Background: Deregulation of the protein kinase MARK has been linked to Alzheimer disease.
Results: Mark-specific inhibitors and a biosensor are identified.
Conclusion: The inhibitors and the biosensor are tools to provide new insights into the role of MARK during polarity establishment and maintenance of neurons.
Significance: The inhibitors might possess therapeutic potential by interfering with abnormal Tau phosphorylation in Alzheimer disease.
Keywords: Alzheimer Disease, Cell Differentiation, Fluorescence Resonance Energy Transfer (FRET), Protein Kinases, Tau, Biosensor, Cell Polarity, Fluorescence Lifetime Imaging (FLIM), Protein Kinase Inhibitors
Abstract
Protein kinases of the microtubule affinity regulating kinase (MARK)/Par-1 family play important roles in the establishment of cellular polarity, cell cycle control, and intracellular signal transduction. Disturbance of their function is linked to cancer and brain diseases, e.g. lissencephaly and Alzheimer disease. To understand the biological role of MARK family kinases, we searched for specific inhibitors and a biosensor for MARK activity. A screen of the ChemBioNet library containing ∼18,000 substances yielded several compounds with inhibitory activity in the low micromolar range and capable of inhibiting MARK activity in cultured cells and primary neurons, as judged by MARK-dependent phosphorylation of microtubule-associated proteins and its consequences for microtubule integrity. Four of the compounds share a 9-oxo-9H-acridin-10-yl structure as a basis that will serve as a lead for optimization of inhibition efficiency. To test these inhibitors, we developed a cellular biosensor for MARK activity based on a MARK target sequence attached to the 14-3-3 scaffold protein and linked to enhanced cyan or teal and yellow fluorescent protein as FRET donor and acceptor pairs. Transfection of the teal/yellow fluorescent protein sensor into neurons and imaging by fluorescence lifetime imaging revealed that MARK was particularly active in the axons and growth cones of differentiating neurons.
Introduction
Microtubule affinity regulating kinase (MARK)3 and its homologues like Par-1 together with Par-5 (14-3-3) and the Par-3·Par-6·aPKC complex belong to a set of conserved proteins that is essential for the establishment of cellular polarity in diverse organisms, worms, flies, and mammalian cells (for reviews, see Refs. 1–4). In humans there are four homologous isoforms of MARK (1–4) that are similar in their catalytic domain but differ mainly by their spacer domain (see Fig. 1A), alternative splicing, and expression pattern; the predominant MARK isoforms in neurons are MARK1 and -2 (5). MARK isoforms have a number of targets, among them the structural microtubule-associated proteins (MAPs) Tau, MAP2, and MAP4 (6), the MAP doublecortin (7), and several proteins that bind to the scaffold protein 14-3-3 after phosphorylation by MARK, for example Cdc25 (8) or KSR1 (kinase suppressor of Ras1) (Ref. 9; for review, see Ref. 10). In the brain one important role of MARK2 is in the control of neuronal migration during brain development, which is in turn linked to the dynamics of the cytoskeleton (11, 12). The structures of the catalytic and the ubiquitin-associated domains and C-terminal “kinase-associated” domain of MARK isoforms have been solved by x-ray crystallography and NMR spectroscopy, respectively (13, 14), whereas the N-terminal header and the central spacer domains appear to be largely disordered (see Fig. 1A). Within the human kinome, MARKs are part of the AMPK subfamily within the calcium/calmodulin-dependent kinase II group of kinases (15), which includes AMPKα1 and -2 and BRSK1 and -2 (also known as SAD-A and -B), some of which are also prominent in neurons and may have partially overlapping functions (16). In cells the catalytic activity of MARK is regulated by intramolecular domain interactions (e.g. ubiquitin-associated domain and kinase associated domains), phosphorylation by other kinases (activating: MARKK, LKB1 (17, 18); inhibiting: GSK3β (19)), interactions with other cytoskeleton-directed kinases (PAK5 (p21-activated kinase 5), TESK (testis-specific kinase) (20, 21)), or by interactions with scaffold proteins such as 14-3-3 (22).
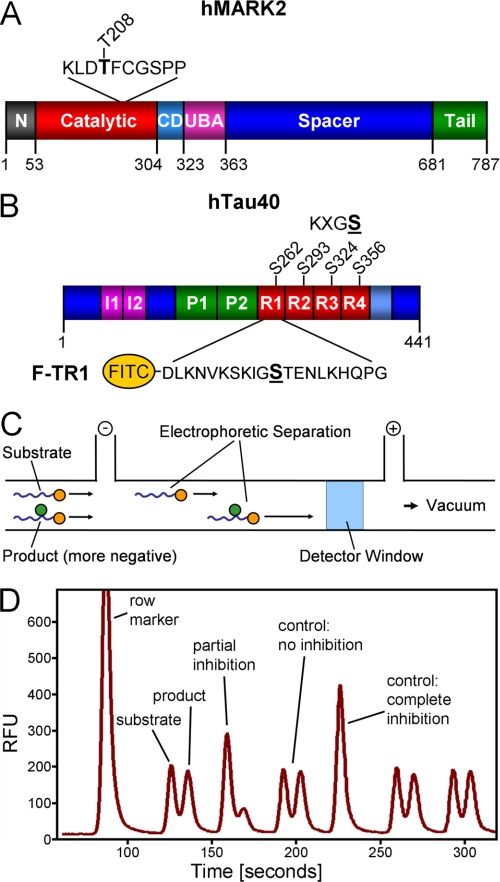
FIGURE 1.
Bar diagrams of MARK and hTau40. A, shown is a bar diagram of MARK2 as a representative for the four mammalian MARK isoforms. All MARKs share a similar architecture with high homologies in the catalytic domain, common docking site (CD), ubiquitin-associated domain (UBA), and kinase-associated domain. Variations are found mostly in the spacer domain and the N-terminal header (N). The activation loop in the catalytic domain contains a critical threonine (Thr-208 in MARK2, indicated above), whose phosphorylation by MARKK or LKB1 is required for full activity. By contrast, phosphorylation of Ser-212 by GSK3β is inhibitory. B, shown is FITC-labeled peptide derived from the first repeat of the Tau protein containing the residue Ser-262 (underlined) that is phosphorylated by MARK isoforms. C, shown is a schematic drawing of the separation of substrate and product by capillary electrophoresis on the LabChip™. The chip holds 12 parallel channels. D, shown is an example of a chromatogram showing substrate and product peaks of sample with no inhibition or partial and complete inhibition. RFU, relative fluorescence units.
One of the prominent targets of MARK is Tau and the related proteins MAP2 and MAP4 within the KXGS motifs of their microtubule binding domains (see Fig. 1B). As a result, the affinity of these MAPs to microtubules is strongly decreased, and their function of stabilizing microtubules can no longer be performed. Although the dynamic instability of microtubules is a desired feature during the process of polarization of a cell and for the outgrowth of cell processes, neurites, and dendritic spines (23), the delicate balance of MARK and the counteracting phosphatases can easily be disturbed, with detrimental effects to the cell. Either the excess of bound Tau can block access of motor proteins to microtubules, which serve as tracks for cellular transport (24), or the phosphorylated Tau is not capable to stabilize the microtubules. In both cases there is transport inhibition, and important supplies of material and energy for the synapses cannot take place, and the cell extensions decay. In addition, Tau molecules not bound to microtubules can undergo missorting and aggregation. These processes represent hallmarks of Alzheimer disease, where MARK-dependent phosphorylation occurs at an early stage (25) and MARK protein itself is elevated in the pathological Tau aggregates (26). This increased MARK-induced phosphorylation of Tau can also be detected in the brains of Tau transgenic mice as an early sign of degeneration (27, 28). However, it is currently not clear where and how the overactivation of MARK is initiated, and there are thus far no specific inhibitors of the kinase. This prompted us to begin a search for low Mr inhibitors of MARK and to develop reagents that enable the observation of MARK activity and MARK function in living neuronal cells.
