the classification, identification, and isolation of target cells (e.g. stem cells and their differentiated progeny). As a practical matter, immunostaining techniques that employ monoclonal antibodies (mAbs) against GSL-glycan epitopes such as the stage-specific embryonic antigens (SSEAs) are frequently used for the identification and isolation of undifferentiated cells from heterogeneous cellular populations (3).
Although immunostaining techniques that employ mAbs are imperative for the identification and localization of intra- and extracellular components, positive immunostaining alone is often not enough to identify a particular entity of GSLs on cells due to the cross-reactivity between glycans. For instance, although the MC813-70 epitope is mostly represented by sialyl Gb5Cer in SSEA-4, this mAb also cross-reacts to different extents with the GM1b and GD1a gangliosides as well as with a common structure in the core 1 O-glycan glycoprotein that carries the sialyl T epitope (4, 5). Therefore, mass spectrometric (MS) analysis, which is often used to determine the detailed structures, is considered to be a complementary technique to conventional immunostaining methodology.
Recently, Liang et al. (6) used MS analyses as well as immunofluorescence and flow cytometry analyses to demonstrate the dynamic structural switching of GSLs from globo- and lacto-series to gangliosides during human embryonic stem (ES) cell differentiation. Whole GSLs were analyzed upon permethylation before matrix-assisted laser desorption-ionization time-of-flight (MALDI-TOF) MS. This approach is valuable in that both the glycan and ceramide moieties of GSLs can be analyzed. However, the structural heterogeneities generated by both glycan and ceramide moieties often makes the MS spectrum complicated and, thus, the analysis of minor GSL components difficult. To reduce this complexity, analysis of glycan moieties released from the ceramide core is a straightforward approach.
The carbohydrate can be cleaved by ozonolysis or oxidation with osmium periodate followed by alkaline degradation of the oxidized lipid (7, 8). However, the cleavage yield requires further improvement because the average percent yield, for instance of gangliosides, is only ∼35% (9). Alternatively, linkages to the ceramide portion of the GSL that occur via a β-d-Glc residue (as is the case for the great majority of linkages) can be cleaved specifically by an endoglycoceramidase to afford intact oligosaccharides and ceramides. For example, pioneering work in regard to the GSL-glycomic analysis of murine tissues was performed by employing ceramide glycanase derived from Macrobdella decora (10). On the other hand, although this enzyme has a relatively broad substrate specificity, it cleaves specific glycans at various rates (e.g. the relative activities toward asialo-GM2, GM2, Gb5, nLc4, and LacCer compared with GM1 are 197, 115, 46, 38, and 19%, respectively (11)). This discrepancy in cleavage rates would make quantitative glycomic analysis difficult. We previously discovered endoglycosidases I and II (EGCase I and II) derived from a culture fluid of the mutant M-750 strain of Rhodococcus sp. and showed that both enzymes are capable of hydrolyzing the glucosylceramide linkage of ganglio-type, lacto-type, and globo-type GSLs by using various standard GSLs (12). Each EGCase has a quantitatively different specificity and complements the other. For instance, globo-type GSLs were completely hydrolyzed by EGCase I, whereas they were very resistant to hydrolysis by EGCase II. Sialosylparagloboside, which belongs to neolacto-type GSLs, was hydrolyzed most by EGCase II. Thus, it should be feasible to employ both EGCase I and II for the analysis of cellular/tissue GSL-glycomes (12).
In this study the conditions were first optimized for the enzymatic reaction of EGCase I and II, employing cells suitable for cellular GSL-glycomics. Several cells known to express predominant GSL types (e.g. the ganglio-, globo-, and (neo)lacto-series) were chosen as models. The advantage of using cellular GSLs as substrates for the optimization of cellular GSL-glycomics is evident; structurally diverse GSLs are readily available, and the optimization can be performed under realistic situations (i.e. in the presence of various impurities derived from cellular components and reagents used for sample preparation). As a reliable method for the purification of glycans in a crude mixture, a recently established chemoselective glycoblotting technique (13) was employed. The feasibility of quantitatively delineating cell state- and property-specific GSL-glycomes based on rhodococcal EGCase-assisted glycan cleavage and glycoblotting-assisted sample preparation followed by MALDI-TOF/TOF MS analysis was demonstrated by applying the optimized protocol to various cell lines including the Chinese hamster ovary (CHO) cell line and its lectin-resistant mutants as well as murine and human embryonic carcinoma cell lines.
EXPERIMENTAL PROCEDURES
Cell Culture
To establish a cellular GSL-glycomic technique, 11 kinds of cells were selected as follows: CHO-K1 cells, a subclone derived from the parental CHO cell line; β1–2-N-acetylglucosaminyltransferase I (GnT-I)-deficient (Lec1) CHO cells; UDP-galactose transporter-deficient (Lec8) CHO cells; murine fibroblast (NIH/3T3) cells; human cervical carcinoma (HeLa) cells; human promyeloleukemic (HL60) cells; human myelogenous leukemia (K562) cells; murine embryonic carcinoma (EC; P19C6, PCC3/A/1 and F9) cells; human EC (NEC8) cells. CHO-K1, Lec1, and Lec8 cells were obtained from the American Type Culture Collection, and the other cells were provided by the RIKEN Bio Resource Center in Japan.
CHO-K1, Lec1, Lec8, HL60, and K562 cells were cultured in RPMI1640 medium (Sigma). EC P19CL6 cells were cultured in α-minimal essential medium (Sigma), and the other cells were cultured in Dulbecco's modified Eagle's medium (Sigma). Antibiotic reagent (Invitrogen) containing penicillin (100 units/ml), streptomycin (100 μg/ml), and 10% fetal calf serum (Nichirei Biosciences Inc., Tokyo, Japan) was used for all cultures. All cells were grown in 10-cm dishes with 10 ml of medium at 37 °C under a 5% CO2 humidified atmosphere. Cells were cultured until they reached ∼100% confluency. The medium was then discarded, and the cultured cells were washed twice with 10 ml of phosphate-buffered saline (PBS). The washed cell layer was scraped into 10 ml of PBS containing 10 mm EDTA and collected by centrifugation at 1000 × g for 10 min. After removing the supernatant, the cell pellet was resuspended in PBS, and the cells were counted. Cells were resuspended to yield a cell solution containing 1 × 106 cells. The solution was dispensed to a fresh tube and centrifuged at 1000 × g for 10 min. The final cell pellet was obtained by removing the supernatant. Cell pellets were stored at −30 °C until use.
For non-adherent HL60 and K562 cells, cultured cells in medium were directly centrifuged, and the precipitated cells were washed twice with PBS. The remainder of the procedure for procuring the final cell pellet was identical to that described above for the adherent cells.
Expression and Purification of EGCases
EGCase I found in Rhodococcus equi was expressed in Rhodococcus erythropolis strain JCM3201 transformed with a pTip LCH 2.2 vector (14) containing egcase I as a C-terminal His-tagged protein. The recombinant EGCase I was purified through the affinity chromatography with Ni2+-Sepharose 6 Fast Flow (GE Healthcare). The fractions containing EGCase I were dialyzed against 20 mm sodium acetate buffer at pH 5.5. Protein quantification was performed with the bicinchoninic acid protein assay (Pierce). The detailed procedure will be described elsewhere. EGCase II was prepared according to the methods described previously (12, 15) or purchased from TAKARA Bio Inc. (Siga).
