Abstract
The Shu complex in yeast plays an important role in the homologous recombination pathway, which is critical for the maintenance of genomic integrity. The identification of human SWS1 (hSWS1) as the homolog of budding yeast Shu2 implicated that the Shu complex is evolutionarily conserved. However, the human counterparts of other components in this complex have not yet been identified and characterized. Here we describe the characterization of a novel human component of this complex, SWSAP1 (hSWS1-associated protein 1)/C19orf39. We show that hSWS1 and SWSAP1 form a stable complex in vivo and in vitro. hSWS1 and SWSAP1 are mutually interdependent for their stability. We further demonstrate that the purified hSWS1·SWSAP1 complex possesses single-stranded DNA-binding activity and DNA-stimulated ATPase activity. Moreover, SWSAP1 interacts with RAD51 and RAD51 paralogs, and depletion of SWSAP1 causes defects in homologous recombination repair. Thus, our results suggest that the human Shu complex (hSWS1·SWSAP1) has an evolutionarily conserved function in homologous recombination.
Keywords: DNA Damage, DNA Recombination, DNA Repair, Homologous Recombination, Protein-protein Interactions
Introduction
The integrity of the genome is continuously challenged by both exogenous and endogenous DNA-damaging agents that induce a wide range of lesions in DNA, such as double strand breaks, single strand breaks, and pyrimidine dimers (1–3). Homologous recombination repair (HRR)4 is a well-conserved cellular process for the repair of single strand DNA (ssDNA) gaps and double strand DNA breaks that arise during DNA synthesis or as a result of replication fork stalling during S-phase (4–6). Accurate homologous recombination repair, using the sister chromatid as a template, is necessary for the maintenance of genome stability, and defects in this process often lead to genomic instability and ultimately initiate cancer development (4–6). Homologous recombination repair is facilitated by a superfamily of recombinases: bacterial RecA, archaeal RadA and Rad51, and eukaryal Rad51 and DMC1 (7–11).
Genetic and biochemical studies in yeast have identified another protein complex, the Shu complex, which has an important role in HRR (12, 13). In Saccharomyces cerevisiae, the Shu complex is comprised of four subunits, Csm2, Psy3, Shu1, and Shu2, which were originally identified as four novel genes within the same epistasis group that, when mutated, can suppress various defects in sgs1 or top3 mutants (13–17). Interestingly, the single csm2, psy3, shu1, or shu2 mutants all demonstrate similar phenotypes (i.e. a mutator phenotype and moderate sensitivity the DNA alkylating agent methylmethane sulfonate (MMS) and cross-linking agents), and mutation of all four does not cause any additive effects (13–17). In addition, all four Shu gene products interact with each other and they fail to function when any one of the members is missing (13–18). These data indicate that these four proteins exist in a multimeric complex and function in the same pathway. Further studies in Schizosaccharomyces pombe revealed Rdl1 and SWS1 are the putative Psy3 and Shu2 homologs, respectively (14). Consistently, like psy3 and shu2 mutations in S. cerevisiae, mutation of rdl1 or sws1 in S. pombe also causes increased sensitivity to MMS and rescues various cellular defects caused by mutation of the recQ helicase gene, rqh1, in S. pombe (14). Furthermore, SWS1 associates in vivo with Rdl1 and a novel protein, Rlp1. Although Rdl1 and Rlp1 share no obvious sequence similarities to Shu1 or Csm2, functional assays reveal that SWS1, Rdl1, and Rlp1 have interdependent functions in controlling recombination (14). Therefore, SWS1, Rdl1, and Rlp1 appear to be part of a multimeric complex in S. pombe, which is similar to the complex proposed to exist in S. cerevisiae that comprises Csm2, Psy3, Shu1, and Shu2 (14). Interestingly, SWS1 is highly conserved in eukaryotes, and ablation of hSWS1 in human cells reduces the number of RAD51 foci, suggesting that Shu-like complexes also exist in human cells and therefore probably perform an evolutionarily conserved role in HRR (14). However, one or more additional subunits of the human Shu complex remains to be identified and characterized.
In this study, we used an affinity purification approach to isolate hSWS1-containing complex and identified a novel protein C19orf39, which we refer to as SWSAP1 (hSWS1-associated protein 1). We demonstrated that hSWS1 and SWSAP1 form a stable complex that maintains the level of both proteins in cells. Similar defects in HRR were observed in both hSWS1- and SWSAP1-depleted cells, suggesting that the human Shu complex (hSWS1·SWSAP1) has an evolutionarily conserved function required for efficient homologous recombination repair.