EXPERIMENTAL PROCEDURES
Kinase Assays
Kinase activities were assayed in buffer B (50 mm Tris-HCl, pH 8.0, 5 mm MgCl2, 2 mm EGTA, 0.5 mm PMSF, 0.5 mm DTT, 0.5 mm benzamidine) for 60 min at 30 °C. Final concentration of [32P]ATP (3.7 × 107 MBq/mol) was 100 μm, and Tau peptide TR1, histone H1, or hTau40 was 100, 7.5, and 2.5 μm, respectively. Reactions were stopped by the addition of sample loading buffer and subjected to SDS-PAGE. After staining with Coomassie (RothiBlueTM), gels were dried, and radioactivity was detected with a BAS3000 phospho-imaging system (Raytest). Activities toward MARK-AR1 were assayed with 2.5 μm recombinant MARK-AR1, and the reaction was followed with a Tecan Safire by a change in the emission ratio of 523 and 476 nm from 1.08 to 1.94 (see Fig. 6, C and D). Inhibitors were used at a final concentration of 5 μm (HD01) and 20 μm (30199 and 39621). For quantification of phosphorylation (see Fig. 6D), an aliquot of the reaction mixture was removed every 3 min and analyzed by SDS-PAGE as described above. Recombinant MARK2 (residues 39–365 containing the catalytic domain with N-terminal His tag; 1 unit = 55 nmol/min/mg) was expressed in Escherichia coli as described (13). A full-length clone of SAD kinase B was obtained from Deutsches Ressourcenzentrum für Genomforschung GmbH (RZPD, Berlin, Germany) and expressed in Sf9 cells. Final concentrations used were 7.3 nm MARK2-(39–365), 4.3 nm SAD-B, 8.3 nm PKA (Promega), 9.3 nm calcium/calmodulin-dependent kinase I (New England Biolabs), 20 nm MAPKAPKII (Invitrogen), 5.3 nm AMPK (α1β1γ1), 4.1 nm ChkI, and 3.8 nm ChkII (all three from Cell Signaling). Data show averages of three experiments (bars = S.E.).
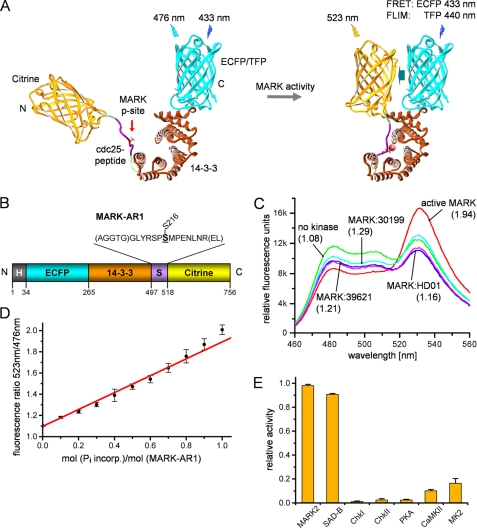
FIGURE 6.
Schematic diagram and in vitro properties of the MARK activity reporter. A, a MARK-specific target peptide (derived from residues 210–223 of Cdc25C, GLYRSPSMPENLNR) containing the phosphorylation site Ser-216 is fused via linkers (in parentheses) to 14-3-3 protein (brown), with ECFP and YFP (citrine) as a FRET pair at both ends. Upon phosphorylation, the target sequence binds into the pocket of 14-3-3, which closes up and generates an enhancement of the FRET signal. TFP, teal fluorescent protein. B, shown is a bar diagram of the MARK activity reporter (MARK-AR1). For in vitro studies, the protein was labeled with a polyhistidine tag (H; gray), allowing simple and rapid purification. N and C denominate the N and C termini of the construct. C, shown is a wavelength scan of the unphosphorylated and phosphorylated MARK-AR1. The fluorescence of the unphosphorylated MARK-AR1 shows a maximum at 476 nm (green curve), whereas the maximum for the phosphorylated protein is shifted to 523 nm due to FRET (red curve). The inhibitors HD01 (blue curve), 30199 (cyan curve), and 39621 (pink curve) prevent the phosphorylation by MARK. Inhibitor concentrations were 5 μm (HD01) and 20 μm (30199 and 39621). Shown in parentheses is the ratio of the emission at 523 nm to the emission at 476 nm. D, the ratio of the emission at 523 nm to the emission at 476 nm can be taken as a measure of activity, as it shows a linear relationship to the phosphorylation of the activity reporter. The x axis displays the amount of incorporated radioactively labeled phosphate into the MARK-AR1, whereas the y axis shows the corresponding emission ratios, ranging from 1.08 (no phosphorylation) to 1.94 (complete phosphorylation of the target site). E, MARK-AR1 can be phosphorylated by MARK and SAD-B. Other kinases reported to phosphorylate Cdc25C cannot effectively phosphorylate this activity reporter.
Compound Screen
MARK2 (7.3 nm) was preincubated for 15 min with 50 μm compound in buffer C (50 mm BES, pH 7.0, 5 mm MgCl2, 100 μm NaCl2, 0.03% Tween 20, 1 mm DTT, 1 μm FITC-labeled Tau peptide). The reaction was started by the addition of 40 μm ATP and stopped after 120 min at 25 °C by the addition of 10 mm EDTA. The products were analyzed with a LabChip3000 (CaliperLifeScience). Hits were verified by the same assay by using an assay with radiolabeled ATP as described (17) and by measuring the dose dependence of the MARK2 activity.
MALDI-TOF Analysis
Compounds were mixed with matrix solution (10 mg/ml 2,5-dihydroxybenzoic acid in water) to give a final concentration of 100 μm, pipetted onto a steel target-plate, and dried in the vacuum. Analysis was performed on a Voyager System 4217 (Applied Biosystems) made available by EMBL in Hamburg. The instrument was run in the linear positive mode with delayed extraction (accelerating voltage, 20,000 V; grid voltage, 94%; extraction delay time, 100 nc).
Cell Culture and Immunofluorescence
CHO Wild Type
CHO wild type cells or cells stably transfected with hTau40 were grown in Ham's F-12 medium with 10% FCS and incubated in a humidified atmosphere containing 5% CO2 at 37 °C. For stable transfection we added G418 (600 μg/ml, Invitrogen) to the culture medium. Cells were seeded at 70% confluency in 24-well plates (1.8 cm2) on coverslips, and MARK2wt, inactive MARK2 mutants, or MARK-AR1 were transfected with Lipofectamine 2000 (Invitrogen) for 4 h followed by the addition of compound for 18 h. Cells were fixed with 3.7% formaldehyde for 15 min at room temperature. The cells were then washed with PBS (3 times). Permeabilization was carried out by adding 80% ice-cold methanol and incubated for 5 min at −20 °C. Cells were washed 3 times with PBS and blocked with 10% goat serum at 37 °C for 45 min. After blocking, the cells were treated with HA-tag antibody (1:250, Sigma) for 1 h or overnight at 37 °C and then washed 3 times with PBS. The appropriate Cy3-tagged secondary antibody was added, and the cells were incubated at 37 °C for 1 h followed by washing 3 times with PBS. Finally the coverslips were mounted for microscopy and analyzed with a LSM510 Meta confocal microscope (Zeiss, Jena, Germany).
