Background: The cytokine function of the danger-associated molecular pattern protein HMGB1 is mediated through RAGE.
Results: HMGB1-RAGE signaling depends on heparan sulfate, but heparan sulfate binding to HMGB1 is dispensable.
Conclusion: Heparan sulfate is essential for HMGB1 signaling because RAGE binds heparan sulfate.
Significance: Perturbing heparan sulfate may be a novel strategy to alter RAGE-dependent signaling.
Keywords: Cytokine, Endothelial Cell, Heparan Sulfate, Inflammation, Signal Transduction, Coreceptor, HMGB1, RAGE
Abstract
In a proteomic search for heparan sulfate-binding proteins on monocytes, we identified HMGB1 (high mobility group protein B1). The extracellular role of HMGB1 as a cytokine has been studied intensively and shown to be important as a danger-associated molecular pattern protein. Here, we report that the activity of HMGB1 depends on heparan sulfate. Binding and competition studies demonstrate that HMGB1 interacts with CHO and endothelial cell heparan sulfate. By site-directed mutagenesis, we identified a loop region that connects the A-box and B-box domains of HMGB1 as responsible for heparan sulfate binding. HMGB1-induced Erk1/2 and p38 phosphorylation is abolished when endothelial heparan sulfate is removed or blocked pharmacologically, resulting in decreased HMGB1-induced endothelial sprouting. However, mutated HMGB1 that lacks the heparan sulfate-binding site retained its signaling activity. We show the major receptor for HMGB1, receptor for advanced glycation end products (RAGE), also binds to heparan sulfate and that RAGE and heparan sulfate forms a complex. Our data establishes that the functional receptor for HMGB1 consists of a complex of RAGE and cell surface heparan sulfate.
Introduction
HMGB1 (high mobility group protein B1) is an abundant DNA-binding protein that normally resides in the nucleus (1). However, inflammatory cells such as monocytes and macrophages can secrete HMGB1 actively through a non-classical, vesicle-mediated secretory pathway (2). During tissue injury, necrotic cells also release HMGB1 as a danger-associated molecular pattern protein (DAMP)3 (3, 4). Extracellular HMGB1 behaves much like a cytokine. It stimulates inflammatory responses in many types of cells, including monocytes, macrophages, neutrophils, dendritic cells, astrocytes, and endothelial cells (1, 4–6), and induces cell migration (4, 5, 7). Much interest exists in developing ways to block HMGB1 function in the context of various diseases such as sepsis, ischemic injury, arthritis, atherosclerosis, and cancer (4, 5, 8).
HMGB1 binds to the receptor for advanced glycation end products (RAGE), a transmembrane protein with three IgG-like extracellular domains and a short cytoplasmic domain. RAGE is expressed at high level in alveolar epithelial cells as well as leukocytes, endothelial cells, neurons, and tumor cells (9). It binds to HMGB1 as well as other ligands, including S100 proteins, AGEs, and amyloids, through its two N-terminal IgG-like domains (9, 10). The extracellular domain of RAGE can be shed by proteolysis, yielding what is commonly referred to as soluble RAGE, or sRAGE. Alternative splicing can give rise to a soluble form of the receptor as well (9, 10). The cytoplasmic domain of RAGE lacks intrinsic kinase activity, suggesting that signal transduction upon ligand binding involves recruitment of a signaling kinase or a kinase scaffolding protein (11, 12). The inflammatory response and migration of target cells mediated by HMGB1-RAGE involve activation of MAPKs, such as Erk1/2 and p38, and NF-κB nuclear translocation (8, 9).
Both HMGB1 and RAGE have been reported to bind heparin, a property that can be exploited for protein purification. In this report, we show that HMGB1 and RAGE also bind to heparan sulfate. We mapped the heparin-binding site on HMGB1 by mutagenesis and created a mutant that lacks the capacity to bind heparan sulfate. Surprisingly, mutant HMGB1 signals normally, but its action still depends on cell surface heparan sulfate. This finding led to the discovery that RAGE is also a heparan sulfate binding protein whose activity depends on binding to cell surface proteoglycans.
EXPERIMENTAL PROCEDURES
Proteomic Screening
U937 cells (2 × 108) were biotinylated with 0.6 mm sulfo-NHS-SS-biotin (Pierce) in 10 ml of PBS, pH 8, at 4 °C for 1 h. The reaction was stopped by adding Tris base to a final concentration of 50 mm. After two washes with cold PBS, cells were resuspended in 3 ml of ice-cold hypotonic buffer (10 mm Tris, 10 mm NaCl, 1.5 mm MgCl2, pH 7.4) supplemented with protease inhibitors. Cells were next subjected to 20 strokes in a Dounce homogenizer and then centrifuged at 1000 × g for 3 min to remove intact nuclei and unbroken cells. The post-nuclear supernatant was solubilized with Nonidet P-40 (final concentration of 1%)in 150 mm NaCl and clarified by centrifugation at 14,000 × g for 15 min.
U937 supernatant (∼ 10 mg of protein) was diluted to 1 mg/ml with heparin column wash buffer (20 mm HEPES, 150 mm NaCl, 1% Nonidet P-40, pH 7) and applied to a heparin fast flow Sepharose column (1 ml; GE Healthcare). The column was washed with 10 ml of wash buffer and eluted sequentially with 5 ml of buffer containing 300 mm NaCl, 3 ml of 500 mm NaCl and 3 ml of 1 m NaCl. The 500 mm and 1 m eluates were combined (∼1 mg of protein).
The sample was mixed with 100 μl of neutravidin beads (Pierce) in a 2-ml disposable column (Bio-Rad) and incubated at room temperature for 30 min with gentle rotation. Once the beads settled, the column was washed twice with 1 ml of cold wash buffer (1% Nonidet P-40, 0.1% SDS in PBS), twice with 1 ml of cold 1 m KCl, and twice with 1 ml of 0.1 m Na2CO3, pH 11.5. The beads were next washed with 3 ml of PBS and transferred into a 1.5-ml centrifuge tube. Bound biotinylated protein was eluted by incubation of the beads with 50 μl of 5% β-mercaptoethanol in PBS for 30 min in a 37 °C water bath. The elution was repeated two more times to maximize protein recovery. Approximately 20 μg of protein was eluted by this method. For LC/MS analysis, 4 μg of protein was digested using an in-gel tryptic digestion kit (Pierce) and analyzed by University of California Davis proteomic core facility.