Extraction of GSLs from Cell Pellets
Total lipids including GSLs were extracted at room temperature by adding chloroform/methanol solution (2/1 (v/v), 450 μl) to the cell pellets, with homogenization using an Ultrasonic Homogenizer (TAITEC, Saitama, Japan). Pellets were sonicated six times with a 10-s pulse at 10-s intervals for a total sonication time of 1 min. After sonication, methanol (150 μl) was added; the composition of the solvent was now chloroform/methanol, 1/1 (v/v). The sonication was repeated in the same manner. Finally, methanol (300 μl) was added (chloroform/methanol, ½ (v/v)), and the sonication was repeated once again. The resulting extracts were subjected to centrifugation at 5000 rpm for 10 min, and the supernatants were transferred to fresh tubes and completely desiccated with a centrifugal evaporator. Desiccated total lipids were directly subjected to release of glycans by employing EGCases, as described below, without further purification.
Release of Glycans from GSLs
Crude cellular lipids were suspended in 50 mm Tris-HCl buffer, pH 7.5 (50 μl), containing 0.1% sodium cholate (Sigma) as a surfactant followed by the addition of EGCase I and/or II (25 milliunits) to release intact glycans from GSLs. The enzymatic digestions were performed at 37 °C for 24 h. To distinguish GSL-derived glycans from contaminating free oligosaccharides, the crude cellular lipids were also suspended in Tris-HCl buffer/sodium cholate in the absence of EGCases. The latter served as a negative control.
Glycoblotting
Samples treated with EGCases were subjected to glycoblotting as previously described with minor modifications (13). In brief, the sample solution (50 μl) was directly applied to the well of a filterplate (MultiScreen Solvinert 0.45 μm Low Binding Hydrophilic PTFE, Millipore, MA) containing BlotGlyco® beads (5.0 mg; Sumitomo Bakelite Co. Ltd., Tokyo). Released glycans were captured in 2% acetic acid in acetonitrile (450 μl) and incubated at 80 °C for 45 min. On-bead acetyl capping of unreacted hydrazide groups was performed using 10% acetic anhydride in MeOH for 30 min at room temperature. Next, on-bead methyl esterification of the carboxyl groups in glycan-derived sialic acid was carried out by incubation with 150 mm 3-methyl-1-p-tolyltriazene in dioxane at 60 °C. The trapped and esterified glycans on the beads were subjected to transiminization by incubation with a mixture of 2% acetic acid in acetonitrile (180 μl) and 20 mm aminooxy-WR (aoWR, 20 μl) (aoWR is a dipeptidic aminooxy compound that was synthesized as described previously (16)). The aoWR-labeled glycans were recovered with distilled water (100 μl), and the collected solution was purified using a hydrophilic interaction liquid chromatography purification plate (MassPrep HILIC μElution plate, Waters, MA) to remove the excess aoWR. To concentrate the aoWR-labeled glycans, the purified solution was desiccated using a rotational evaporator and subsequently dissolved in distilled water (10 μl).
Glycan Digestion with β-Galactosidases
aoWR-labeled glycans were digested with β1,3-galactosidase or β1,4-galactosidase for determination of the isomeric structure. One-tenth of the final solution obtained from glycoblotting (1 μl) was diluted with 50 mm acetate buffer, pH 5.0 (8 μl), and incubated at 37 °C for 3 h in the presence of β1,3-galactosidase (100 units) derived from (Xanthomonas manihotis) or β1,4-galactosidase (3 milliunits) derived from Streptococcus pneumoniae (Calbiochem). Digested glycans were subjected to MS analysis without further purification.
MALDI-TOF/TOF MS Analysis
Purified GSL-glycan solutions were mixed with 2,5-dihydrobenzoic acid solution (10 mg/ml in 30% acetonitrile) and subsequently subjected to MALDI-TOF MS analysis. All measurements were performed using an Ultraflex II TOF/TOF mass spectrometer equipped with a reflector and controlled by the FlexControl 3.0 software package (Bruker Daltonics GmbsH, Bremen, Germany) according to general protocols. All spectra were obtained using reflectron mode with an acceleration voltage of 25 kV, a reflector voltage of 26.3 kV, and a pulsed ion extraction of 160 ns in the positive ion mode. Masses were annotated using the FlexAnalysis 3.0 software package (Bruker Daltonics GmbsH, Bremen, Germany). The GlycoSuiteDB and SphinGOMAP (17) online databases were used for structural identification of GSL-glycans. Absolute quantification was performed by comparative analyses between areas of MS signals derived from each GSL-glycan and 5 pmol of the internal standard N,N′,N′′,N′′′-tetraacetyl chitotetraose (GN4), which was added to the sample solution before glycoblotting. In TOF/TOF mode measurements for fragment ion analysis, precursor ions were accelerated to 8 kV and selected in a timed ion gate. Fragment ions generated by laser-induced decomposition of the precursor were further accelerated by 19 kV in the LIFT cell. Fragment masses were analyzed after passing the ion reflector.
Hierarchical Cluster Analysis
Cluster analyses were performed with software Cluster 3.0 developed by the Eisen et al. (18) with a hierarchical clustering algorithm. The calculated dendrogram and correlation matrix were visualized using TreeView 1.6 software downloaded from the same site.
RESULTS
Glycoblotting-assisted Sample Preparation for Cellular GSL-glycomics
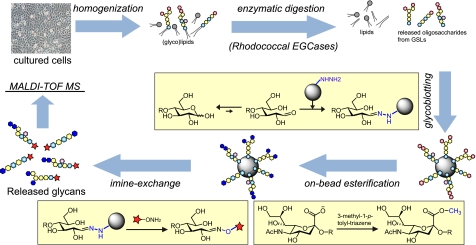
To employ cellular GSL-glycans as substrates for the optimization of rhodococcal EGCase I and II-assisted glycan cleavage for cellular GSL glycomics, a glycoblotting procedure followed by MALDI-TOF(/TOF) MS analysis was adopted (Fig. 1). Briefly, GSLs were extracted from cells by homogenization via sonication and lysis with methanol and chloroform followed by centrifugation (10). After enzymatic digestion, glycans were selectively captured onto high density hydrazide beads (BlotGlyco®) for highly efficient purification of oligosaccharides from complex biological samples. The captured oligosaccharides were subjected to on-bead methyl esterification to stabilize sialic acid(s) for the simultaneous quantitation of neutral and sialylated oligosaccharides by MALDI-TOF/TOF MS. The oligosaccharides were finally recovered as aoWR derivatives via imine exchange, which enables highly sensitive and quantitative analysis by MALDI-TOF (16).
FIGURE 1.
Schematic diagram of the established procedure for quantitative cellular GSL-glycomics. Total lipids were extracted by ultrasonic homogenization in a chloroform and methanol solvent, and glycan moieties were released in the presence of a mixture of EGCase I and II (25 milliunits of each). Because of the high purification capability of glycoblotting, no further purifications were required. GSL-derived glycans were recovered as derivatives of aoWR and subjected to MALDI-TOF MS analysis.
Naven and Harvey (19) demonstrated that oligosaccharides with masses greater than 1000 Da exhibit similar signal strengths irrespective of structure when examined by MALDI-TOF MS. A few of the GSL-glycans analyzed in this study have molecular masses less than 1000 Da, and thus the quantitative values obtained for these glycans may be underestimated by MALDI-TOF MS. However, because the analysis for all of the glycans was reproducible, as described below, estimation of accurate absolute abundance of lower moleculare mass glycans should be carried out by applying correction factors.
When the MALDI-TOF MS spectra were compared between the samples with or without deglycosylation treatment by EGCases, several signals were observed that were designated as oligosaccharide signals even when enzymatic digestion was omitted (supplemental Fig. S1). Free oligosaccharides inherently present in cells were responsible for these signals, and some free oligosaccharides have the same molecular mass as GSL-glycans. Therefore, analyses were done both in the presence and absence of EGCases, and the quantitative values for GSL-glycans were corrected by subtraction of the values for free oligosaccharides.