For quantification of Tau phosphorylated at the KXGS motifs and for the determination of cell shape and size, cells stably transfected with hTau40 were treated as described above and then stained with 12E8 antibody (1:200) for detection of pTau and with YL1/2 for tubulin (1:1000, Serotec). Approximately 100 fields were scanned with a Leica DMI 4000B Metamorph-based microscope using Metamorph scanslide application. ECFP-MARK2wt or ECFP-MARK2A2-transfected cells were selected, and average fluorescence intensities, and cell size were measured using Metamorph software. 12E8 background signals from MARK2A2-transfected cells were subtracted from MARK2wt-transfected cells. 35–45 cells were analyzed for each transfection and treatment. Statistical analysis was done using analysis of variance with post-hoc Dunnett's Multiple Comparisons Test.
PC12 Cells
PC12 cells were grown in a medium containing Dulbecco's modified Eagle's medium (DMEM), 4500 mg/liter glucose, 1% l-glutamine, 10% FCS, 15% HS, and 100 μg/ml penicillin/streptomycin. 2 × 104 cells per coverslip (2 cm2; precoated overnight with poly-d-lysine) were incubated at 37 °C with 5% CO2. MARK-AR1 was transfected by treatment with Lipofectamine 2000 (Invitrogen) for 4 h followed by the incubation in growth medium for 18 h. The differentiation of PC12 cells was carried out in differentiation medium (DMEM:F-12 = 1:1) containing 0.1% serum and 100 ng/ml NGF for the depicted period of time. Living or fixed cells were analyzed by fluorescence lifetime microscopy (FLIM).
Cortical Neurons
Cortex tissue dissected from E18 rat embryos was digested with 0.05% trypsin for 7 min. Plating medium was added, and cells were allowed to settle and then were resuspended in HBS and mechanically dissociated. Neurons were plated at a density of 100 neurons/mm2 on coverslips (2 cm2) precoated overnight with poly-d-lysine. After culturing for 4 h, the medium was changed to neuronal culture medium (Neurobasal medium with B-27 and l-glutamine), and the cells were grown for 8 days. MARK-AR1 was transfected by treatment with Lipofectamine 2000 (Invitrogen) for 1 h followed by incubation in culturing medium for 48 h. Living cells were analyzed by FLIM before and after treatment with compounds.
FLIM Assay
A pulsed diode laser driver PDL 800-B (PicoQuant, Berlin, Germany) was used to drive a picosecond 440-nm diode laser (PicoQuant) to excite the donor (ECFP, mTFP (mono-teal fluorescent protein)) at a rate of 40 MHz. A dual channel SPAD (Single Photon Avalanche Diodes, MPD, Bolzano, Italy) detection unit (PicoQuant) equipped with an emission filter (480/40 nm) was used to detect the emitted photons. Pico Harp 300 (PicoQuant) was applied as the TCSPC (time-correlated single photon counting) instrument. The scan of the image was controlled by an Olympus FluoView FV1000 confocal laser scanning microscope (Olympus Deutschland, Hamburg, Germany). The images were focused using the YFP channel (which is less sensitive to bleaching). To increase the signal to noise ratio, the FLIM data were recorded with a widened pinhole (400 μm). This allowed us to reduce the time of measurement to 30–60 s, which in turn reduced phototoxicity and photobleaching especially during live cell imaging. The reconstruction of time-resolved fluorescence decays and the data analysis were performed using SymphoTime software (PicoQuant).
RESULTS
Identification and Verification of MARK Inhibitors
Because of the general similarity of kinase catalytic domains, inhibitors developed against a given kinase often react with other kinases as well. For example, earlier studies (in collaboration with L. Meijer, Roscoff, France; see Ref. 41) had shown that some compounds that were initially developed to inhibit proline-directed kinases such as GSK3β, Cdc2, or Cdk5 turned out to be potent inhibitors of MARK. In particular, compound HD01 (developed as an inhibitor of GSK3β) potently inhibits MARK with an IC50 value of 0.67 μm (23). Similarly, the GSK3β inhibitors SB-216763 and SB-415286 show cross-reactivity with MARK and its activating kinase MARKK (Ref. 29 and own observations). Thus, for a better understanding of the role of MARK during neuronal cell polarity, we sought to develop more specific modifiers of MARK activity, particularly those that distinguish different kinases that phosphorylate Tau.
A primary screen was performed with the ChemBioNet compound collection (17951 compounds, Mr 350–500 Da), designed at the Leibniz Institute for Molecular Pharmacology after analysis of the World Drug Index for privileged substructures or scaffolds (30). For the screening, MARK was incubated with a fluorescence-tagged Tau-derived peptide (20-mer, corresponding to residues 252–271 containing the target Ser-262, with FITC conjugated to the N terminus; Fig. 1B) in the presence or absence of inhibitor compounds in a 384-well format. Reactions were terminated by the addition of EDTA, limiting the turnover of the substrate to one-tenth of the total amount. This was followed by a chip-based analysis of the kinase reaction using a LapChip™3000 system (31). The charge difference between the phosphorylated and unphosphorylated peptides leads to a separation into two peaks (Fig. 1, C and D), which were analyzed quantitatively with the HTS Well AnalyzerTM software (Caliper Life Science).
When applying a threshold level of 75% inhibition, 96 hits were found. These potential inhibitors were verified in a second run and characterized by measuring their IC50 values. Among the 96 compounds, 12 exhibited potent inhibition of MARK2, with IC50 values ranging from 0.4 to 10 μm in the presence of 100 μm ATP (for dose-response curves, see Fig. 2A; for IC50 values, see Table 1). All compounds are ATP competitors, as their efficiency declines with increasing concentrations of ATP. By contrast, increasing the concentration of the substrate peptide TR1 did not interfere with inhibition (Fig. 2C). The identities of the 12 substances were confirmed by MALDI-TOF analysis.
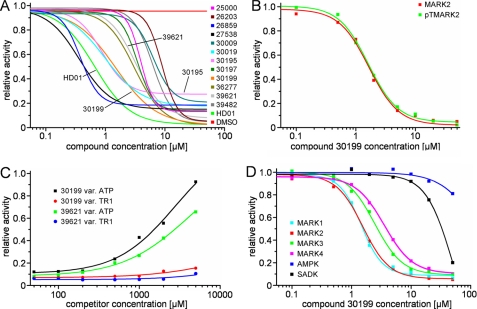
FIGURE 2.
Dose-response curves of the 12 best compounds in comparison with compound HD01 (green line) and DMSO (red line). A, dose-response curves were measured at an ATP concentration of 100 μm. Note that some compounds (e.g. 30195) do not completely inhibit MARK, even at 50 μm (probably due to limited solubility in aqueous solutions). B, inhibition efficiency is not altered by full activation of MARK with MARKK (shown here for compound 30199). C, inhibition, shown here for compounds 30199 and 39621 at a concentration of 20 μm, is released with increasing concentrations of ATP (black and green curves) but not with increasing concentrations of the substrate peptide TR1 (red and blue curves). This indicates an ATP competitive mode of action of the compounds. D, compound 30199 inhibits MARKs but not SADK and AMPK. The compound does not discriminate between the individual MARK-isoforms (cyan, red, green, and magenta) but is far less effective against AMPK and SADK-B (blue and black).
TABLE 1.