HMGB1 and sRAGE
U937 cells (1.2 × 109) were chilled at 4 °C for 6 h, sedimented by centrifugation, and lysed in cold lysis buffer (1% Triton X-100, 20 mm Tris, pH 8, 150 mm NaCl) supplemented with protease inhibitors (Sigma). The cell lysate was centrifuged at 14,000 × g for 20 min, and the supernatant was diluted with 50 mm Tris (pH 8.1) to achieve a final NaCl concentration of 60 mm. The diluted lysate was applied to a DEAE HiTrap column (2 × 1 ml, GE Healthcare). HMGB1 eluted between 150 to 240 mm salt at pH 8.1. The pooled fractions were applied to a 1.3-ml UNO-Q column (Bio-Rad) after diluting the NaCl concentration to 80 mm. HMGB1 eluted between 200 to 325 mm salt at pH 8.1. The partially purified HMGB1 was then applied to a 1-ml heparin-Sepharose column, and HMGB1 was eluted between 550 mm to 600 mm salt at pH 7.1. In the last step of purification, pooled heparin fractions was concentrated and resolved by a G2000SW gel permeation column (Tosoh). Approximately 50 μg of highly pure HMGB1 (at least > 95% pure as judged by silver staining of PAGE gels after analysis of 200 ng of material) was obtained.
In experiments employing mutants of HMGB1, recombinant protein was generated in Escherichia coli. The complete open reading frame of human HMGB1 (Open Biosystems) was cloned into pET45b (Novagen) using the PshA I site. Expression was carried out in Origami-B cells (Novagen) carrying the pGro7 (Takara) plasmid expressing chaperonin proteins GroEL and GroES of E. coli. Transformed cells were grown in LB medium supplemented with 12.5 μg/ml tetracycline, 15 μg/ml kanamycin, 35 μg/ml chloramphenicol, and 50 μg/ml carbenicillin at 37 °C. When the absorbance at 600 nm reached 0.4–0.7, isopropyl β-d-thiogalactopyranoside (0.15 mm) and l-arabinose (1 mg/ml) were added to induce the expression of HMGB1 and chaperonin proteins, respectively. The cells were allowed to shake overnight at 22 °C. Purification was carried out using a Ni-Sepharose 6 fast flow column, followed by gel permeation chromatography with HiLoad 16/60 Superdex 75 and anion exchange chromatography with MonoQ 5/50 (GE Healthcare). Mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Agilent).
The extracellular domain (Ala-23–Glu-326) of RAGE was cloned into pET45b using PshA I site. Expression was carried out in the same way as described for HMGB1. Purification was performed by using a Ni-Sepharose 6 fast flow column followed by gel filtration chromatography with Superdex 200 10/300.
Biotinylation of HMGB1 and sRAGE
HMGB1 or sRAGE was diluted in 3 ml of PBS and was loaded onto a 200-μl heparin-Sepharose column. Sulfo-NHS-LC-biotin in PBS (500 μl of a 1 mm solution, pH 8) was then applied to the column, and biotinylation was allowed to proceed for 30 min at room temperature. The reaction was stopped by applying 600 μl of PBS containing 100 mm glycine, pH 7. After washing the column with 1 ml of 20 mm HEPES buffer (pH 7.1) containing 150 mm NaCl, the column was eluted sequentially with 600 mm NaCl and 1 m NaCl in 20 mm HEPES buffer (800 μl). Protein eluted by 1 m NaCl was used for all binding experiments.
HMGB1 and sRAGE Binding to Immobilized Heparin and Heparan Sulfate
Porcine mucosal heparin (SPL Scientific Protein Laboratories) and bovine kidney heparan sulfate (Sigma) were immobilized (0.1 ml of 10 μg/ml) on 96-well heparin binding plates (BD Biosciences) according to the manufacturer's instructions. Plates were blocked with 5% BSA in PBS and incubated with various concentrations of biotinylated HMGB1 or sRAGE. Bound ligand was quantitated with streptavidin-HRP (Jackson Immunology) followed by the addition of HRP substrate (Thermo Scientific).
HMGB1 and sRAGE Binding to CHO Cells
Wild-type CHO-K1 cells or heparan sulfate-deficient mutants pgsD, pgsE, and pgsF (5 × 105 cells) (13) were incubated with biotinylated HMGB1 (1 μg/ml) or sRAGE (2 μg/ml) in 100 μl of PBS for 1 h at 4 °C. After rinsing, the cells were stained with streptavidin-phycoerythrin (eBioscience) at a dilution of 1:1000 for 15 min and analyzed by flow cytometry. In some experiments, cells were treated with heparin lyases I (2 milliunits/ml), II (2 milliunits/ml), and III (5 milliunits/ml) for 15 min at room temperature prior to binding experiments. In some experiments, heparin, 2-O-desulfo-heparin and 6-O-desulfo-heparin and N-desulfo/re-N-acetylated heparin (Neoparin, Inc.) were included (10−11 to 10−5 mol/liter). The geometric means of binding histograms obtained in the presence of heparin were normalized to the one obtained in the absence of heparin and expressed as a percentage of maximum binding. The inhibition curve was fitted to one site competition, and IC50 values were calculated in Prism.
Endothelial Cell Sprouting Assay
Isolation of murine brain and lung microvascular endothelial cells and a sprouting assay using rat tail type I collagen gel (BD Biosciences) were performed essentially as described (14). Highly purified U937 cell endogenous HMGB1 or FGF2 (Shenandoah Biotechnology) was diluted in DMEM to the desired concentration. The net length of endothelial sprouts per (100×) microscopic field was measured after 24 h. Data were normalized to the response of unstimulated cells and expressed as fold stimulation.
Immunoblotting
Human cardiac microvascular endothelial cells (HMVEC-c) cells (Lonza, cc-7030) were serum-starved in DMEM for 5 h prior to treatment with U937-derived HMGB1. In some experiments, HMVEC-c were incubated with a mixture of heparin lyases I (2 milliunits/ml), II (2 milliunits/ml), and III (5 milliunits/ml), or with 10 μg/ml protamine (Sigma), or with 10 μg/ml blocking antibody to RAGE (R&D Systems) in serum-free DMEM at 37 °C for 15 min. Cells were then stimulated with 50 to 2500 ng/ml HMGB1 at 37 °C for 10 min. Cells were lysed in radioimmune precipitation assay buffer (Cell Signaling) containing protease and phosphatase inhibitors (Pierce). Samples (10 μg) were resolved on 4–12% Bis-Tris NuPage gel and blotted onto polyvinylidene difluoride membrane. After incubation with primary and secondary antibodies to Erk1/2, phospho-Erk1/2, and phospho-p38 (Cell Signaling), reactive bands were visualized by West Pico chemiluminescent substrate (Pierce).