Glycoblotting has been proven to uncompromisingly remove impurities and to improve the ionization efficiency of glycans upon aoWR derivatization. As such, glycoblotting drastically improves the signal-to-noise ratio. Indeed, a pilot study showed that 2 × 105 cells were sufficient to qualitatively analyze cellular glycomics (data not shown). GSLs could be directly subjected to EGCase digestion without further fractionation, such as the traditional Folch separation. Likewise, it was not necessary to separate the GSLs into acidic and neutral fractions by ion exchange chromatography because the standard glycoblotting protocol includes methyl esterification of sialic acid to render sialylated oligosaccharides chemically equivalent to neutral oligosaccharides (20). It is worth noting that the omission of the Folch separation step improved the recovery yield of GSL-glycans (data not shown). Thus, the fact that there was no need for prefractionation of GSLs before glycan release and glycoblotting not only simplified the analysis but also avoided the risk of sample loss.
Optimization of Conditions for EGCase Treatment
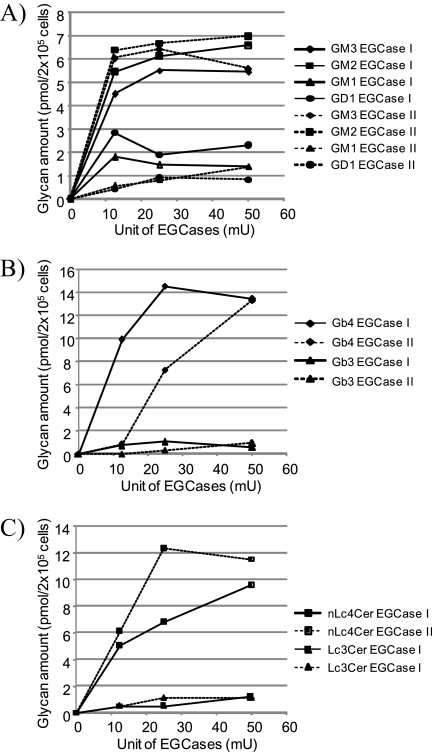
Highly efficient glycan release from GSLs is indispensable for quantitative GSL-glycomics; therefore, an attempt was first made to optimize the conditions for the enzymatic reactions of Rhodococcal EGCase I and II so as to render their application to cellular GSL-glycomics experimentally practical. NIH/3T3, HL60, and F9 cells were selected as model cells for the optimization of the enzymatic conditions. This selection was based on previous observations that these cells are relatively rich in the GSL ganglio-, lacto- and globo-series, respectively (21–25), representing all major classes of GSLs. All investigations were performed at 37 °C for 24 h using 2 × 105 cells. As shown in Fig. 2A, 12.5 milliunits of either EGCase I and II was sufficient to reach the release plateau for the ganglio-series GSLs (GM3, GM2, GM1 and GD1). As shown in Fig. 2B, 25 milliunits of EGCase I was sufficient to reach the release plateau for the globo-series GSLs (Gb3 and Gb4), whereas 50 milliunits of EGCase II was required to release the same amount of Gb3 and Gb4. In contrast, the (neo)lacto-series (Lc3 and nLc4) was digested more efficiently by EGCase II; 25 milliunits of the enzyme was sufficient to reach the release plateau, whereas 50 milliunits of EGCase I was required to release the same amount of Lc3 and nLc4 (Fig. 2C). The observed differences in the rate of GSL hydrolysis between EGCase I and II are essentially consistent with kinetic studies of EGCases using various model GSLs (12).
FIGURE 2.
Optimization of EGCase I and II enzymatic reactions. The optimization of cellular GSL-glycomics was performed using cellular GSLs as enzymatic substrates. Total lipids extracted from 2 × 105 NIH/3T3 (A), F9 (B), or HL60 (C) cells were directly digested using various amounts of EGCase I or II in the presence of 0.1% sodium cholate. Ganglio-series GSLs (i.e. GM3, GM2, GM1, and GD1a) in NIH/3T3 cells (A), globo-series GSLs (i.e. Gb3 and Gb4) in F9 cells (B), and (neo)lacto-series GSLs (i.e. nLc4 and Lc3) in HL60 cells (C) were used as cellular GSL substrates. mU, milliunits.
Based on the results described above, a mixture of enzymes consisting of 25 milliunits of both EGCase I and EGCase II was employed for further cellular GSL-glycomic analysis. The reproducibility and accuracy of the technique were investigated by analysis of NIH/3T3 cells in three separate experiments (sample preparation, glycoblotting, and MALDI-TOF MS analysis). Fairly good coefficients of variance (<10%) were obtained for glycan species whose relative proportion was >1% for both the absolute and relative abundance of each oligosaccharide (supplemental Fig. S2). Notably, glycoblotting can be performed in a highly parallel manner (i.e. in 96-well microplate format), indicating that this new analytical procedure provides truly high throughput GSL-glycomics.
Cellular GSL-glycan Profiling
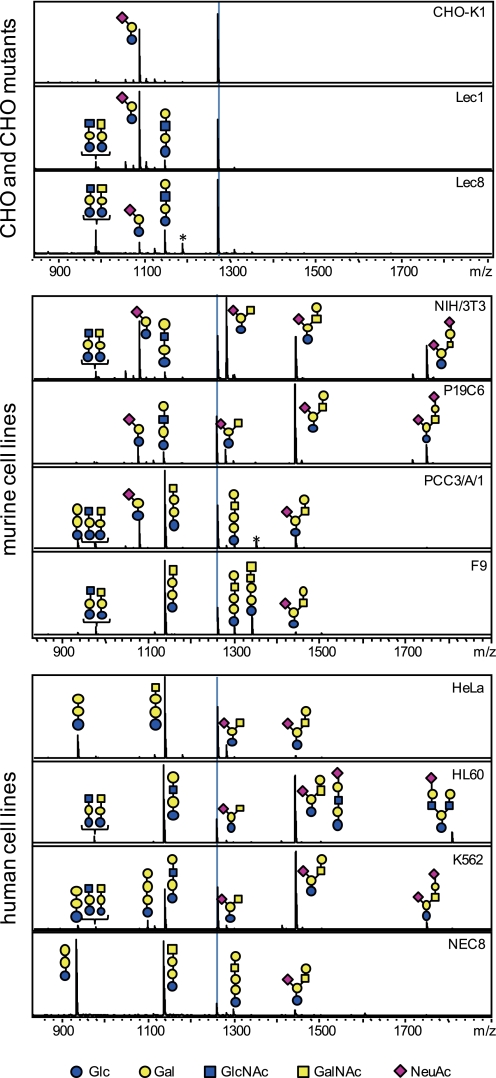
Based on the successful establishment of the techniques for analysis of the cellular GSL-glycomics, quantitative GSL-glycomics were analyzed using a total of 11 cell lines. The cell lines consisted of CHO-K1 cells and lectin-resistant CHO mutants (Lec1 and Lec8 cells, corresponding to GnT-I-deficient and UDP-galactose transporter-deficient CHO cells, respectively), four murine cell lines (NIH/3T3, P19C6, PCC3/A/1, and F9), and four human cell lines (HeLa, HL60, K562, and NEC8). The MALDI-TOF MS spectra corresponding to the GSL-glycomic profile of each cell line is shown in Fig. 3. Up to 37 different kinds of GSL-derived glycans were quantitatively observed.
FIGURE 3.
Representative MALDI-TOF MS spectra of GSL-derived glycans derived from 11 kinds of cells. All signals were detected as aoWR-glycan derivatives. The dotted line on each panel indicates the internal standard, GN4 (5 pmol). Estimated structures are shown. Blue circles, yellow circles, blue squares, yellow squares, and purple diamonds represent Glc, Gal, GlcNAc, GalNAc, and Neu5Ac, respectively. Peaks labeled by asterisks stem from contaminating free oligosaccharides.