IC50 values of inhibitory compounds
| Compound ID | IC50 value |
|---|---|
| μm | |
| 25000 | 4.2 |
| 26203 | 9.4 |
| 26859 | 0.5 |
| 27538 | 0.4 |
| 30009 | 7.8 |
| 30019 | 1.4 |
| 30195 | 1.3 |
| 30197 | 3.5 |
| 30199 | 1.6 |
| 36277 | 3.4 |
| 39621 | 3.6 |
| 39482 | 6.6 |
| HD01 | 0.7 |
Cross-reactivity of MARK Inhibitors with Other Tau-related Kinases
To ascertain the specificity, the compounds were subjected to a test of cross-reactivity with selected kinases that also are known effectors of Tau protein (Fig. 3). First, certain compounds inhibiting GSK3β show cross-reactivity with MARK (HD01, SB-216763, SB-415286 (23, 29)). Second, SAD kinase B (also known as BRSK1), a close relative of MARK, lies in the same phylogenetic branch of the calcium/calmodulin-dependent kinase II group of kinases (15) and shows cross-reactivity with some MARK inhibitors (at elevated concentrations; see below and Fig. 2D). The third example of cross-reactivity is seen with MARKK (also known as TAO-1), the activating kinase of all MARK isoforms (17). When testing cross-reactivity, we also included Cdc2/cyclin B, Cdk5/p25, and p38/SAPK as representatives of kinases involved in the regulation of important cellular processes capable of phosphorylating Tau at Ser/Thr-Pro motifs. The compounds were tested at a concentration of 50 μm, and most of them exhibited no or very mild inhibition toward MARKK, p38/SAPK, and Cdc2/CycB (except compound 26859), whereas HD01 affected all kinases tested, even MARKK and p38/SAPK (inhibition of 50%). GSK3β was only affected by compounds 25000, 39482, and HD01 (and slightly by compound 39621). In contrast, SAD kinase B (also known as BRSK1) was effectively inhibited by compounds HD01, 27538, 30009, 36277, and 39482 but not by 26203 and 39621. Both compounds turned out to be specific for MARK compared with the other set of kinases tested. The other compounds showed only a mild effect on SAD kinase B activity.
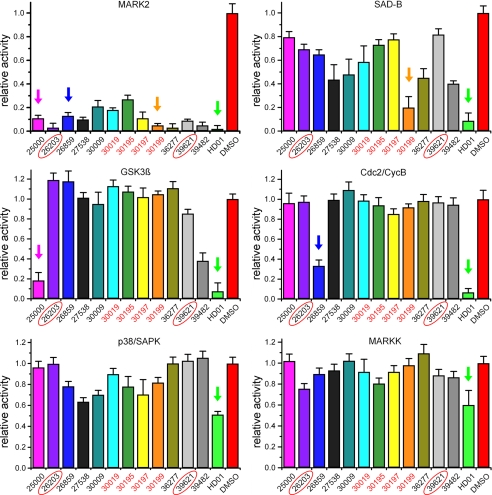
FIGURE 3.
Cross-reactivity of compounds with selected kinases. Although HD01 inhibits all kinases tested (green arrows), the identified compounds exhibit distinct specificities. Thus, compound 25000 not only inhibits MARK (IC50 = 4.2 μm, Table 1) but also GSK3β (IC50 = 7.5 μm, pink arrow). The compounds 30019, 30195, 30197, and 30199 (red) share a similar structure and show only small effects on SAD-B (orange arrow), a close relative of MARK. Substance 26859 has a completely different structure and has only moderate effects on the other kinases except Cdc2 (blue arrow), whereas substances 26203 and 39621 are specific for MARK (red circles).
Next we tested whether the MARK-selective substances could discriminate between the four MARK isoforms. The dose-response curves for compound 30199 (Fig. 2D) showed inhibition of all MARK isoforms at a low (1.6–3.0) micromolar concentration. By contrast, the IC50 values for SAD-B and AMPK (both relatives of MARK and involved in the regulation of the energy level in cells (32)) were far greater than 30 μm, emphasizing the distinct specificities of these kinases.
The compounds 30019, 30195, 30197, and 30199 share a 9-oxo-9H-acridin-10-yl as a functional group (Fig. 4). None of them affected GSK3β, and they showed only little cross-reactivity with SAD-B and AMPK. Therefore, this common functional group structure is a promising basis for optimization of compounds by combinatorial chemistry.
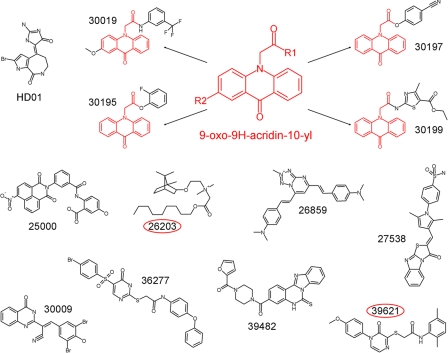
FIGURE 4.
Structures of the identified inhibitory compounds. Shown is a comparison of the structures of HD01 and the identified inhibitors. Four of the compounds (30019, 30195, 30197, and 30199) share a 9-oxo-9H-acridin-10-yl functionality (structure shown in red). The red circles highlight the MARK-specific compounds 26203 and 39621.
Inhibition of MARK Activity in Cells
The compounds showing low cross-reactivity with other kinases were tested in a cellular assay. Chinese hamster ovary (CHO) cells were transfected with ECFP-MARK2 and kept in medium for 18 h supplemented either with 10 μm concentrations of inhibitor compounds, compound HD01, or DMSO as a control. As shown previously (5), expression of active MARK in CHO cells leads to the disruption of the microtubule network, and subsequently, the cells shrink, round up, and finally die (Fig. 5A). The effect is explained by the destabilization of microtubules after the phosphorylation of MAPs; this can be counteracted by microtubule stabilizers (taxol) or by increasing the effective concentration of MAPs (6). Consistent with this, in the present experiments, the MARK2-dependent toxicity was rescued by treatment of the cells with the inhibitor 39621 (Fig. 5C), which suppressed MARK2 activity and phosphorylation of MAPs, and thus, prevented microtubule breakdown as shown by almost normal cell size (Fig. 5D) and a reduced level of Tau phosphorylation at KXGS sites (Fig. 5E). As a control, the transfection of an inactive mutant of MARK2 (altered by changing the regulatory loop, T208A/S212A (18)) had no effect on the microtubule network and toxicity, confirming that the effect was indeed due to phosphorylation by MARK2 (Fig. 5B). Treatment of MARK2-transfected cells with the inhibitors described above revealed that the four compounds with similar structure (30019, 30195, 30197, 30199; Fig. 4) were the most potent ones in protecting the microtubule network along with compound 39621 (Fig. 5, C–E). Compound 26203 (not shown) was highly specific for MARK in the in vitro assay yet rescued the microtubule network in cells only partially, possibly because of its relatively high IC50 value (9.4 μm) or poor cell penetration.
FIGURE 5.