Proximity Ligation Assay
Proximity ligation was measured using a mouse monoclonal antibody (10E4, US Biological) to heparan sulfate and an affinity-purified rabbit polyclonal antibody to human RAGE extracellular domain (Millipore), diluted to 1 μg/ml, and processed as described previously (15).
RESULTS
Identification of Monocyte Surface-expressed Heparin-binding Proteins
A four-step proteomic approach was designed to discover heparin-binding proteins present on cells. U937 human monocytic cells were biotinylated with the impermeant reagent, Sulfo-NHS-SS-Biotin. Biotinylated proteins were then purified by heparin-Sepharose and streptavidin affinity chromatography. Analysis by liquid chromatography/mass spectrometry yielded 21 heparin-binding proteins, many with high sequence coverage (Table 1). Twelve of the identified proteins have been shown previously to bind to heparin. Interestingly, nine proteins are classified as nuclear and 11 are classified as cytoplasmic proteins. We were surprised at first that so many of the tagged proteins were cytoplasmic or nuclear residents. Contamination by cell lysis during the biotinylation step seemed unlikely because more abundant heparin-binding nuclear proteins, such as histones, were not identified in any of the preparations. An in-depth literature search revealed that many of the identified nuclear proteins can be secreted, including HMGB1, lupus La protein, hepatoma-derived growth factor, nucleolin, and nucleophosmin (16–19). None of the three heterogeneous nuclear ribonucleoproteins (hnRNPs Q, L, and R) have been shown to be expressed extracellularly, but surface expression is likely because a related protein hnRNP U has been observed at the cell surface (20). In subsequent studies, we focused on the heparin-binding properties of HMGB1 as a paradigm for future studies of the other proteins discovered in this proteomic screen.
TABLE 1.
Extracellular heparin-binding proteins identified by mass spectrometry
| NCBI Protein ID | Protein | Subcellular locationa | Heparin bindingb | Peptide coverage |
||
|---|---|---|---|---|---|---|
| 1st | 2nd | 3rd | ||||
| % | ||||||
| P23284 | Cyclophilin B | Ex/Cp | Yes | 62 | 66 | 70 |
| P09429 | High mobility group protein B1 | Nu/Cp/Ex | Yes | 60 | 34 | 55 |
| P26038 | Moesin | Cp/Ex | Yes | 48 | 56 | 57 |
| P04075 | Fructose-bisphosphate aldolase A | Cp/Ex | Yes | 58 | 22 | 54 |
| Q06830 | Peroxiredoxin-1 | Cp/Ex | NA | 24 | 45 | 50 |
| P05455 | Lupus La protein | Nu/Ex | NA | 31 | 17 | 45 |
| P51858 | Hepatoma-derived growth factor | Ex/Nu | Yes | 19 | 46 | 19 |
| P19338 | Nucleolin | Nu/Ex | Yes | 31 | 26 | 22 |
| P08567 | Pleckstrin | Cp | NA | 17 | 30 | 15 |
| Q13765 | Nascent polypeptide-associated complex subunit α | Cp | NA | 27 | 26 | 14 |
| P06748 | Nucleophosmin | Nu/Ex | Yes | 31 | 9 | 17 |
| P30533 | α-2-Macroglobulin receptor-associated protein | Ex | Yes | 23 | 12 | 18 |
| P62937 | Cyclophilin A | Ex/Cp | Yes | 9 | 13 | 43 |
| O60506 | Heterogeneous nuclear ribonucleoprotein Q | Nu | NA | 33 | 7 | 9 |
| P14866 | Heterogeneous nuclear ribonucleoprotein L | Nu | NA | 10 | 6 | 22 |
| P08238 | Heat shock protein HSP 90-β | Cp/Ex | Yes | 9 | 16 | 12 |
| Q14444 | Caprin-1 | Cp | NA | 10 | 6 | 10 |
| O43390 | Heterogeneous nuclear ribonucleoprotein R | Nu | NA | 9 | 4 | 10 |
| Q12906 | Interleukin enhancer-binding factor 3 | Nu/Cp | NA | 11 | 6 | 5 |
| P07900 | Heat shock protein HSP 90-α | Cp/Ex | Yes | 6 | 4 | 10 |
| P11021 | 78-kDa glucose-regulated protein | Cp/Ex | Yes | 8 | 6 | 5 |
a Nu, Nuclear; Cp, cytoplasmic; Ex, extracellular.
b NA, data not available.
Binding of HMGB1 to CHO Cells Depends on Heparan Sulfate
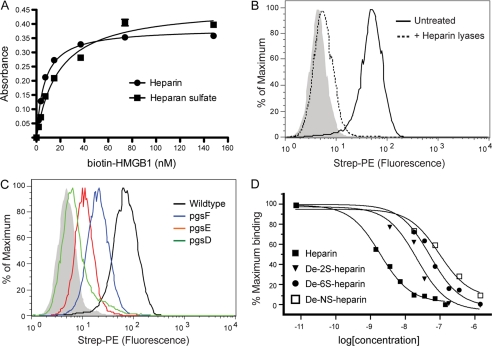
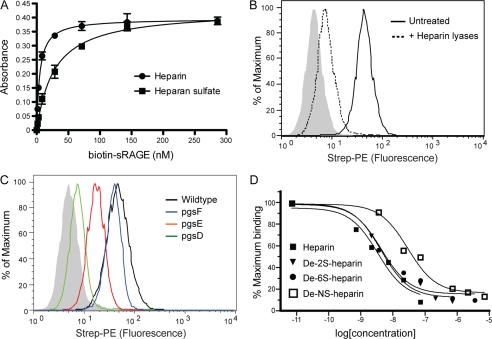
HMGB1 was identified originally as a DNA-binding protein, and its capacity to bind heparin was exploited for its purification (21). HMGB1 also can bind heparan sulfate proteoglycans, but the biological significance of this interaction has not been explored (22). Incubation of biotinylated HMGB1 with immobilized heparin or bovine kidney heparan sulfate yielded Kd values of 6.8 ± 0.4 nm and 18.4 ± 2 nm for heparin and heparan sulfate, respectively (Fig. 1A).
FIGURE 1.