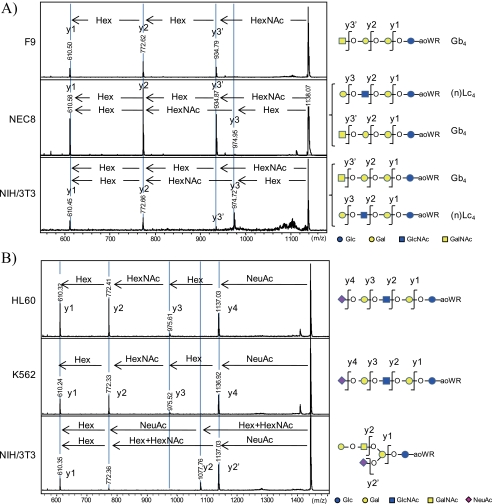
The isomeric structures of GSL-glycans were determined by the investigation of aoWR-labeled glycans by MALDI-TOF/TOF MS analysis and/or by enzymatic digestion using linkage-specific glycosidases (i.e. β1,3- and β1,4-galactosidases). The MALDI-TOF/TOF MS measurements for the aoWR-labeled glycans allowed the determination of the GSL-glycan structure by sequential assignment of fragment signals. Because of the high proton affinity of aoWR, only aoWR-labeled fragments (y-ions) could be detected on the TOF/TOF spectra. This feature is advantageous in simplifying the TOF/TOF spectra and makes the structural elucidation of GSL-glycans straightforward. For instance, the signal corresponding to (Hex)3(HexNAc)1 was found in all cells analyzed and could be due to either (n)Lc4 or Gb4. The representative TOF/TOF spectra for the fragment analyses of (Hex)3(HexNAc)1 in NIH/3T3, F9, and NEC8 cells are shown in Fig. 4A. F9 cells yielded intense fragments of (Hex)1+aoWR, (Hex)2+aoWR, and (Hex)3+aoWR, whereas no fragments corresponding to (HexNAc)1+aoWR, (Hex)1(HexNAc)1+aoWR, or (Hex)2(HexNAc)1+aoWR were observed. This result strongly suggests that (Hex)3(HexNAc)1+aoWR in F9 cells makes up a structure corresponding to GalNAcβ1–4Galα1–4Galβ1–4Glc (Gb4). In the case of NEC8 and NIH/3T3 cells, fragments of both (Hex)2(HexNAc)1+aoWR and (Hex)3+aoWR were observed in the TOF/TOF spectra, indicating that (Hex)3(HexNAc)1+aoWR in NIH/3T3 and NEC8 cells was present in a mixture of (n)Lc4 and Gb4 glycans. As another example, the fragment analyses of the glycan (Hex)3(HexNAc)1(NeuAc)1 found in HL60, K562, and NIH/3T3 cells are shown in Fig. 4B. A liner structure of the glycan structure (sialosyl paragloboside (SPG)) was suggested for HL60 and K562 cells. The fragment analysis of (Hex)3(HexNAc)1(NeuAc)1 in NIH/3T3 cells yielded an additional fragment, indicating the loss of (Hex)1(HexNAc)1 and the presence of GM1.
FIGURE 4.
Representative MALDI-TOF/TOF spectra for the structural determination of glycan isoforms. The isoforms of (Hex)3(HexNAc)1 obtained from F9, NEC8, and NIH/3T3 cells (A) and (Hex)3(HexNAc)1(NeuAc)1 observed in HL60, K562, and NIH/3T3 cells (B) could be distinguished by TOF/TOF analysis. In the case of (Hex)3(HexNAc)1, this glycan corresponded to only Gb4 in the globo-series in F9 cells. On the other hand, TOF/TOF analyses for the glycan in NEC8 and NIH/3T3 cells showed a mixture consisting of Gb4 and (n)Lc4. In similar manner the glycan (Hex)3(HexNAc)1(NeuAc)1 possessed a linear structure (SPG) in HL60 and K562 cells as opposed to a branched structure with a (Hex)1(HexNAc)1 unit at the non-reducing terminal in NIH 3T3 cells, characteristic of GM1.
For further structural determination and relative quantification of isomers, enzymatic digestion employing β1,3 or β1,4-galactosidase was performed. The absolute quantities of the Lc4, nLc4, and Gb4 glycans in NIH 3T3 cells, whose composition is commonly (Hex)3(HexNAc)1, was found to be 10, 13, and 46 fmol/2 × 105 cells, respectively. This conclusion was drawn because enzymatic digestion with β1,3- or β1,4-galactosidase digested 14.2 or 18.6%, respectively, of (Hex)3(HexNAc)1 in NIH/3T3 cells. By contrast, (Hex)3(HexNAc)1 in HL60 cells was completely destroyed by digestion with β1,4-galactosidase but was resistant to β1,3-galactosidase, indicating that the signal was solely due to nLc4. Likewise, (Hex)3(HexNAc)1(NeuAc)1 in NIH/3T3 cells was completely digested by β1,3-galactosidase, indicating that the signal was solely due to GM1. Because the corresponding signal in HL60 cells was resistant to both galactosidases, it was assigned to SPG as determined by TOF/TOF analysis. Supporting data for the structural determination of other examples are shown in supplemental Fig. S3.
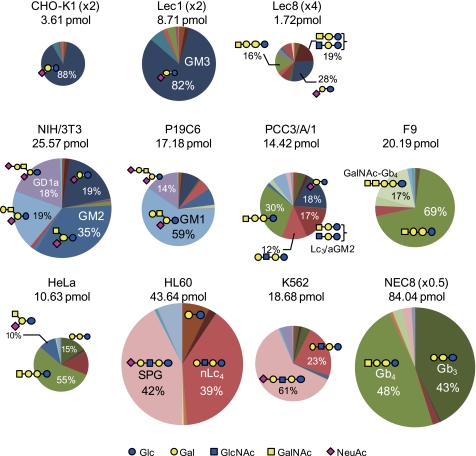
The estimated composition/structure and the quantitative value of each GSL-glycan are summarized in Table 1. To visually compare the GSL-glycomes among cell types, the quantified values are also shown as pie charts (Fig. 5) in which the size of the circle and the colors reflect the absolute quantity of GSL-glycans and the GSL-glycan structure, respectively. Pie charts in Fig. 5 clearly showed that the GSL-glycomes differed substantially among cell types, indicating that the cellular GSL-glycome is highly cell type-specific.
TABLE 1.
Summary of the absolute quantification of GSL-derived glycome (pmol/2 × 105 cells)
Cellular GSL-glycome analysis using the procedure of EGCase- and glycoblotting-assisted sample preparation in combination with MALDI-TOF(/TOF) MS measurements identified 37 kinds of GSL-glycans from 11 kinds of cells in subpicomol order (ND, not detected).