Effects of MARK2 inhibitors in cellular assays. A and B, MARK2 wild type or MARK2 inactive mutant (A2) was expressed in stably hTau40-transfected cells. The cyan fluorescent protein channel highlights MARK-transfected cells, Ser(P)-262-Tau (Tau-pS262; 12E8 epitope, KXGS phosphorylation) is shown in green, and tubulin (Yl1/2) is in red. A, transfection of active wild type MARK2 results in increased Ser(P)-262-Tau signals (see arrows; compare with non-transfected cells), breakdown of microtubules, and reduced cell size. B, transfection of the inactive mutant of MARK2 (MARK2A2) does not increase KXGS phosphorylation and does not alter the cell shape (see the arrows). C, treatment of cells transfected by active MARK with the inhibitor 39621 results in the restoration of the microtubule network and reduction of phosphorylation at KXGS sites (12E8 signals), thus, suppressing the toxicity caused by MARK2 (see the arrows). D, quantification of cell size relative to MARK2A2-transfected cells is shown. E, quantification of pTau (12E8 immunofluorescence intensities units) after background subtraction (12E8 signals in MARK2A2-transfected cells) is shown. Error bars: S.E. from 35–45 cells for each condition. * indicates p < 0.05; *** indicates p < 0.001 (analysis of variance with post-hoc Dunnett's Multiple Comparisons Test versus control (MARK2wt)). Scale bars: 10 μm.
Construction of a FRET-based Reporter of MARK Activity in Cells
The distribution and activity of neuronal signaling molecules is usually not homogeneous, and therefore, we sought to develop a reporter that would reflect the activation state as a function of time and location within living cells. To study the effects of the inhibitors on endogenous levels of MARK in living cells, especially neurons, we decided to create a biosensor of MARK activity. We adapted the principle of the PKA activity reporter (33) that uses FRET as a readout of phosphorylation of a specific substrate. As a target substrate we chose the peptide 210–223, derived from Cdc25C and containing Ser-216, which is efficiently phosphorylated by all MARK isoforms (8).4 The reporter was constructed by linking (from N to C) ECFP, the phosphopeptide binding domain of 14-3-3, the substrate peptide, and citrine (a stable variant of YFP). The resulting protein, therefore, contains ECFP and citrine on opposing ends of the fusion construct (Fig. 6B). Upon phosphorylation the phosphopeptide binds into the pocket of 14-3-3. This in turn induces a conformational change in 14-3-3, which brings the two fluorescent end ligands into close proximity, resulting in a FRET signal by transfer of energy from ECFP to citrine (Fig. 6A). Free ECFP is characterized by excitation/emission maxima of 433/476 nm (Fig. 6C, green curve), but when citrine moves into close vicinity, the emission of CFP is used to excite citrine, which can be observed by emission at 523 nm. Thus, phosphorylation of the target peptide can be observed by an increase of citrine fluorescence at 523 nm (Fig. 6C, red curve).
To ascertain the mode of operation of the reporter construct, we performed the assay in the presence of MARK inhibitors, e.g. HD01 (5 μm; IC50 value is 0.67 (23)) and 30199 and 39621 (both 20 μm). All compounds efficiently suppressed the FRET effect as expected (Fig. 6C, blue, cyan, and pink curves). The FRET efficiency correlated well with the extent of phosphorylation of the activity reporter, as measured by incorporation of radioactively labeled phosphate (Fig. 6D). Thus, the reporter system is suitable as a fluorescence tool to screen for MARK inhibitors (data not shown). Furthermore, to examine the specificity of the substrate peptide for MARK activity, we assayed several kinases known to phosphorylate Cdc25C at Ser-216, such as checkpoint kinases 1 and 2, PKA, calcium/calmodulin-dependent kinase II, and mitogen-activated protein kinase-activated protein kinase 2 (MAPKAPK2) (34); in addition, we tested SAD kinase B/BRSK1, a kinase closely related to MARK. Among these, only SAD-B could induce FRET in the activity reporter to a similar extent as MARK (Fig. 6E), illustrating that the reporter discriminates between different kinases.
The next step was to test the functionality of the activity reporter in cells. CHO cells were co-transfected with MARK-AR1 and the kinase MARK2. Instead of observing intensity-based FRET, we decided to detect FRET by FLIM, which records the time between fluorophore excitation and photon emission. In contrast to the sensitized emission approach, FLIM is less dependent on the expression level of the donor-acceptor pair because the lifetime is independent of the number of fluorescing molecules. Changes in the lifetime can be solely attributed to changes in the environment of the donor fluorophore, such as proximity to the acceptor, ion concentrations, and temperature (35). Cells were fixed after 20 h and imaged by FLIM. If energy transfer occurs from the donor (ECFP) to the acceptor (citrine), the lifetime of the donor is reduced, which is related to FRET efficiency and the fraction of donor-acceptor interactions. In cells transfected with the activity reporter alone (Fig. 7, A–D) the donor (ECFP) has a long lifetime of ∼2.42 ns (Fig. 7I), whereas in cells co-transfected with MARK (Fig. 7, E–H), it has a much shorter lifetime, averaging ∼2.18 ns (Fig. 7I).
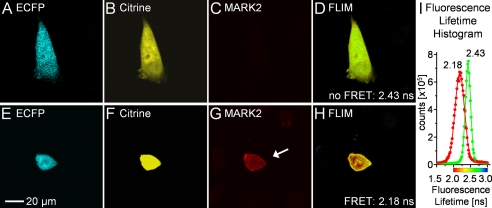
FIGURE 7.
Functionality test of MARK-AR1 in CHO cells. CHO cells were transfected with MARK-AR1 and HA-tagged MARK2. 20 h after transfection cells were fixed and imaged. The upper panel shows both channels of the fluorescence intensity image (A and B) of a cell transfected with MARK-AR1-only, hence, no HA/Cy3-staining of MARK2 in C and the pseudo-colored fluorescence lifetime (FLIM) in D. The green color in D corresponds to a long fluorescence lifetime of 2.43 ns (no FRET). The cell shown in the lower panel is co-transfected with MARK-AR1 (E and F) and HA-MARK2 (G). The short (2.18 ns) fluorescence lifetime in the presence of active MARK is shown as red (high FRET) in H. The graph (I) displays the averaged histograms of cells showing FRET (red dots, n = 8) or no FRET (green dots; n = 8) and gaussian fits of the data. The peaks are centered at 2.182 ± 0.001 ns (FRET) and 2.427 ± 0.001 ns (no FRET).
Endogenous MARK Activity in Differentiating Neuronal Cells
A key goal of developing the activity reporter(s) was to observe the endogenous activity of MARK in differentiating neurons, as kinases of the MARK/Par1 are thought to play a pivotal role in neuronal development. Fig. 8 illustrates this for the case of PC12 cells differentiated with NGF. Cells were transfected with the activity reporter, differentiated after 18 h with NGF for 1 h, and fixed with formaldehyde. The fluorescence intensity image shows the overexpression and overall distribution of MARK-AR1 within the cell and the clear neurite outgrowth in the transfected cell after 1 h of NGF differentiation (Fig. 8). The FLIM image from the same cell shows the shortest donor lifetime in the terminals of the neurite (Fig. 8C, inset), indicating strong local activity of endogenous MARK. The fluorescence lifetime histogram ranges from 2.4 ns throughout the cell to 2.2 ns at the neurite terminals. The results demonstrate that MARK/Par1 activity is related to the elaboration of neurites.
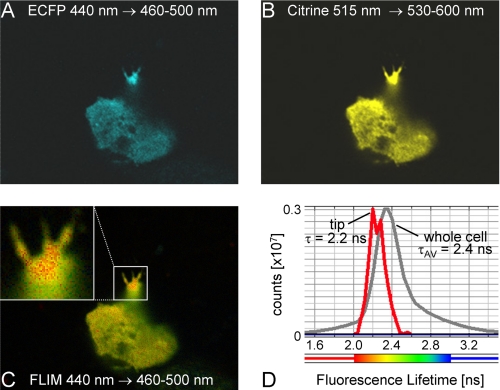
FIGURE 8.