EXPERIMENTAL PROCEDURES
Plasmids
The full-length and deletion/point mutants of human sws1 and swsap1 were generated by PCR and subcloned into the pDONR201 vector using Gateway technology (Invitrogen). The corresponding fragments in entry vectors were transferred into a Gateway-compatible destination vector, which harbors an N-terminal triple-epitope tag (S protein tag, FLAG epitope tag and Streptavidin-binding peptide tag) or Myc epitope tag for expression in mammalian cells. The full-length human rad51, rad51 paralogs, and soss-c were described previously (6, 19). The siRNA-resistant wild-type and mutant swsap1 constructs were generated by changing five nucleotides in the swsap1 siRNA #2 targeting region (G465C, A468C, G471A, C474T, and G477A substitutions).
Cell Cultures and Transfection
293T and HeLa cells were maintained in RPMI supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. SF9 insect cells were maintained in Grace's medium supplemented with 10% fetal bovine serum. Cell lines of human origin were maintained in a 37 °C incubator with 5% CO2, whereas insect cells were maintained at 27 °C. U2OS cells with DR-GFP integration were kindly provided by Maria Jasin at Memorial Sloan-Kettering Cancer Center (New York). Cell transfection was performed using Lipofectamine 2000 (Invitrogen) following the manufacture's protocol.
Antibodies
Rabbit polyclonal antibodies specifically recognizing hSWS1 or SWSAP1 were raised against peptides SSPSGRRVYQVLGSSSKTYTC and CWLQPDAPGPGEHG, respectively. Anti-GST and anti-Myc antibodies were purchased from Santa Cruz Biotechnology and anti-FLAG (M2) antibody from Sigma. Antibodies recognizing RAD51, RAD51C, and GAPDH were described previously (6, 20).
Baculoviruses
DNA fragment containing full-length/point mutants of hsws1, swsap1, or soss-c in pDONR201 vector were transferred to pDEST20, pDEST10, SFB-tagged, and pDEST8 vectors for the expression of GST-, His-, SFB-tagged, and non-tagged proteins in insect cells, respectively. Transposition occurred in DH10Bac-competent cells, and correct bacmids confirmed by PCR were transfected into SF9 cells for baculovirus production. After viral amplification, SF9 cells were infected with desired baculovirus, and cell lysates were collected 48 h later for the purification of various recombinant proteins.
Gel Filtration Chromatography
Sf9 cells were co-infected with baculovirus stocks expressing His-tagged SWSAP1 and non-tagged hSWS1. Forty-eight hours later, cells were harvested, washed with 1× PBS, and resuspended in lysis buffer (20 mm Tris-HCl (pH 8.0), 200 mm NaCl, 0.01% Nonidet P-40, 1 mm DTT, and 1 μg/ml each of leupeptin and aprotinin). The lysate was centrifuged for 10 min at 12,000 rpm. The supernatant was incubated at 4 °C with 200 μl of nickel-nitrilotriacetic acid beads for 4 h. Nickel-nitrilotriacetic acid beads were washed with wash buffer (lysis buffer containing 20 mm imidazole) and eluted with elution buffer (lysis buffer containing 200 mm imidazole). The eluted protein was dialyzed in buffer A (20 mm Tris-HCl (pH 8.0), 200 mm NaCl, 0.01% Nonidet P-40) and resolved on Superdex 200 gel filtration column against buffer A. Indicated fractions were analyzed on 12.5% SDS-PAGE.
ATPase Assays
An Innova Biosciences ATPase assay kit was used following the manufacturer's instructions. The assay mixture (20 μl) contained 1 μm purified protein, 50 mm Tris-HCl, 2.5 mm MgCl2, 0.5 mm ATP, either in the absence of DNA or with 10 μm d40T. After incubation at 37 °C for the indicated time, the reaction was stopped by the addition of 5 μl of Gold mix for 2 min; 2 μl of stabilizer was then added, and the mixture was incubated for another 30 min at room temperature. The absorbance was measured at 650 nm using Nanodrop2000. For protein purification, Sf9 cells were infected with baculovirus stocks expressing GST-SWS1, GST-SWSAP1-WT, GST-SWSAP1-K18A, GST-SWSAP1-D96A, or GST-SWSAP1·SFB-SWS1 complex. Forty-eight hours later, cells were harvested, washed with 1× PBS, and resuspended in lysis buffer (20 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, 1 mm DTT and 1 μg/ml each of leupeptin and aprotinin). The lysate was centrifuged for 15 min at 10,000 rpm. The supernatant was incubated at 4 °C with 200 μl of glutathione-Sepharose 4B for 4 h. The resin was washed with lysis buffer, and protein was eluted with elution buffer (50 mm Tris-HCl (pH 8.0) and 20 mm glutathione).
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assays were done using a Lightshift chemiluminescent electrophoretic mobility shift assay kit (Pierce) as instructed by the manufacturer. Reaction mixture (20 μl) contained 1× binding buffer, 50 ng/μl poly(dI-dC), 10 mm MgCl2, 100 ng/μl BSA, 5′-biotin end-labeled ssDNA (d58T) (20 fmol) or dsDNA (d58A-T; d58T annealed to its complimentary d58A) (20 fmol), and indicated concentrations of purified protein. After incubated for 30 min at 37 °C, the reaction was terminated by the addition of 5 μl of gel loading buffer. Samples were separated by 5% native polyacrylamide gel and transferred to PVDF membrane on ice. After cross-linking DNA to membrane using a UV-light cross-linker (120 mJ/cm2), blocking, washing, and detection were performed using the Chemiluminescent Nucleic Acid Detection Module according to the manufacturer's instructions.