| # | m/z | Composition | Common name | Class | CHO-K1 | Lec1 | Lec8 | NIH/3T3 | HeLa | HL60 | K562 | P19C6 | PCC3/A/1 | F9 | NEC8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 772.3 | (Hex)2 | LacCer | Lac | 0.109 | 0.222 | 0.062 | 0.223 | 0.060 | 3.171 | 0.221 | 0.229 | 0.327 | 0.178 | 0.224 |
| 2 | 934.4 | (Hex)3 | Gb3 | Gb | 0.005 | 0.012 | 0.022 | ND | 1.573 | 0.001 | 0.265 | 0.063 | 0.566 | 0.227 | 35.837 |
| 3 | 975.4 | (Hex)2(HexNAc)1 | Lc3/aGM2 | Lc/Ga | ND | 0.161 | 0.328 | 0.260 | 0.059 | 0.903 | 0.168 | 0.101 | 0.252 | 0.000 | 0.084 |
| 4 | 1077.4 | (Hex)2(Neu5Ac)1 | GM3 | Ga | 3.187 | 7.125 | 0.489 | 5.814 | 0.064 | 0.024 | 0.041 | 1.277 | 2.626 | 0.006 | 0.195 |
| 5 | 1093.5 | (Hex)2(Neu5Gc)1 | GM3(Gc) | Ga | 0.181 | 0.546 | ND | 0.124 | ND | ND | ND | 0.014 | 0.025 | ND | ND |
| 6 | 1096.4 | (Hex)4 | Gb | 0.030 | ND | 0.042 | ND | 0.007 | 0.013 | 0.693 | 0.099 | 0.045 | 0.013 | 0.086 | |
| 7 | 1137.5 | (Hex)3(HexNAc)1 | Lc4/aGM1 | Lc/Ga | ND | ND | 0.128 | 0.010 | 1.544 | ND | ND | ND | 2.397 | ND | 1.549 |
| 8 | 1137.5 | (Hex)3(HexNAc)1 | nLc4 | nLc | 0.066 | 0.184 | 0.065 | 0.013 | ND | 17.155 | 4.261 | 0.889 | 1.754 | ND | ND |
| 9 | 1137.5 | (Hex)3(HexNAc)1 | Gb4 | Gb | ND | 0.316 | 0.268 | 0.046 | 5.808 | ND | ND | ND | 4.348 | 13.965 | 39.951 |
| 10 | 1178.5 | (Hex)2(HexNAc)2 | 0.011 | 0.012 | 0.094 | 0.051 | 0.062 | 0.012 | ND | 0.018 | 0.009 | 0.020 | 0.163 | ||
| 11 | 1280.5 | (Hex)2(HexNAc)1(Neu5Ac)1 | GM2 | Ga | ND | 0.015 | 0.032 | 10.379 | 1.087 | ND | 0.291 | 1.248 | 0.209 | ND | ND |
| 12 | 1283.5 | (Hex)3(HexNAc)1(Fuc)1 | Fuc-Lc4/SSEA-1 | (n)Lc | ND | ND | ND | ND | ND | 0.338 | 0.040 | ND | ND | 0.012 | 0.763 |
| 13 | 1296.5 | (Hex)2(HexNAc)1(Neu5Gc)1 | GM2(Gc) | Ga | ND | ND | ND | 0.333 | ND | ND | ND | 0.021 | ND | ND | ND |
| 14 | 1299.5 | (Hex)4(HexNAc)1 | Gaλα13-(n)Lc4 | (n)Lc | ND | ND | ND | 0.085 | ND | ND | ND | ND | ND | 0.356 | ND |
| 15 | 1299.5 | (Hex)4(HexNAc)1 | Galβ13-(n)Lc4 | (n)Lc | 0.012 | 0.104 | 0.148 | 0.005 | 0.078 | ND | ND | 0.211 | 0.181 | 0.515 | 0.462 |
| 16 | 1299.5 | (Hex)4(HexNAc)1 | Gb5 (SSEA-3) | Gb | ND | ND | ND | ND | ND | 0.109 | 0.037 | ND | 0.135 | 0.752 | 1.884 |
| 17 | 1340.5 | (Hex)3(HexNAc)2 | nLc5 | nLc | 0.004 | 0.006 | 0.030 | 0.016 | 0.009 | 0.155 | 0.012 | 0.017 | 0.019 | ND | 0.026 |
| 18 | 1340.5 | (Hex)3(HexNAc)2 | Forrsman | Gb | ND | ND | ND | ND | ND | ND | ND | ND | ND | 3.505 | ND |
| 19 | 1429.6 | (Hex)3(HexNAc)1(Fuc)2 | diFuc-Lc4 | Lc | ND | ND | ND | 0.014 | ND | 0.015 | 0.013 | 0.032 | ND | ND | ND |
| 20 | 1442.6 | (Hex)3(HexNAc)1(Neu5Ac)1 | GM1 | Ga | ND | ND | 0.004 | 2.074 | 0.195 | ND | ND | 10.142 | 1.010 | 0.343 | ND |
| 21 | 1442.6 | (Hex)3(HexNAc)1(Neu5Ac)1 | SPG | nLc | ND | ND | ND | ND | ND | 18.419 | 11.445 | ND | 0.332 | ND | 2.021 |
| 22 | 1458.6 | (Hex)3(HexNAc)1(Neu5Gc)1 | GM1(Gc)/SPG(Gc) | Ga/nLc | ND | ND | ND | 0.125 | ND | 0.073 | 0.129 | 0.233 | 0.010 | ND | 0.031 |
| 23 | 1486.6 | (Hex)3(HexNAc)2(Fuc)1 | Lc | ND | ND | ND | 0.046 | ND | 0.013 | 0.007 | 0.019 | ND | ND | 0.022 | |
| 24 | 1502.6 | (Hex)4(HexNAc)2 | Lc | ND | 0.007 | 0.005 | ND | 0.067 | 0.260 | 0.067 | 0.022 | 0.025 | 0.263 | 0.016 | |
| 25 | 1543.6 | (Hex)3(HexNAc)3 | Lc | ND | ND | ND | 0.005 | ND | ND | ND | ND | ND | 0.006 | ND | |
| 26 | 1604.6 | (Hex)4(HexNAc)1(Neu5Ac)1 | nLc | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0.683 | |
| 27 | 1648.7 | (Hex)4(HexNAc)2(Fuc)1 | (n)Lc | ND | ND | ND | ND | ND | 0.064 | ND | ND | ND | ND | 0.007 | |
| 28 | 1664.7 | (Hex)5(HexNAc)2 | nLc | ND | ND | 0.004 | 0.014 | ND | ND | 0.009 | 0.006 | 0.011 | 0.015 | ND | |
| 29 | 1747.7 | (Hex)3(HexNAc)1(Neu5Ac)2 | GD1a | Ga | ND | ND | ND | 5.731 | 0.007 | 0.010 | 0.896 | 2.480 | 0.092 | 0.006 | 0.011 |
| 30 | 1763.7 | (Hex)3(HexNAc)1(Neu5Ac)1(Neu5Gc)1 | GD1a(Gc) | Ga | ND | ND | ND | 0.189 | ND | ND | 0.006 | 0.050 | ND | ND | ND |
| 31 | 1791.7 | (Hex)3(HexNAc)2(Fuc)1(Neu5Ac)1 | Lc/Ga | ND | ND | ND | 0.012 | ND | ND | ND | 0.009 | ND | ND | ND | |
| 32 | 1807.7 | (Hex)4(HexNAc)2(Neu5Ac)1 | nLc | ND | ND | ND | 0.006 | 0.005 | 2.841 | 0.073 | 0.004 | 0.050 | 0.003 | 0.003 | |
| 33 | 1867.7 | (Hex)5(HexNAc)3 | nLc | ND | ND | ND | ND | 0.004 | 0.005 | 0.003 | ND | ND | ND | 0.008 | |
| 34 | 1953.8 | (Hex)4(HexNAc)2(Fuc)1(Neu5Ac)1 | (SLex) | nLc | ND | ND | ND | ND | ND | 0.003 | ND | ND | ND | ND | ND |
| 35 | 2013.8 | (Hex)5(HexNAc)3(Fuc)1 | (n)Lc | ND | ND | ND | ND | ND | 0.009 | ND | ND | ND | ND | 0.004 | |
| 36 | 2172.8 | (Hex)5(HexNAc)3(Neu5Ac)1 | nLc | ND | ND | ND | ND | ND | 0.045 | 0.003 | ND | ND | ND | 0.007 | |
| 37 | 2318.9 | (Hex)5(HexNAc)3(Fuc)1(Neu5Ac)1 | nLc | ND | ND | ND | ND | ND | 0.001 | ND | ND | ND | ND | ND | |
| total | 3.61 | 8.71 | 1.72 | 25.57 | 10.63 | 43.64 | 18.68 | 17.18 | 14.42 | 20.19 | 84.03 |
FIGURE 5.