Endogenous MARK activity is localized in the termini of the neurites in differentiated PC12 cells. PC12 cells were transfected with MARK-AR1. 18 h after transfection cells were differentiated with 100 ng/ml NGF for 1 h and fixed. Fluorescence intensity image shows the expression and distribution of transfected MARK-AR1 (A, ECFP; B, citrine). The fluorescence lifetime image (C) reveals the strongest FRET signal in the termini of the neurite, indicating the presence of local MARK activity. The whole cell has an average lifetime of 2.4 ns (D, gray curve), whereas the tip of the neurite has an average lifetime of 2.2 ns (C, inset; D, red curve).
Further improvement of the activity reporter was achieved by circumventing some of the disadvantages of ECFP. This fluorophore is robust for in vitro studies and for measuring FRET by sensitized emission, but it is not optimal for FLIM. ECFP has two excitation (433 and 451 nm) and two emission maxima (475 and 504 nm), that require a multiexponential fit of the fluorescence lifetimes and thus a loss in readout accuracy. By comparison, cerulean can be fitted adequately as a single exponential decay but suffers from decreased photostability (36). We, therefore, replaced the ECFP with the turquoise variant mTFP, which is one of the brightest and most photostable fluorescent proteins reported to date (43). In addition, the fluorescence is insensitive to physiologically relevant pH changes, and the fluorescence lifetime decay is best fitted as a single exponential. Its single transition excitation and emission maxima are 462 and 492 nm, respectively.
In mammalian neurons MARK phosphorylates the axon specific microtubule-associated protein Tau and other related MAPs at serines within KXGS motifs in the microtubule binding domain, causing the detachment of MAPs and the disassembly of microtubules. The proper regulation of this type of phosphorylation is important for the dynamic behavior of axonal microtubules in neurons. To detect the endogenous MARK activity in living neurons, we transfected primary cortical neurons prepared from E18 rats with MARK-AR1 at 8 DIV and performed live cell FLIM at 10 DIV. MARK activity can be observed in cell bodies, dendrites, and proximal regions of axons (Fig. 9A); however, the most remarkable observation is the gradient of the endogenous MARK activity in the distal region of the axons, with the highest activity in the growth cones. Note that in the cell shown here, only one growth cone exhibits high MARK activity (≤1.9 ns), whereas the other two, regarded as “inactive,” have lower MARK activities (≥2.0 ns, for details, see Table 2). In contrast, cells that were transfected with a non-phosphorylate-able mutant of the activity reporter (Fig. 9B1) show a single lifetime of 2.2 ns in the cell body (apart from a few aggregates), axon shafts, and in the growth cones (Fig. 9, B2 and B3).
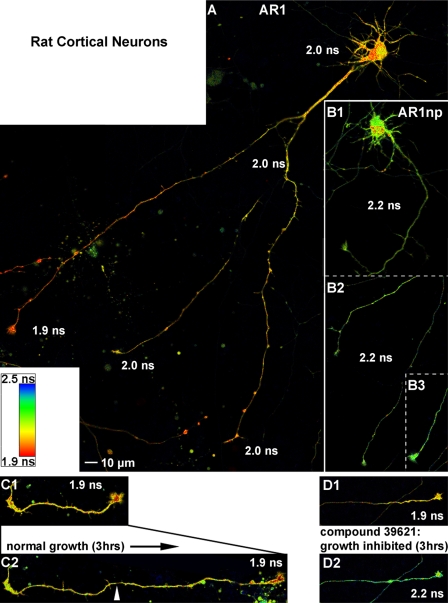
FIGURE 9.
The MARK activity reporter detects endogenous MARK activity in living neurons. A, primary cortical neurons (8 DIV) from E18 rats were transfected with MARK-AR1. Live cell FLIM images were taken at 10 DIV. Strongest MARK activity is detected in fast migrating, “active” growth cones (1.9 ns) with a decreasing gradient of the kinase activity down the axon. A second group of growth cones displays lower activity (2.0 ns) and migration speed (inactive). There is also endogenous MARK activity in the cell body, dendrites, and the proximal region of the axon. B, cells expressing the non-phosphorylate-able mutant MARK-AR1np display a homogeneous higher lifetime (2.2 ns) of the donor, representing the situation of a non-FRET activity reporter. This demonstrates that the FRET signals shown in cells with the wild type activity reporter are dependent on phosphorylation and not on other intra- or intermolecular interactions. In addition it simulates the situation of no MARK-activity in the cells. C and D, MARK-AR1 displays the effect of MARK inhibitors in living cells. 8 DIV primary cortical neurons from E18 rats were transfected with MARK-AR1 and incubated either with DMSO as control (C1 and C2) or with the specific MARK-inhibitor 39621 (D1 and D2). Cells were imaged before (upper images) and 180 min after addition of chemicals (lower images). In the control cell the active growth cone advanced during incubation. The activity of MARK, indicated by a short lifetime of fluorescence (1.9 ns) in the growth cone, was not reduced (C1 and C2). Treatment with the specific inhibitor 39621 resulted in a stop of growth accompanied by an increase of lifetime from 1.9 to 2.2 ns in the growth cone, shown by a shift of the pseudo-colored image from red (D1) to green (D2). For detailed fluorescence lifetimes and migration speeds, see Table 2.
TABLE 2.
Fluorescence lifetime in different parts of the neurons and effect of treatment with the MARK inhibitor 39621
| Fluorescence lifetime (ns) | Active growth cones | Inactive growth cones | Axon shaft | Cell body |
|---|---|---|---|---|
| Activity reporter | 1.897 ± 0.007 | 2.016 ± 0.017 | 2.020 ± 0.010 | 1.974 ± 0.013 |
| Non-phosphorylate-able reporter (ARnp) | 2.219 ± 0.007 | 2.223 ± 0.009 | 2.199 ± 0.011 | |
| After 3 h | Active growth cones | Inactive growth cones | 39621-treated growth cones | n = 15–25 |
| Fluorescence lifetime (ns) | 1.910 ± 0.006 | 2.018 ± 0.010 | 2.166 ± 0.013 | |
| Migration speed (μm/h) | 32.553 ± 2.515 | 4.209 ± 1.851 | 5.773 ± 1.195 | |
MARK-AR1 as a Biosensor to Characterize the Inhibitors of MARK
Considering the sensitivity of MARK-AR1 and FLIM in detecting the endogenous activity of MARK in cells, it is possible to utilize this biosensor to characterize chemical inhibitors of the kinase. Primary cortical neurons prepared from E18 rats were transfected with MARK-AR1 at 8 DIV. At 10 DIV, live cells were imaged using FLIM. Cells were then treated with the compounds at a concentration of 20 μm for 180 min, and the same cells were imaged again. We chose compound 39621 because of its high MARK selectivity and found that it efficiently reduces the endogenous MARK activity (Fig. 9, D1 and D2). This effect is best visible in the tips of neurites where the average lifetime of fluorescence rises from 1.9 to 2.2 ns. In addition, the growth of the tip is reduced to 5.7 μm/h, a value similar to inactive tips (Table 2). If no compound is added to the cells, the lifetime of fluorescence remains constant at 1.9 ns (Fig. 9C1), and the axon displays rapid growth of 32.5 μm/h (compare C2 with C1 and D2 with D1).