Binding of HMGB1 to CHO cells depends on heparan sulfate. A, binding of biotinylated recombinant HMGB1 to immobilized heparin (circles) and heparan sulfate (squares) was measured. B, binding of biotinylated HMGB1 (1 μg/ml) to CHO-K1 cells was measured by flow cytometry with or without heparin lyase pretreatment. The control sample was incubated only with streptavidin-phycoerythrin (Strep-PE) and is shown as a filled gray histogram. C, binding to wild-type CHO-K1 is shown in black, to 2-O-sulfation-deficient mutant pgsF in blue, to N-sulfation-deficient mutant pgsE in red, and to heparan sulfate-deficient mutant pgsD in green. D, HMGB1 binding was performed by flow cytometry in the presence of 10−11 to 10−6 m heparin or chemically desulfated heparins. Shown are the relative fluorescence units normalized to maximum binding.
Heparin lyase treatment of CHO cells showed that binding of biotinylated recombinant HMGB1 strictly depends on heparan sulfate and not other glycosaminoglycans (Fig. 1B). CHO pgsD-677 cells, which lack heparan sulfate but make elevated amounts of chondroitin/dermatan sulfate (supplemental Fig. S1A), also fail to bind HMGB1 (Fig. 1C, relative fluorescence units (RFU) = 2 versus 64 in mutant and wild-type cells, respectively). Binding decreased in pgsE-606 (RFU = 8) and in pgsF-17 (RFU = 17) cells that lack glucosamine N-sulfotransferase and uronyl 2-O-sulfotransferase activities, respectively (Fig. 1C). Heparin, a highly sulfated form of heparan sulfate, blocked binding to wild-type cells with an IC50 of 1.8 ± 0.2 nm (Fig. 1D). Competition studies using various desulfated versions of heparin confirmed that loss of sulfate groups from any position significantly reduced the capacity of heparin to block binding (Fig. 1D, IC50 values = 21 ± 6, 49 ± 8, and 90 ± 30 nm for 2-O-desulfo-, 6-O-desulfo, and N-desulfo heparins). The dramatic reduction of potency when N-sulfate groups were removed is consistent with the equally dramatic loss of binding to pgsE cells. The combined genetic and competition studies suggests the order of importance of sulfate groups in binding of HMGB1 to heparin/heparan sulfate is N-sulfation > 6-O-sulfation > 2-O-sulfation. This pattern differs significantly from the behavior of FGF2, which depends more on 2-O-sulfation than N-sulfation in this assay (supplemental Fig. S1B).
Location of Heparan Sulfate-binding Site of HMGB1
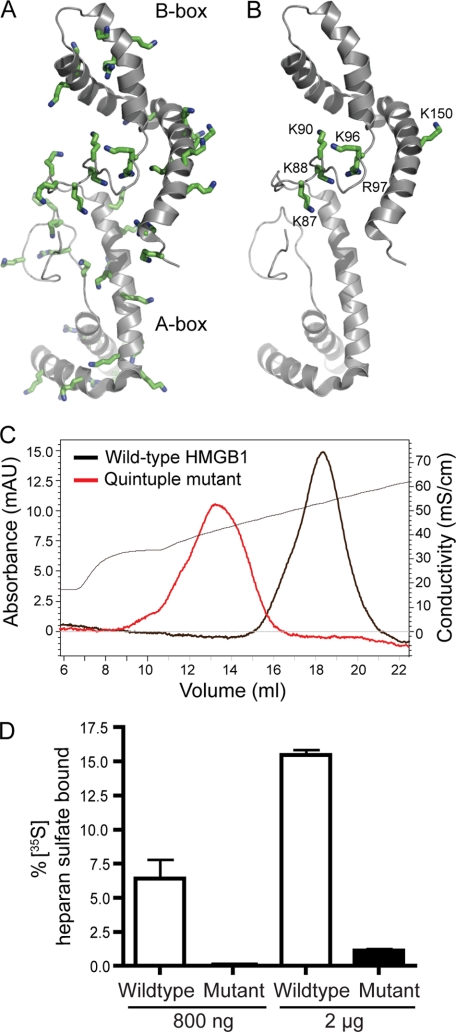
Most ligands bind to heparin/heparan sulfate by way of electrostatic interactions of the negatively charged sulfate groups and uronic acids in the chains with positively charged lysine and arginine residues in the ligand. HMGB1 contains a large number of basic residues, 43 lysines and 8 arginines, which account for 24% of the amino acids (Fig. 2A and supplemental Fig. S2). To determine the relevant lysine residues for binding, we used a mapping strategy that combines biotinylation, heparin affinity chromatography, and mass spectrometry. Briefly, HMGB1 was bound to heparin-Sepharose, biotinylated in situ, and then eluted stepwise with a salt gradient. Unmodified HMGB1 eluted as a sharp peak centered ∼555 mm NaCl. After biotinylation, the majority of the HMGB1 eluted at significantly lower salt concentration (supplemental Fig. S3), suggesting that lysine residues relevant to binding had been modified. In theory, molecules that elute with lower salt should contain more biotinylated lysines residues involved in binding than molecules that bind at higher salt, whereas the latter should contain biotinylated lysines dispensable for binding. Assisted by mass spectrometry, we found that most lysine residues located within the major box domains (A-box domain, residues 13–79, and B-box, residues 99–163) were biotinylated in HMGB1 eluting in the high salt fractions 14–15, suggesting they were not essential for heparin binding (supplemental Table S1). In contrast, none of the lysine residues within the long loop region that connects the A- and B-boxes (residues 79–99) were biotinylated in high salt fractions, whereas many of them (Lys-82, Lys-87, Lys-88, and Lys-90) were biotinylated in lower salt fractions.
FIGURE 2.
A cluster of basic residues located at the loop region of HMGB1 contributes to heparan sulfate binding. A, schematic representation of a solution structure of HMGB1 (Protein Data Bank code 2YRQ). The HMGB1 A-box is shown in light gray, and the B-box is shown in dark gray. The side chains of all basic residues are shown as green sticks. B, basic residues that contribute to heparan sulfate binding as determined by site-directed mutagenesis. C, heparin-Sepharose chromatography of wild-type HMGB1 and quintuple mutant K87A/K88A/K96A/R97A/K150A. The conductivity trace is shown as a thin black line. D, binding of wild-type HMGB1 and K87A/K88A/K96A/R97A/K150A mutant (800 ng or 2 μg) to 35S-labeled endothelial heparan sulfate as determined by filter assay (n = 3, error bar represents S.E.). mAU, milliabsorbance units.