Cellular delineation/characterization through quantitative GSL-glycomic analyses. Both the absolute quantity of GSL-glycans (i.e. the size of the glycome) and the relative GSL-glycan composition are represented for each cell type. The areas of the circles represent the amount of total GSL-glycans per 2 × 105 cells. The circle size of CHO-K1 and Lec1 cells are magnified 2-fold, that of Lec8 cells is magnified 4-fold, and that of NEC8 is reduced to one-half size to facilitate visualization.
CHO-K1, Lec1, and Lec8 cells expressed GM3-glycan as a major component. This glycan represented 88, 82, and 28% of the total cellular GSL-glycome, respectively. No gangliosides further elongated than GM3 were observed in CHO-K1 cells, reflecting the nature of these cells, which are characterized by a lack of ability to synthesize GM2 and more complex gangliosides (26). By contrast, Lec1 and Lec8 cells expressed a small amount of GM2 and/or GM1, presumably because these cells retain the property of the parental CHO cell line to synthesize gangliosides that are more elongated than GM3. The glycomic profiles of CHO-K1 and Lec1 cells were quite similar qualitatively, whereas quantitatively, the total amount of GSL-glycans detected in Lec1 cells was unexpectedly more than twice the amount detected in CHO-K1 cells. Unlike in CHO-K1 and Lec1 cells, Lec8 cells expressed a relatively high proportion of glycans derived from globo- and lacto-series GSLs. The proportion of glycans having galactose at the non-reducing end was extremely high in Lec8 cells (27.8%) compared with CHO-K1 and Lec1 cells (6.2 and 6.1%, respectively).
Among the murine cell lines, the GSL-glycomes of NIH/3T3 and P19C6 cells were predominantly dominated by glycans derived from ganglio-series GSLs (94 and 90% of total detected GSL-glycans, respectively). In case of NIH/3T3 cells, the most abundant GSL-glycan was found to be GM2 (40.6% of total GSLs). GM3, GM1, and GD1a were the next most abundant glycans (22.7, 8.1, and 22.4%, respectively). In P19C6 cells, the predominant ganglioside was found to be GM1 (59%), with GD1a, GM3, and GM2 being quantitatively the next most abundant, at 14.4, 7.4, and 7.3%, respectively. On the other hand, PCC3/A/1 and F9 cells mostly contained glycans derived from globo-series GSLs (56 and 96% of total GSL-glycans, respectively). In F9 cells, the major ganglioside was Gb4 (69.2%), with the Forssman antigen, widely known to be expressed during the processes of growth and differentiation by murine embryos (27), being the next most abundant (17.4%). Gb4 was also identified as the major component in PCC3/A/1 cells (30.1%), whereas a relatively high proportion of ganglio- and (neo)lacto-series GSL-glycans were also observed (e.g. GM3 and nLc4-glycans represented 18.2 and 12.2% of total GSL-glycans, respectively). Overall, the total amount of cellular GSL-glycans was highest in NIH/3T3 cells (29.91 pmol/2 × 105 cells) compared with the other cells (14.4–20.2 pmol/2 × 105 cells).
Among the human cell lines, HL60 and K562 cells, both derived from human leukemia cells, gave fairly similar GSL-glycan profiles. In both cases, (Hex)3(HexNAc)1(NeuAc)1 (assigned to NeuAcα2–6Galβ1–4GlcNAcβ1–3Galβ1–4Glc known as SPG by enzyme-assisted structural determination) was found to be the major component (42.2% in HL60 and 61.3% in K562 cells). nLc4 was the next most abundant GSL-glycan (39.3% in HL60 and 22.8% in K562 cells). The observed high content of SPG in HL60 and K562 cells is consistent with previous observations based on histochemical analyses (28, 29). Gb4 was found to be the major component in HeLa and NEC8 cells (54.5% in HeLa and 47.5% in NEC8 cells), with Gb3 as the next most abundant (14.8% in HeLa and 42.6% in NEC8 cells). Thus, HeLa and NEC8 cells both predominantly expressed glycans derived from globo-series GSLs (73 and 95% of total GSL-glycans, respectively). The major difference between the two cells was the relatively high content of ganglio-series GSL-glycan (10% of GM2) in HeLa cells, whereas the ganglio-series was almost negligible in NEC8 cells.
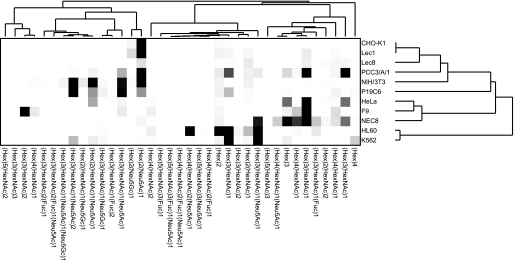
Hierarchical clustering analyses (17) were conducted using absolute quantitative GSL-glycomic data. The obtained dendrogram appeared to adequately classify the cells. CHO-K1 cells and CHO-derived mutants were classified into a group, whereas the glycomic profile of Lec8 cells differed from those of CHO-K1 and Lec1 cells (Fig. 6). HL60 and K562 cells were also classified into a group. Human cervical carcinoma cells (HeLa) and human EC cells (NEC8) were revealed to be close relations, but unexpectedly the three murine EC cells (P19C6, PCC3/A/1 and F9) analyzed in this study showed poor correlation in the hierarchical clustering analysis.
FIGURE 6.
Hierarchical clustering analysis of GSL-glycan expression profiles. Graphic representation of hierarchical clustering results based on expression profiles of 37 GSL-glycans in 11 cell types is shown. Rows, cell types; columns, GSL-glycans. Color intensity from white to red indicates the magnitude of differential expression.
DISCUSSION
Many studies analyzing cellular GSLs have been carried out throughout several decades by a variety of techniques (e.g. thin layer chromatography and immunostaining). However, the correlation between gene transcript levels and actual GSL expression profiles has been investigated less extensively. Therefore, both qualitative and quantitative analyses of cellular GSL-glycomes are important to delineate cellular properties. In this study a novel protocol was established for the analysis of qualitative and quantitative cellular GSL-glycomics. This protocol aimed to characterize/delineate cells based on efficient glycan release by rhodococcal EGCases followed by glycoblotting-assisted sample preparation using as few as 2 × 105 cells. Glycoblotting not only substantially simplified the sample preparation protocol but also improved the recovery yield by reducing the risk of sample loss. In-depth structural elucidation was also achieved by MALDI-TOF/TOF analysis combined with linkage-specific glycosidase digestion. In addition, it is worth noting that the defatted pellet, a fraction that does not contain GSLs, was the starting material for the analysis of cellular glycosaminoglycomics, as recently described.4 Thus, the analysis of cellular glycomics of GSLs and glycosaminoglycans was efficiently streamlined.
Three different types of endoglycoceramidases are known to be derived from Rhodococcus strains (EGCase I, II, and III). Type I and II can release glycans of ganglio-, lacto-m and globo-type GSLs, yielding oligosaccharides and ceramide. However, types I and II are inactive in the hydrolysis of gala-type GSLs, cerebrosides, sulfatides, and glycoglycerolipids. A mixture of EGCase I and II (25 milliunits of each) was employed in the current study to maximize the cleavage efficiency against cellular GSLs such that the releasing efficiency of GSL-glycans reached a plateau. This criterion does not necessarily indicate that the release of glycans is quantitative. However, considering our previous observation that ∼70% of cell surface GM3 on intact erythrocytes was hydrolyzed by EGCase II (20 milliunits) in the absence of detergent (30), fairly good releasing efficiency, if not quantitative, can be expected under the conditions of this study where detergent was present (a preferred condition for the hydrolysis by EGCases).