DISCUSSION
MARK Inhibitors
Hyperphosphorylation of Tau protein is a key hallmark of Alzheimer disease and other neurodegenerative diseases (37, 38). Tau contains many potential phosphorylation sites but only some of them have a strong impact on Tau physiological function of binding and stabilizing microtubules. This includes in particular the KXGS motifs in the repeat domain (especially Ser-262) phosphorylated by MARK family kinases (5, 39). This occurs at an early stage of the disease, and MARK colocalizes with neurofibrillary tangles (25, 26). Apart from its role in disease, MARK (also known as Par-1) plays a role in cell differentiation and establishment of polarity in various cell types and organisms (10, 40). For these reasons we were interested to identify inhibitors of MARK and to develop reporters of MARK activity applicable to live cells.
About 18,000 compounds were initially screened as inhibitors of MARK, yielding 96 inhibitors. Among these, 12 were effective in vitro with IC50 values below 10 μm (Table 1). However, in the case of kinase inhibitors, specificity needs special consideration. The protein family of kinases is large (518 members in the human kinome (15) and well conserved in the catalytic domain, and most of the known kinase inhibitors are ATP competitors; thus, there is considerable overlap in inhibitory activity (29). In earlier work we had tested a number of kinases and corresponding inhibitors that affect Tau phosphorylation (e.g. MAPK, GSK3β, and cyclin-dependent kinases). Some of them were potent inhibitors but not specific; a case in point is the marine sponge constituent HD01, which strongly inhibits the proline-directed kinases Cdk2, Cdk5, GSK3β (41), but also non-proline-directed kinases such as MARK; the latter has a pronounced effect on neuronal differentiation (23). Therefore, the MARK inhibitors described in this paper were tested against a variety of Tau kinases and shown to be not inhibitory or only weakly inhibitory to them (Fig. 3). Furthermore, the described compounds were much more effective toward MARKs than they were toward the related AMPK or SAD-B (also known as BRSK1) (Fig. 2). The MARK inhibitors are effective not only in vitro but also in a cellular assay (5); when MARK activity is increased in CHO cells, the microtubule network is destroyed. This effect is enhanced by MARK activation (e.g. upstream kinases (17), but it is abolished by the compounds tested here without showing toxic side effects at the effective concentrations (Fig. 5). Finally, four compounds were particularly notable because they have a common scaffold, exhibited effective inhibition of Tau phosphorylation in cells, and showed little or no cross-reactivity toward SAD kinase and AMPK, close relatives of MARK. These substances (30019, 30195, 30197, and 30199) share a 9-oxo-9H-acridin-10-yl as a functional group and provide an excellent basis for further chemical modification by combinatorial chemistry (Fig. 4).
MARK Biosensors
In earlier studies we investigated the role of MARK by transfection of active or inactive mutants, immunofluorescence using antibodies against phosphorylated targets (e.g. Tau) and antibodies against MARK in different states of activity, or downstream effects on the microtubule cytoskeleton, synaptic integrity, transport processes etc. (5, 12, 42). All of these methods are indirect as the activity is not visible in living cells. To overcome this limitation and to study the action of MARK, effects on MARK targets, and effects of MARK inhibitors in neuronal cells, we constructed a MARK activity reporter MARK-AR1. This activity reporter allows us to observe the activity of MARK with high spatial and temporal resolution in living cells. The principles are based on the reporter construct designed for observing PKA activity in cells (33), i.e. using the adaptor protein 14-3-3 coupled to a phosphorylate-able peptide (in this case, the MARK target sequence from Cdc25) and embedded in a FRET pair of GFP derivatives (e.g. ECFP and YFP; Fig. 6). The conformational change induced by binding of the phosphopeptide to 14-3-3 changes the distance between FRET donor and acceptor and thus renders the reaction visible. In a later step for an optimized FLIM signal, we switched to the modified donor teal fluorescent protein, which is more suitable for observations in cells because of its singular excitation/emission peaks (which allows the data fit with a single exponential decay) and its enhanced photostability (43).
This arrangement enables one to observe MARK activity directly in neurons. We found high MARK activity in primary cortical neurons at the growth cones of axons, consistent with our previous observations that MARK and its activating kinase co-localize in the tips of neurites of differentiating PC12 cells or cortical neurons (17). This locally enhanced activity supports the view that the microtubule cytoskeleton must be dynamic near advancing growth cones to allow actin-based protrusions to take place (for review, see Ref. 44) (Fig. 9). The activity of MARK had been shown in the tips of neurites in differentiated and fixed PC12 cells by staining with antibodies specific for active MARK (Thr(P)-208) and for its phosphorylated primary substrate Tau (Ser(P)-262) (19). However, the reporter now allowed us to observe MARK activity in living cells in a time-resolved manner and to follow the response to a variety of cues, such as activators or treatments with kinase inhibitors. For example, the time-lapse observations show that the new MARK inhibitors are able to suppress MARK activity in living neurons, which results in the termination of axon elongation (Fig. 9). This effect can clearly be attributed to MARK and not to the related kinase SAD-B. This kinase has also been reported to play a role in polarity development of neuronal cells (16). However, even though SAD-B is able to phosphorylate the activity reporter, it does not respond to the MARK-specific inhibitor 39621. Thus, the strong reduction of activity revealed by the activity reporter indicates that MARK, not SAD-B, is involved in the growth and elongation of axons in neuronal cells. We conclude that the specificity of the compounds and their potent inhibition of MARK in cells will make them useful tools for elucidating the role of endogenous MARK in polarized cells. The inhibitors are, therefore, promising reagents for protecting neurons against the hyperphosphorylation of Tau and ensuing degeneration.
Acknowledgements
We thank Carola Seyffarth (FMP Berlin) for performing the screen of the compound library and Annika Eikhof (Hamburg) for technical assistance in preparing primary cortical cultures. We are grateful to Dr. Jochen Müller-Dieckmann (EMBL-Hamburg) for providing access to the MALDI and Dr. Roger Yonchien Tsien (University of California, San Diego, CA) for the PKA activity reporter.
This work was supported by grants from the Institute for the Study of Aging (ISOA, New York), the Deutsche Forschungsgemeinschaft, and the EU-FP7 (Memosad project).
T. Timm and G. Drewes, unpublished data.
- MARK1–4
- microtubule-associated protein (MAP)/microtubule affinity-regulating kinase 1–4 (the term MARK is used where no distinction of isoforms is required)
- AMPK
- ATP-activated kinase
- BRSK
- brain-specific protein kinase (also known as SAD A/B (synapses of amphids-deficient)
- Cdk5
- cyclin-dependent kinase 5
- Cdc
- cell division control protein 2 (also known as Cdk1)
- Cdc25C
- cell division control protein 25C
- ECFP/TFP/YFP
- enhanced cyan/teal/yellow fluorescent protein
- FLIM
- fluorescence lifetime imaging microscopy
- GSK3β
- glycogen synthase kinase 3β
- HD01
- hymenialdisine
- hTau40
- largest Tau isoform in the human central nervous system (441 residues)
- MARKK
- MARK activating kinase
- Par-1
- partitioning-defective mutant 1
- BES
- 2-[bis(2-hydroxyethyl)amino]ethanesulfonic acid
- mTFP
- mono-teal fluorescent protein
- DIV
- days in vitro
- MARK-AR1
- MARK activity reporter.