Based on this information and phylogenetic comparisons indicating conservation of a subset of lysine and arginine residues (supplemental Fig. S2), we selected a number of residues for mutagenesis (Table 2). Most of the selected mutations did not alter the elution of HMGB1 from heparin-Sepharose, except those located within the loop region (K87A–K88A, K90A, K96A–R97A). Various combinations of mutations showed that residues Lys-87, Lys-88, Lys-96, and Lys-97 from the loop region and Lys-150 from the B-box were most relevant to binding (Table 2 and Fig. 2B). The quintuple mutant bound to heparin less avidly, eluting at 410 mm NaCl (Fig. 2C). The quintuple mutant at physiological salt concentration essentially lost the capacity to bind [35S]heparan sulfate prepared from endothelial cells (Fig. 2D), which presumably reflects the lower overall sulfation of heparan sulfate compared with heparin.
TABLE 2.
Salt concentration for elution of HMGB1 mutants from heparin-Sepharose
| Mutant | Peak NaCl |
|---|---|
| Wild-type HMGB1 | 555 mm |
| R10A/K12A | 545 mm |
| K68A/R70A | 555 mm |
| K82A | 555 mm |
| K87A/K88A | 500 mm |
| K90A | 515 mm |
| K96A/R97A | 500 mm |
| R110A/K112A | 550 mm |
| K141A | 550 mm |
| K152A/K154A | 545 mm |
| R163A | 550 mm |
| 167stop | 550 mm |
| K87A/K88A/K96A/R97A | 445 mm |
| K87A/K88A/K96A/R97A/K150A | 410 mm |
| K87A/K88A/K96A/R97A/K8A | 435 mm |
| K87A/K88A/K96A/R97A/K90A | Not soluble |
| K87A/K88A/K96A/R97A/K90Q | 425 mm |
| K87A/K88A/K96A/R97A/K86Q | 445 mm |
HMGB1-induced Sprouting Is Substantially Reduced in Ndst1-deficient Endothelial Cells
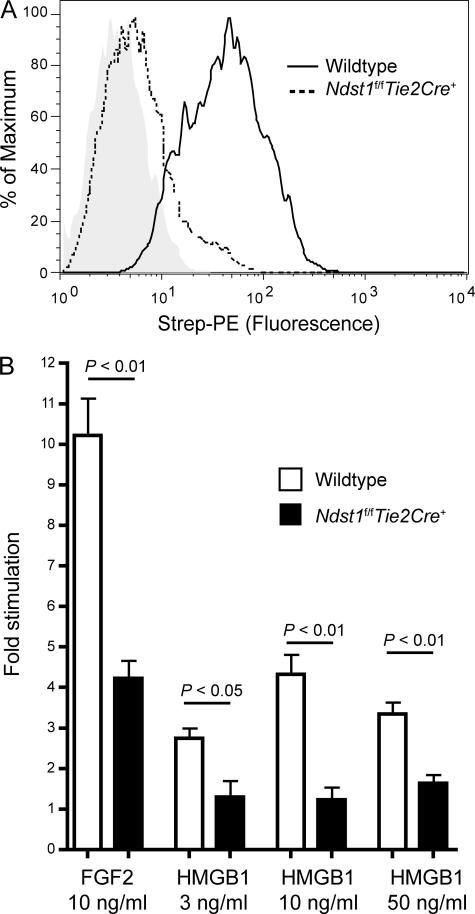
To determine whether the interaction of HMGB1 with heparan sulfate affected its biological activity, we derived primary microvascular endothelial cells from Ndst1f/fTie2Cre+ mice, which, similar to CHO pgsE cells, are deficient in glucosaminyl N-deacetylase-N-sulfotransferase-1 (15). Binding of HMGB1 to mutant endothelial cells was reduced to 5% of wild-type (Fig. 3A), which agrees well with reduced HMGB1 binding to CHO pgsE cells (Fig. 1B). Sprouting of primary endothelial cells on type I collagen was stimulated by HMGB1 as much as 4.3-fold above untreated control cells reaching a maximum at 10 ng/ml (Fig. 3B). On a molar basis, HMGB1 was ∼60% as potent as FGF2 in inducing endothelial sprouting. Sprouting of Ndst1-deficient endothelial cells was greatly reduced when challenged with either HMGB1 or FGF2 (Fig. 3B), demonstrating the importance of heparan sulfate.
FIGURE 3.
HMGB1-induced sprouting in Ndst1-deficient endothelial cells. A, binding of biotinylated HMGB1 (2 μg/ml) to wild-type or Ndst1-deficient mouse microvascular endothelial cells was measured by flow cytometry. B, sprouting of mutant and wild-type mouse endothelial cells on collagen in response to HMGB1 after 24 h treatment. Data are mean values ± S.E. for net length of endothelial sprouts per 100× microscopic field, normalized to the response of unstimulated wild-type cells (n = 4). Strep-PE, streptavidin-phycoerythrin.
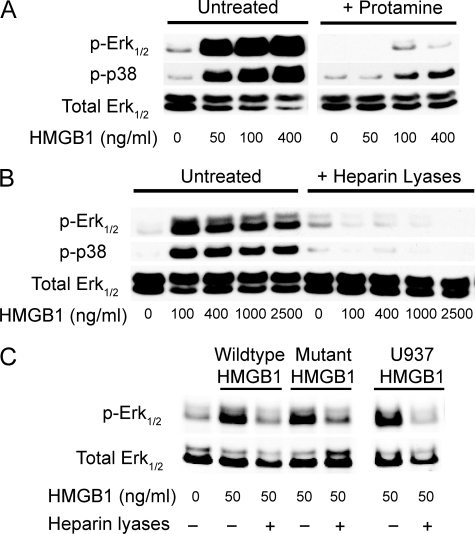
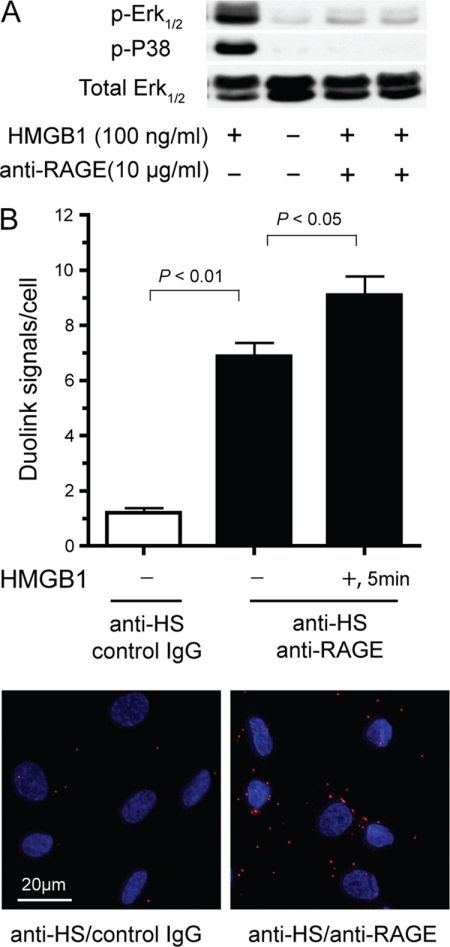
Stimulation of primary HMVEC-c with HMGB1 for 10 min resulted in phosphorylation of both Erk1/2 and p38 (Fig. 4A). Protamine, which is used to neutralize therapeutic heparin, or brief treatment with heparin lyases blocked phosphorylation of Erk1/2 and p38 (Fig. 4, A and B). Remarkably, heparin lyase-treated cells remained unresponsive to HMGB1 even at concentrations as high as 2500 ng/ml (Fig. 4B). This finding indicates that loss of heparan sulfate cannot be overcome by merely increasing the ligand concentration, which differs from the behavior of other heparin-dependent growth factors such as FGF2 (14). Attempts to measure signaling responses in murine endothelial cells were met with very high background, preventing analysis of heparan sulfate-deficient cells.