Techniques were first established for the analysis of cellular-GSL glycomics and GSL-glycomes of 11 kinds of cells were next successfully unveiled both qualitatively and quantitatively. Both predictable and unpredictable results could be found in the glycomic study of CHO-K1 cells and their lectin-resistant mutants. For instance, the total amount of GSL-glycans detected in Lec1 cells was more than twice the amount found in CHO-K1 cells despite the fact that GnT-I is not directly involved in the biosynthesis of GSLs. The total amount of GSL-glycans in Lec8 cells was less than half that in CHO-K1 cells, which can be explained by a previous report demonstrating that galactose in glycoproteins and glycolipids is reduced by 80–90% in mutant cells compared with wild-type cells (31, 32). Our analysis revealed the presence of a wide variety of GSL-glycans corresponding to all classes of GSLs (i.e. ganglio-, globo-m and (neo)lacto-series GSLs) in Lec8 cells, whereas CHO-K1 cells predominantly expressed glycans derived from ganglio-series GSLs (∼93.4% of total GSL-glycans present). Interestingly, glycans having a galactose residue(s) at the non-reducing terminus were increased in Lec8 cells (0.5 pmol/2 × 105 cells) compared with CHO-K1 cells (0.2 pmol/2 × 105 cells). These unexpected GSL-glycan expression profiles could be explained by compensation for a perturbation of N-glycan biosynthesis due to the deficiency of the GnT-I and UDP-Gal transporter in Lec1 and Lec8 cells, respectively.
Oligosaccharides containing N-glycolylneuraminic acid (Neu5Gc) as well as N-acetylneuraminic acid (Neu5Ac) were observed in 8 of the 11 cell types analyzed in this study. The relative proportion of Neu5Gc/Neu5Ac was highest in CHO-K1 cells and their lectin-resistant mutants (∼5–6%). The relative proportion was ∼1–4% in murine cells, except for F9 cells, in which sialylated species themselves were scarce. Neu5Gc was also detected in human cells (HL60 and K562 cells), where the relative proportion of Neu5Gc/Neu5A was less than 1.1%. Importantly, humans are genetically unable to synthesize Neu5Gc, and therefore, Neu5Gc was possibly taken up by the human cells from the culture medium, as reported previously (33).
NEC8 (human EC) cells displayed an extremely high proportion (>93%) of glycans derived from globo-series GSLs and a relatively small proportion (4.3 to 6.2%) of (neo)lacto-series GSL-glycans, with an almost negligible amount of ganglio-series GSL-glycans (0.2–1.8%). This profile appeared to be similar to the GSL profile for human ES cells recently unveiled by Liang et al. (6). It is well documented that human EC cells can in some circumstances act as a malignant surrogate for normal ES cells. Structures rich in glycans derived from globo-series GSLs are a common feature of both EC and ES cells in human and murine species; although murine EC cells are reported not to express SSEA3 and SSEA4, they strongly express the Forsman antigen that is absent in human EC cells (34).
The three murine EC cell types (P19C6, PCC3/A/1, and F9) analyzed in this study showed distinct GSL-glycomic profiles. The GSL-glycome of F9 cells appeared to share similar features with the GSL-glycomes of NEC8 and human ES cells; that is, a high proportion of globo-series GSL-glycans (92%) and a small proportion of lacto-series GSL-glycans (5.7%) with a lower proportion of ganglio-series GSL-glycans (1.8%). In contrast, the GSL-glycomic profile of P19C6 cells was characterized by a high proportion of ganglio-series GSL-glycans (90%) and a small proportion of lacto-series GSL-glycans (5.3%) with a lower proportion of globo-series GSL-glycans (2.2%). The GSL-glycomic profile of PCC3/A/1 cells was somewhere in between the two and was characterized by comparative proportions of globo-, ganglio-, and (neo)lacto-series GSL-glycans (37, 28–46, and 15–33%, respectively). The relative amount of SSEA-3 (Gb5), known as an undifferentiated cell marker, was fairly constant (0.9–3.7%) in PCC3/A/1 and F9 cells and was comparable with the amount in NEC8 cells (2.2%). No expression of SSEA-3 was observed in P19C6 cells. All three of the murine EC cell lines (PC19C6, PCC3/A/1, and F9) maintain self-renewal capacity in serum-containing medium in the absence of leukemia inhibitory factor or a feeder layer of cells, and they show a limited differentiation capacity into restricted lineage and/or mature cell types (35). Therefore, the observed differences in GSL-glycomic profiles may correlate with different developmental potentials for each EC cell type. Hierarchical cluster analysis yielded a close relation between NEC8 and HeLa cells, which may not be surprising considering that cancer cells often possess traits reminiscent of those ascribed to early embryonic cells.
In conclusion, the protocol described herein demonstrates the usefulness of qualitative and quantitative GSL-glycomics as a cellular descriptor. This protocol is anticipated to facilitate further studies to delineate characteristics of cell lines derived from normal and diseased tissues. The high sensitivity and ability to gain considerable structural information from minor components in a lipid mixture make this method an exciting advance in systems biology glycomics.
This work was supported in part by the Project for Developing Innovation Systems of the Ministry of Education, Culture, Sports, Science, and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
N. Fujitani, Y. Takegawa, Y. Ishibashi, K. Araki, J. Furukawa, S. Mitsutake, Y. Igarashi, M. Ito, and Y. Shinohara, submitted for publication.
- GSL
- glycosphingolipid
- SSEA
- stage-specific embryonic antigen
- Cer
- ceramide
- EGCase
- endoglycoceramidase
- GN4
- N,N′,N′′,N′′′-tetraacetyl chitotetraose
- Neu5Ac
- N-acetylneuraminic acid
- GlcNAc
- N-acetylglucosamine
- Neu5Gc
- N-glycolylneuraminic acid
- GnT-I
- β1–2-N-acetylglucosaminyltransferase I
- EC
- embryonic carcinoma
- aoWR
- aminooxy-WR
- SPG
- sialosyl paragloboside
- ES
- embryonic stem
- GM3
- NeuAcα 2,3Galβ1,4Glc-ceramide
- GM3(Gc)
- NeuGcα 2,3Galβ1,4Glc-ceramide
- GM2
- GalNAcβ1,4[Neu5Acα2,3]Galβ1,4Glc-ceramide
- GM2(Gc)
- GalNAcβ1,4[Neu5Gcα2,3]Galβ1,4Glc-ceramide
- GM1a
- Galβ1,3GalNAc[NeuAcα2,3]Galβ1,4Glc-ceramide
- GM1b
- Neu5Acα2,3Galβ1,3GalNAcβ1,4Galβ1,4Glc-ceramide
- GD1a
- NeuAcα2,3Galβ1,3GalNAcβ1,4[NeuAcα2,3]Galβ1,4Glc-ceramide
- GD1a(Gc)
- NeuGcα2,3Galβ1,3GalNAcβ1,4[NeuAcα2,3]Galβ1,4Glc-ceramide or NeuAcα2,3Galβ1,3GalNAcβ1,4[NeuGcα2,3]Galβ1,4Glc-ceramide
- LacCer
- Galβ1,4Glc-ceramide
- Gb3
- Galα1,4Galβ1,4Glc-ceramide
- Lc3
- GlcNAcβ1,3Galβ1,4Glc-ceramide
- aGM2
- GalNAcβ1,4Galβ1,4Glc-ceramide
- Lc4
- Galβ1,3GlcNAcβ1,3Galβ1,4Glc-ceramide
- aGM1
- Galβ1,3GalNAcβ1,4Galβ1,4Glc-ceramide
- nLc4
- Galβ1,4GlcNAcβ1,3Galβ1,4Glc-ceramide
- Gb4
- GalNAcβ1,3Galα1,4Galβ1,4Glc-ceramide
- Fuc-Lc4
- Galβ1,4[Fucα1,3]GlcNAcβ1,3Galβ1,4Glc-ceramide or Fucα1,2Galβ1,4GlcNAcβ1,3Galβ14Glc-ceramide
- Galα13-(n)Lc4
- Galα1,3Galβ1,3GlcNAcβ1,3Galβ1,4Glc-ceramide or Galα1,3Galβ1,4GlcNAcβ1,3Galβ1,4Glc-ceramide
- Galβ13-(n)Lc4
- Galβ1,3Galβ1,3GlcNAcβ1,3Galβ1,4Glc-ceramide or Galβ1,3Galβ1,4GlcNAcβ1,3Galβ1,4Glc-ceramide
- Gb5
- Galβ1,3GalNAcβ1,3Galα1,4Galβ1,4Glc-ceramide
- nLc5
- GlcNAcβ1,3Galβ1,4GlcNAcβ1,3Galβ1,4Glc-ceramideForrsman, Forrsman antigen with a structure of GalNAcα1,3GalNAcβ1,3Galα1,4Galβ1,4Glc-ceramide
- diFuc-Lc4
- difucosyl-Lc4 with a structure of Fucα1,2Galβ1,4[Fucα1,4]GlcNAcβ1,3Galβ1,4Glc-ceramide
- SPG
- Neu5Acα2,3Galβ1,4GlcNAcβ1,4Galβ1,4Glc-ceramide
- SPG(Gc)
- Neu5Gcα2,3Galβ1,4GlcNAcβ1,4Galβ1,4Glc-ceramide
- WR
- tryptophanyl arginine
- HexNAc
- N-acetylhexosamine.