REFERENCES
- 1. Kemphues K. (2000) Cell 101, 345–348 [DOI] [PubMed] [Google Scholar]
- 2. Nance J. (2005) Bioessays 27, 126–135 [DOI] [PubMed] [Google Scholar]
- 3. Shulman J. M., Benton R., St. Johnston D. (2000) Cell 101, 377–388 [DOI] [PubMed] [Google Scholar]
- 4. Cohen D., Brennwald P. J., Rodriguez-Boulan E., Müsch A. (2004) J. Cell Biol. 164, 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Drewes G., Ebneth A., Preuss U., Mandelkow E. M., Mandelkow E. (1997) Cell 89, 297–308 [DOI] [PubMed] [Google Scholar]
- 6. Ebneth A., Drewes G., Mandelkow E. M., Mandelkow E. (1999) Cell Motil. Cytoskeleton 44, 209–224 [DOI] [PubMed] [Google Scholar]
- 7. Schaar B. T., Kinoshita K., McConnell S. K. (2004) Neuron 41, 203–213 [DOI] [PubMed] [Google Scholar]
- 8. Peng C. Y., Graves P. R., Ogg S., Thoma R. S., Byrnes M. J., 3rd, Wu Z., Stephenson M. T., Piwnica-Worms H. (1998) Cell Growth Differ. 9, 197–208 [PubMed] [Google Scholar]
- 9. Müller J., Ory S., Copeland T., Piwnica-Worms H., Morrison D. K. (2001) Mol. Cell 8, 983–993 [DOI] [PubMed] [Google Scholar]
- 10. Matenia D., Mandelkow E. M. (2009) Trends Biochem. Sci. 34, 332–342 [DOI] [PubMed] [Google Scholar]
- 11. Sapir T., Shmueli A., Levy T., Timm T., Elbaum M., Mandelkow E. M., Reiner O. (2008) J. Neurosci. 28, 13008–13013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mandelkow E. M., Thies E., Trinczek B., Biernat J., Mandelkow E. (2004) J. Cell Biol. 167, 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Panneerselvam S., Marx A., Mandelkow E. M., Mandelkow E. (2006) Structure 14, 173–183 [DOI] [PubMed] [Google Scholar]
- 14. Tochio N., Koshiba S., Kobayashi N., Inoue M., Yabuki T., Aoki M., Seki E., Matsuda T., Tomo Y., Motoda Y., Kobayashi A., Tanaka A., Hayashizaki Y., Terada T., Shirouzu M., Kigawa T., Yokoyama S. (2006) Protein Sci. 15, 2534–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 16. Barnes A. P., Lilley B. N., Pan Y. A., Plummer L. J., Powell A. W., Raines A. N., Sanes J. R., Polleux F. (2007) Cell 129, 549–563 [DOI] [PubMed] [Google Scholar]
- 17. Timm T., Li X. Y., Biernat J., Jiao J., Mandelkow E., Vandekerckhove J., Mandelkow E. M. (2003) EMBO J. 22, 5090–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lizcano J. M., Göransson O., Toth R., Deak M., Morrice N. A., Boudeau J., Hawley S. A., Udd L., Mäkelä T. P., Hardie D. G., Alessi D. R. (2004) EMBO J. 23, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Timm T., Balusamy K., Li X., Biernat J., Mandelkow E., Mandelkow E. M. (2008) J. Biol. Chem. 283, 18873–18882 [DOI] [PubMed] [Google Scholar]
- 20. Matenia D., Griesshaber B., Li X. Y., Thiessen A., Johne C., Jiao J., Mandelkow E., Mandelkow E. M. (2005) Mol. Biol. Cell 16, 4410–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johne C., Matenia D., Li X. Y., Timm T., Balusamy K., Mandelkow E. M. (2008) Mol. Biol. Cell. 19, 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benton R., Palacios I. M., St, Johnston D. (2002) Dev. Cell 3, 659–671 [DOI] [PubMed] [Google Scholar]
- 23. Biernat J., Wu Y. Z., Timm T., Zheng-Fischhöfer Q., Mandelkow E., Meijer L., Mandelkow E. M. (2002) Mol. Biol. Cell 13, 4013–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stamer K., Vogel R., Thies E., Mandelkow E., Mandelkow E. M. (2002) J. Cell Biol. 156, 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Augustinack J. C., Schneider A., Mandelkow E. M., Hyman B. T. (2002) Acta Neuropath. 103, 26–35 [DOI] [PubMed] [Google Scholar]
- 26. Chin J. Y., Knowles R. B., Schneider A., Drewes G., Mandelkow E. M., Hyman B. T. (2000) J. Neuropath. Exp. Neurol. 59, 966–971 [DOI] [PubMed] [Google Scholar]
- 27. Eckermann K., Mocanu M. M., Khlistunova I., Biernat J., Nissen A., Hofmann A., Schönig K., Bujard H., Haemisch A., Mandelkow E., Zhou L., Rune G., Mandelkow E. M. (2007) J. Biol. Chem. 282, 31755–31765 [DOI] [PubMed] [Google Scholar]
- 28. Mocanu M. M., Nissen A., Eckermann K., Khlistunova I., Biernat J., Drexler D., Petrova O., Schönig K., Bujard H., Mandelkow E., Zhou L., Rune G., Mandelkow E. M. (2008) J. Neurosci. 28, 737–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lisurek M., Rupp B., Wichard J., Neuenschwander M., von Kries J. P., Frank R., Rademann J., Kühne R. (2010) Mol. Divers. 14, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Esterman A. L., Cohen S. P. (2006) Gen. Eng. News 26, 34–36 [Google Scholar]
- 32. Bright N. J., Thornton C., Carling D. (2009) Acta Physiol. (Oxf) 196, 15–26 [DOI] [PubMed] [Google Scholar]
- 33. Zhang J., Ma Y., Taylor S. S., Tsien R. Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14997–15002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hutchins J. R., Clarke P. R. (2004) Cell Cycle 3, 41–45 [PubMed] [Google Scholar]
- 35. Clegg R. M., Holub O., Gohlke C. (2003) Methods Enzymol. 360, 509–542 [DOI] [PubMed] [Google Scholar]
- 36. Rizzo M. A., Springer G. H., Granada B., Piston D. W. (2004) Nat. Biotechnol. 22, 445–449 [DOI] [PubMed] [Google Scholar]
- 37. Buée L., Troquier L., Burnouf S., Belarbi K., Van der Jeugd A., Ahmed T., Fernandez-Gomez F., Caillierez R., Grosjean M. E., Begard S., Barbot B., Demeyer D., Obriot H., Brion I., Buée-Scherrer V., Maurage C. A., Balschun D., D'hooge R., Hamdane M., Blum D., Sergeant N. (2010) Biochem. Soc. Trans. 38, 967–972 [DOI] [PubMed] [Google Scholar]
- 38. Holtzman D. M., Morris J. C., Goate A. M. (2011) Sci. Transl. Med. 3, 77sr1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Biernat J., Mandelkow E. M. (1999) Mol. Biol. Cell 10, 727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goldstein B., Macara I. G. (2007) Dev. Cell 13, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meijer L., Thunnissen A. M., White A. W., Garnier M., Nikolic M., Tsai L. H., Walter J., Cleverley K. E., Salinas P. C., Wu Y. Z., Biernat J., Mandelkow E. M., Kim S. H., Pettit G. R. (2000) Chem. Biol. 7, 51–63 [DOI] [PubMed] [Google Scholar]
- 42. Thies E., Mandelkow E. M. (2007) J. Neurosci. 27, 2896–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ai H. W., Henderson J. N., Remington S. J., Campbell R. E. (2006) Biochem. J. 400, 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Geraldo S., Gordon-Weeks P. R. (2009) J. Cell Sci. 122, 3595–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]