FIGURE 4.
HMGB1-induced Erk1/2 and p38 phosphorylation. A, immunoblot analysis of Erk1/2 and p38 phosphorylation in HMVEC-c after stimulation with U937-derived HMGB1. HMVEC-c were preincubated with or without 1 μm protamine and then treated with 50 to 400 ng/ml of HMGB1 for 10 min. B, HMVEC-c were pretreated with or without heparan lyases I, II, and III for 15 min and then stimulated with 100 to 2500 ng/ml of HMGB1. All blots are representative of at least three similar experiments. C, immunoblot analysis of Erk1/2 phosphorylation in HMVEC-c after stimulation with 50 ng/ml of E. coli expressed wild-type HMGB1, K87A/K88A/K96A/R97A/K150A mutant or U937-derived HMGB1.
We next examined whether the quintuple mutant of HMGB1 was able to induce phosphorylation of Erk1/2. To our surprise, despite complete loss of heparan sulfate binding (Fig. 2D), mutant HMGB1 induced Erk1/2 phosphorylation as well as the control (Fig. 4C). Nevertheless, induction of Erk1/2 phosphorylation depended on cell surface heparan sulfate because heparin lyase treatment abolished phosphorylation (Fig. 4C). This finding indicated that heparan sulfate must be essential for the function of certain cell surface receptors critical for HMGB1 signaling.
Heparan Sulfate Interacts with RAGE
The cytokine function of extracellular HMGB1 is mediated primarily through RAGE. Like HMGB1, sRAGE binds to heparin and heparan sulfate with relatively high affinity (4.8 ± 0.4 nm and 28 ± 2.4 nm for heparin and heparan sulfate, respectively) (Fig. 5A, compare with Fig. 1A). sRAGE also binds to CHO cells in a heparan sulfate-dependent manner based on heparin lyase treatment (RFU = 3 versus 43, Fig. 5B) and studies of pgsD cells (RFU = 2.5 versus 43, Fig. 5C). Binding to pgsE cells was reduced to 33% (RFU = 14 versus 43, Fig. 5C), suggesting that N-sulfation was important for binding. Interestingly, binding was only very slightly reduced in pgsF cells (RFU = 35 versus 43, Fig. 5C), suggesting that 2-O-sulfation was dispensable. Heparin blocked binding to wild-type cells with an IC50 of 3.2 ± 1.1 nm. 2-O-desulfo- and 6-O-desulfo-heparin competed almost as well as unmodified heparin (IC50 = 4.1 ± 0.9 and 4.1 ± 1.4 nm, respectively), which suggests that sulfation at these positions was not required for sRAGE binding (Fig. 5D). In contrast, N-desulfo-heparin competed poorly compared with heparin (IC50 = 28 ± 8.1 nm; Fig. 5D). Thus, both RAGE and HMGB1 critically depend on N-sulfation for binding to heparan sulfate.
FIGURE 5.
RAGE interacts with heparan sulfate. A, binding of biotinylated sRAGE to immobilized heparin (circles) and heparan sulfate (squares) was measured. B, binding of biotinylated sRAGE (2 μg/ml) to wild-type CHO-K1 cells was measured by flow cytometry with or without heparin lyase pretreatment. C, binding of biotinylated sRAGE to various CHO mutants. D, heparin and various desulfated heparins compete for sRAGE binding to CHO cells. Strep-PE, streptavidin-phycoerythrin.
RAGE is a functional receptor for HMGB1 on HMVEC-c because a blocking antibody totally abolished the phosphorylation of Erk1/2 and p38 (Fig. 6A). We predicted that RAGE and heparan sulfate might form a complex at the cell surface. Using two antibodies, one against heparan sulfate and the other against RAGE, we showed by proximity ligation that complexes exist on the surface of endothelial cells. Interesting, the RAGE/heparan sulfate interaction was readily detected in the absence of HMGB1 (Fig. 6B), suggesting that there were preformed complexes of heparan sulfate and RAGE at endothelial surface. A time-dependent increase of association occurred after stimulation with HMGB1, increasing by 35% at 5 min (Fig. 6B), possibly as a result of receptor complex stabilization upon ligand binding.
FIGURE 6.
RAGE-dependent signaling. A, immunoblot analysis of Erk1/2 and p38 phosphorylation in HMVEC-c after stimulation with HMGB1. Some wells were preincubated with a blocking antibody (10 μg/ml) to RAGE for 20 min prior to stimulation. B, interaction between endogenous RAGE and heparan sulfate at the cell surface as measured by a proximity ligation assay. HMVEC-c cells were either unstimulated or stimulated with 100 ng/ml HMGB1 for 5 min, followed by fixation and incubation with antibodies to heparan sulfate, the extracellular domain of RAGE, and proximity ligation assay reagents. Each red dot indicates an interaction between heparan sulfate and RAGE. Nuclei are shown in blue. The data are presented as red dots (duolink signals) per cell. As a negative control, cells were incubated with anti-heparan sulfate antibody and nonspecific rabbit IgG. Error bars represent S.E. (n = 4 separate experiments). Representative images of unstimulated samples were shown in the lower panel.