REFERENCES
- 1. Hakomori S. I. (2008) Biochim. Biophys. Acta 1780, 325–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hakomori S. I. (2010) FEBS Lett. 584, 1901–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lanctot P. M., Gage F. H., Varki A. P. (2007) Curr. Opin. Chem. Biol. 11, 373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kannagi R., Cochran N. A., Ishigami F., Hakomori S., Andrews P. W., Knowles B. B., Solter D. (1983) EMBO J. 2, 2355–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kannagi R., Levery S. B., Ishigami F., Hakomori S., Shevinsky L. H., Knowles B. B., Solter D. (1983) J. Biol. Chem. 258, 8934–8942 [PubMed] [Google Scholar]
- 6. Liang Y. J., Kuo H. H., Lin C. H., Chen Y. Y., Yang B. C., Cheng Y. Y., Yu A. L., Khoo K. H., Yu J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 22564–22569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wiegandt H., Bücking H. W. (1970) Eur. J. Biochem. 15, 287–292 [DOI] [PubMed] [Google Scholar]
- 8. Hakomori S. I. (1966) J. Lipid Res. 7, 789–792 [PubMed] [Google Scholar]
- 9. Yowler B. C., Stoehr S. A., Schengrund C. L. (2001) J. Lipid Res. 42, 659–662 [PubMed] [Google Scholar]
- 10. Parry S., Ledger V., Tissot B., Haslam S. M., Scott J., Morris H. R., Dell A. (2007) Glycobiology 17, 646–654 [DOI] [PubMed] [Google Scholar]
- 11. Zhou B., Li S. C., Laine R. A., Huang R. T., Li Y. T. (1989) J. Biol. Chem. 264, 12272–12277 [PubMed] [Google Scholar]
- 12. Ito M., Yamagata T. (1989) J. Biol. Chem. 264, 9510–9519 [PubMed] [Google Scholar]
- 13. Furukawa J., Shinohara Y., Kuramoto H., Miura Y., Shimaoka H., Kurogochi M., Nakano M., Nishimura S. (2008) Anal. Chem. 80, 1094–1101 [DOI] [PubMed] [Google Scholar]
- 14. Nakashima N., Tamura T. (2004) Biotechnol. Bioeng. 86, 136–148 [DOI] [PubMed] [Google Scholar]
- 15. Izu H., Izumi Y., Kurome Y., Sano M., Kondo A., Kato I., Ito M. (1997) J. Biol. Chem. 272, 19846–19850 [DOI] [PubMed] [Google Scholar]
- 16. Uematsu R., Furukawa J., Nakagawa H., Shinohara Y., Deguchi K., Monde K., Nishimura S. (2005) Mol. Cell. Proteomics 4, 1977–1989 [DOI] [PubMed] [Google Scholar]
- 17. Sullards M. C., Allegood J. C., Kelly S., Wang E., Haynes C. A., Park H., Chen Y., Merril A. H., Jr. (2007) Methods Enzymol 432, 83—115 [DOI] [PubMed] [Google Scholar]
- 18. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Naven T. J., Harvey D. J. (1996) Rapid Commun. Mass Spectrom. 10, 1361–1366 [DOI] [PubMed] [Google Scholar]
- 20. Miura Y., Shinohara Y., Furukawa J., Nagahori N., Nishimura S. (2007) Chem. Eur. J. 13, 4797–4804 [DOI] [PubMed] [Google Scholar]
- 21. Yogeeswaran G., Sheinin R., Wherrett J. R., Murray R. K. (1972) J. Biol. Chem. 247, 5146–5158 [PubMed] [Google Scholar]
- 22. Visco V., Lucania G., Sansolini T., Dolo V., Garofalo T., Sorice M., Frati L., Torrisi M. R., Pavan A. (2000) Histochem. Cell Biol. 113, 43–50 [DOI] [PubMed] [Google Scholar]
- 23. Macher B. A., Klock J. C. (1980) J. Biol. Chem. 255, 2092–2096 [PubMed] [Google Scholar]
- 24. Stroud M. R., Handa K., Salyan M. E., Ito K., Levery S. B., Hakomori S., Reinhold B. B., Reinhold W. N. (1996) Biochemistry 35, 758–769 [DOI] [PubMed] [Google Scholar]
- 25. Krupnick J. G., Damjanov I., Damjanov A., Zhu Z. M., Fenderson B. A. (1994) Int. J. Cancer 59, 692–698 [DOI] [PubMed] [Google Scholar]
- 26. Rosales, Fritz V. M., Daniotti J. L., Maccioni H. J. (1997) Biochim. Biophys. Acta 1354, 153–158 [DOI] [PubMed] [Google Scholar]
- 27. Willison K. R., Stern P. L. (1978) Cell 14, 785–793 [DOI] [PubMed] [Google Scholar]
- 28. Nojiri H., Takaku F., Tetsuka T., Motoyoshi K., Miura Y., Saito M. (1984) Blood 64, 534–541 [PubMed] [Google Scholar]
- 29. Kannagi R., Papayannopoulou T., Nakamoto B., Cochran N. A., Yokochi T., Stamatoyannopoulos G., Hakomori S. (1983) Blood 62, 1230–1241 [PubMed] [Google Scholar]
- 30. Ito M., Ikegami Y., Tai T., Yamagata T. (1993) Eur. J. Biochem. 218, 637–643 [DOI] [PubMed] [Google Scholar]
- 31. Briles E. B., Li E., Kornfeld S. (1977) J. Biol. Chem. 252, 1107–1116 [PubMed] [Google Scholar]
- 32. Stanley P. (1980) ACS Symp. Ser. 128, 213–221 [Google Scholar]
- 33. Martin M. J., Muotri A., Gage F., Varki A. (2005) Nat. Med. 11, 228–232 [DOI] [PubMed] [Google Scholar]
- 34. Willison K. R., Karol R. A., Suzuki A., Kundu S. K., Marcus D. M. (1982) J. Immunol. 129, 603–609 [PubMed] [Google Scholar]
- 35. Kawazoe S., Ikeda N., Miki K., Shibuya M., Morikawa K., Nakano S., Oshimura M., Hisatome I., Shirayoshi Y. (2009) Dev. Growth Differ. 51, 81–93 [DOI] [PubMed] [Google Scholar]