DISCUSSION
In this study, we show that cell surface biotinylation coupled with heparin affinity chromatography tags many traditionally nuclear and cytoplasmic proteins. Although lysis of some cells could render these proteins susceptible to biotinylation, many other abundant heparin-binding proteins such as histones and various transcription factors, RNA binding proteins, and proteins involved in DNA replication were not detected. Other studies have shown that several of the tagged proteins are secreted via non-conventional pathways or during necrosis (17). Regardless of the mechanism, the release of nuclear and cytoplasmic proteins represents a danger signal that tissue damage has occurred. That so many of these proteins bind to heparin suggested that their extracellular activities might depend on cell surface heparan sulfate, which unlike heparin is expressed ubiquitously by virtually all animal cells. Indeed, among the identified proteins, cyclophilin A (23), cyclophilin B (24), and hepatoma-derived growth factor (25) have been shown to interact with heparan sulfate and cause a cellular response. Our attention was drawn to HMGB1 because it consistently showed very high peptide coverage in three proteomic screens, it is one of the major DAMPs released by necrotic cells during tissue injury, and it induces angiogenic and inflammatory responses in endothelial cells. We showed genetically and biochemically that HMGB1 is indeed a heparan sulfate dependent signaling factor, but to our surprise, the requirement for heparan sulfate is manifested at the level of the receptor RAGE rather than HMGB1.
Three other proteins identified in the screen belong to the heat shock protein (HSP) family (Table 1). Binding between HSPs and heparin has been documented as well, but whether the interaction is physiologically relevant has not been studied (26). Similar to HMGB1, HSPs are DAMPs released from necrotic cells during tissue injury and can trigger inflammatory responses (17, 27). This raises the interesting possibility that heparan sulfate, by interacting with multiple DAMPs, might serve as a central coreceptor that actuates the inflammatory response in injured tissue.
The interaction of heparan sulfate with FGF2-FGFR has become a paradigm for the action of heparan sulfate as a coreceptor (28, 29). However, the dependence of HMGB1 activity on heparan sulfate differs in several ways from the interaction of heparan sulfate with FGF2. First, N-sulfation plays a more prominent role in mediating heparan sulfate binding to HMGB1 than to FGF2 (Fig. 1C and supplemental Fig. S1B). Although the sprouting response of Ndst1-deficient endothelial cells to HMGB1 is completely abolished, its response to FGF2, albeit substantially reduced, remained significant (4.2-fold over untreated cells, Fig. 3B). Second, the requirement of heparan sulfate for signal transduction can be overcome by increasing the concentration of FGF2 (14), whereas even a 50-fold excess of HMBG1 (2.5 μg/ml) failed to induce signal transduction in the absence of heparan sulfate (Fig. 4B). The complete dependence of HMGB1 on heparan sulfate coreceptors suggests a fundamental difference in the way that RAGE and FGF receptors function.
HMGB1 bears an exceptionally large number of basic residues (24% of total amino acids). Mutagenesis revealed that only six basic residues make significant contribution to heparan sulfate binding. Five of these residues, Lys-87, Lys-88, Lys-90, Lys-96, Arg-97, are located within the second half of a long loop region that connects the A-box and B-box domains. A single residue in the last helix of B-box, Lys-150, also contributes significantly to binding. Combined, these residues form a contiguous string of positive charges that lie nearly perpendicular to the long axis of the HMGB1 (Fig. 2B). Four of the six residues, Lys-87, Lys-90, Lys-96, and Arg-97, also contribute to DNA binding (30, 31). However, DNA binding requires at least four additional charged residues in the A-box and six in the B-box (32). Therefore, it appears that HMGB1 uses overlapping but distinct basic residues for heparan sulfate and DNA binding.
The quintuple mutant K87A/K88A/K96A/K97A/K150A of HMGB1 represents an ideal tool to study the physiological consequences of blunted HMGB1/heparan sulfate interactions. By analogy, the quintuple mutant is similar to the non-heparin binding form of VEGF, VEGF121, which is able to bind and stimulate VEGF receptors (15). Similarly, the quintuple mutant of HMGB1 signals independently of its capacity to bind heparan sulfate (Fig. 4C). However, genetic studies have shown that mutant animals expressing only VEGF121 do not survive due to misregulated angiogenesis (33). This phenomenon can be explained by failure of VEGF121 to establish the correct spatial localization, whereas the heparin binding forms of VEGF can interact with heparan sulfate in the extracellular matrix and form the proper gradients needed to guide angiogenesis. A similar scenario may be relevant to HMGB1. When HMGB1 is released by necrotic tissue after damage, it presumably binds to heparan sulfate in the matrix, which in turn restricts the cellular responses to the area of local damage. We can test this hypothesis by injection or transgenic expression of the quintuple mutant in vivo.
During the inflammatory response, chemokines and cytokines secreted by monocytes or macrophages act directly on endothelial cells, and many of them function in a heparan sulfate-dependent manner (34). Endothelial heparan sulfate has profound effects on chemokine presentation, oligomerization, and transcytosis (35, 36). Similarly, heparan sulfate serves as a critical co-receptor for the proangiogenic factors FGF and VEGF by interacting with both ligands and receptors (14, 15, 37–39). We can now add HMGB1 and RAGE to the growing list of inflammatory mediators and receptors that depend on cell surface heparan sulfate. Although we used only endothelial cells in our studies, the importance of heparan sulfate in RAGE signaling may extrapolate to other cell systems expressing RAGE and other ligands that signal through this receptor.
Acknowledgments
We thank Geetha Srikrishna and Hudson Freeze for helpful discussions and providing reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant HL57345 (to J. D. E.). This work was also supported by Postdoctoral Fellowship 0825274F from the American Heart Association (to D. X.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- DAMP
- danger-associated molecular pattern protein
- RAGE
- the receptor for advanced glycation end products
- RFU
- relative fluorescence units
- sRAGE
- soluble RAGE
- HMVEC-c
- human cardiac microvascular endothelial cells.
REFERENCES
- 1. Lotze M. T., Tracey K. J. (2005) Nat. Rev. Immunol. 5, 331–342 [DOI] [PubMed] [Google Scholar]
- 2. Gardella S., Andrei C., Ferrera D., Lotti L. V., Torrisi M. R., Bianchi M. E., Rubartelli A. (2002) EMBO Rep. 3, 995–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kono H., Rock K. L. (2008) Nat. Rev. Immunol. 8, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klune J. R., Dhupar R., Cardinal J., Billiar T. R., Tsung A. (2008) Mol. Med. 14, 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rauvala H., Rouhiainen A. (2011) Biochim. Biophys. Acta 1799, 164–170 [DOI] [PubMed] [Google Scholar]
- 6. Fiuza C., Bustin M., Talwar S., Tropea M., Gerstenberger E., Shelhamer J. H., Suffredini A. F. (2003) Blood 101, 2652–2660 [DOI] [PubMed] [Google Scholar]
- 7. Mitola S., Belleri M., Urbinati C., Coltrini D., Sparatore B., Pedrazzi M., Melloni E., Presta M. (2006) J. Immunol. 176, 12–15 [DOI] [PubMed] [Google Scholar]
- 8. Sims G. P., Rowe D. C., Rietdijk S. T., Herbst R., Coyle A. J. (2010) Annu. Rev. Immunol. 28, 367–388 [DOI] [PubMed] [Google Scholar]
- 9. Yan S. F., Ramasamy R., Schmidt A. M. (2010) Circ. Res. 106, 842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Basta G. (2008) Atherosclerosis 196, 9–21 [DOI] [PubMed] [Google Scholar]
- 11. Hudson B. I., Kalea A. Z., Del Mar Arriero M., Harja E., Boulanger E., D'Agati V., Schmidt A. M. (2008) J. Biol. Chem. 283, 34457–34468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ishihara K., Tsutsumi K., Kawane S., Nakajima M., Kasaoka T. (2003) FEBS Lett. 550, 107–113 [DOI] [PubMed] [Google Scholar]
- 13. Zhang L., Lawrence R., Frazier B. A., Esko J. D. (2006) Methods Enzymol. 416, 205–221 [DOI] [PubMed] [Google Scholar]
- 14. Fuster M. M., Wang L., Castagnola J., Sikora L., Reddi K., Lee P. H., Radek K. A., Schuksz M., Bishop J. R., Gallo R. L., Sriramarao P., Esko J. D. (2007) J. Cell Biol. 177, 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu D., Fuster M. M., Lawrence R., Esko J. D. (2011) J. Biol. Chem. 286, 737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Christian S., Pilch J., Akerman M. E., Porkka K., Laakkonen P., Ruoslahti E. (2003) J. Cell Biol. 163, 871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harris H. E., Raucci A. (2006) EMBO Rep. 7, 774–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McArthur C., Wang Y., Veno P., Zhang J., Fiorella R. (2002) Arch Oral Biol. 47, 443–448 [DOI] [PubMed] [Google Scholar]
- 19. Brandt R., Nawka M., Kellermann J., Salazar R., Becher D., Krantz S. (2004) Biochim. Biophys. Acta 1670, 132–136 [DOI] [PubMed] [Google Scholar]
- 20. Jordan P., Heid H., Kinzel V., Kübler D. (1994) Biochemistry 33, 14696–14706 [DOI] [PubMed] [Google Scholar]
- 21. Rauvala H., Pihlaskari R. (1987) J. Biol. Chem. 262, 16625–16635 [PubMed] [Google Scholar]
- 22. Salmivirta M., Rauvala H., Elenius K., Jalkanen M. (1992) Exp. Cell Res. 200, 444–451 [DOI] [PubMed] [Google Scholar]
- 23. Saphire A. C., Bobardt M. D., Gallay P. A. (1999) EMBO J. 18, 6771–6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Allain F., Vanpouille C., Carpentier M., Slomianny M. C., Durieux S., Spik G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2714–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang C. H., Davamani F., Sue S. C., Lee S. C., Wu P. L., Tang F. M., Shih C., Huang T. H., Wu W. G. (2011) Biochem. J. 433, 127–138 [DOI] [PubMed] [Google Scholar]
- 26. Ménoret A., Bell G. (2000) J. Immunol. Methods 237, 119–130 [DOI] [PubMed] [Google Scholar]
- 27. Millar D. G., Garza K. M., Odermatt B., Elford A. R., Ono N., Li Z., Ohashi P. S. (2003) Nat. Med. 9, 1469–1476 [DOI] [PubMed] [Google Scholar]
- 28. Yayon A., Klagsbrun M., Esko J. D., Leder P., Ornitz D. M. (1991) Cell 64, 841–848 [DOI] [PubMed] [Google Scholar]
- 29. Rapraeger A. C., Krufka A., Olwin B. B. (1991) Science 252, 1705–1708 [DOI] [PubMed] [Google Scholar]
- 30. Stros M., Muselíková E. (2000) J. Biol. Chem. 275, 35699–35707 [DOI] [PubMed] [Google Scholar]
- 31. Taudte S., Xin H., Kallenbach N. R. (2000) Biochem. J. 347, 807–814 [PMC free article] [PubMed] [Google Scholar]
- 32. Stott K., Tang G. S., Lee K. B., Thomas J. O. (2006) J. Mol. Biol. 360, 90–104 [DOI] [PubMed] [Google Scholar]
- 33. Carmeliet P., Ng Y. S., Nuyens D., Theilmeier G., Brusselmans K., Cornelissen I., Ehler E., Kakkar V. V., Stalmans I., Mattot V., Perriard J. C., Dewerchin M., Flameng W., Nagy A., Lupu F., Moons L., Collen D., D'Amore P. A., Shima D. T. (1999) Nat. Med. 5, 495–502 [DOI] [PubMed] [Google Scholar]
- 34. Handel T. M., Johnson Z., Crown S. E., Lau E. K., Proudfoot A. E. (2005) Annu. Rev. Biochem. 74, 385–410 [DOI] [PubMed] [Google Scholar]
- 35. Wang L., Fuster M., Sriramarao P., Esko J. D. (2005) Nat. Immunol. 6, 902–910 [DOI] [PubMed] [Google Scholar]
- 36. Proudfoot A. E., Handel T. M., Johnson Z., Lau E. K., LiWang P., Clark-Lewis I., Borlat F., Wells T. N., Kosco-Vilbois M. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pellegrini L., Burke D. F., von Delft F., Mulloy B., Blundell T. L. (2000) Nature 407, 1029–1034 [DOI] [PubMed] [Google Scholar]
- 38. Schlessinger J., Plotnikov A. N., Ibrahimi O. A., Eliseenkova A. V., Yeh B. K., Yayon A., Linhardt R. J., Mohammadi M. (2000) Mol. Cell 6, 743–750 [DOI] [PubMed] [Google Scholar]
- 39. Jakobsson L., Kreuger J., Holmborn K., Lundin L., Eriksson I., Kjellén L., Claesson-Welsh L. (2006) Dev. Cell 10, 625–634 [DOI] [PubMed] [Google Scholar]