RNA Interference
Human sws1 and swsap1 siRNA duplexes (SMARTpool of four oligonucleotides) were purchased from Dharmacon Research (Lafayette, CO). The annotations and sequences of the Human sws1 and swsap1 siRNA oligonucleotides were as follows (sense strands): #1, 5′-GAUGAAGUUUCGACGAGUA-3′; #2, 5′-UCUGAGACGCUUCGGUAAA-3′; #3, 5′-GAGAGUUAGUAUCGAGAAC-3′; #4, 5′-CAAAGAAGCAAUAAUACGU-3′, and #1, 5′-AGAAGAGUACCUAGCGGAA-3′; #2, 5′-UGGCACUGCUCCAGCGGUA-3′; #3, 5′-GGAGAAAUGAUGAUCGCUC-3′, and #4, 5′-CUUAUUUACUGCUGCGAAC-3′, respectively. The sequence of RAD51C siRNA was 5′-G UACAGCACUGGAACUUCU-3′. siRNAs transfection was performed with 100 nm siRNA duplexes using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer's instruction.
The Establishment of Stable Cell Lines and Affinity Purification of S-FLAG-SBP(SFB)-tagged Protein Complexes
293T cells were transfected with plasmids encoding SFB-tagged proteins. Cell lines stably expressing tagged proteins were selected by culturing in the medium containing puromycin (2 μg/ml) and confirmed by immunoblotting and immunostaining. For affinity purification, 293T cells stably expressing tagged proteins were lysed with NETN buffer for 20 min. Crude lysates were removed by centrifugation at 14,000 rpm at 4 °C for 10 min, and the pellet was sonicated for 40 s in high salt solution (20 mm HEPES (pH 7.8), 0.4 m NaCl, 1 mm EDTA, 1 mm EGTA, and protease inhibitor) to extract chromatin-bound protein fractions. The supernatants were cleared at 14,000 rpm to remove debris and then incubated with streptavidin-conjugated beads (Amersham Biosciences) for 2 h at 4 °C. The beads were washed three times with NETN buffer, and then bead-bound proteins were eluted with NETN buffer containing 1 mg/ml biotin (Sigma). The elutes were incubated with S protein beads (Novagen). The beads were again washed three times with NETN buffer and subjected to SDS-PAGE. Protein bands were excised and digested, and the peptides were analyzed by mass spectrometry.
Co-immunoprecipitation and Western Blotting
For co-immunoprecipitation assays, constructs encoding SFB-tagged and Myc-tagged proteins were transiently co-transfected into 293T cells. Cells were lysed with NETN buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40) containing 20 mm NaF, and 1 μg/ml pepstatin A and aprotinin on ice for 20 min. After removal of cell debris by centrifugation, the soluble fractions were collected and incubated with S-protein beads for 3 h at 4 °C. Beads were washed four times with NTEN buffer, boiled in 2× SDS loading buffer, and resolved on SDS-PAGE. Membranes were blocked in 5% milk in TBST buffer and then probed with antibodies as indicated.
Immunofluorescence Staining
To visualize damage-induced foci, cells cultured on coverslips were treated with 1 mm of MMS and then allowed to recover. Cells were then washed with PBS and fixed using 3% paraformaldehyde solution for 10 min at room temperature and then extracted with buffer containing 0.5% Triton X-100 for 5 min. Samples were blocked with 5% goat serum and incubated with primary antibody for 30 min. Samples were washed and incubated with secondary antibody for 30 min. Cells were then counterstained with DAPI to visualize nuclear DNA.
Cell Survival Assays
Cells (1 × 103) were seeded onto 60-mm dish in triplicates. At 24 h after seeding, cells were treated with MMS or CPT. The medium was replaced 24 h later, and cells were then incubated for 14 days. Resulting colonies were fixed and stained with Coomassie Blue, and numbers of colonies were counted.
Gene Conversion Assay
A U2OS derivative clone stably expressing HR reporter DR-GFP was described previously (21). 1 × 106 U2OS-DR-GFP cells were electroporated with 12 μg of pCBASce plasmid at 270 V, 975 microfarads using a Bio-Rad GenePulser II. Cells were plated onto 10-cm dishes and incubated for 48 h prior to FACS analyses using a BD Biosciences FACScan on a green (FL1) versus orange (FL2) fluorescence plot. Results were the averages of data obtained from three independent experiments.
RESULTS
Identification of SWSAP1/C19orf39 as a New Component of Human Shu Complex
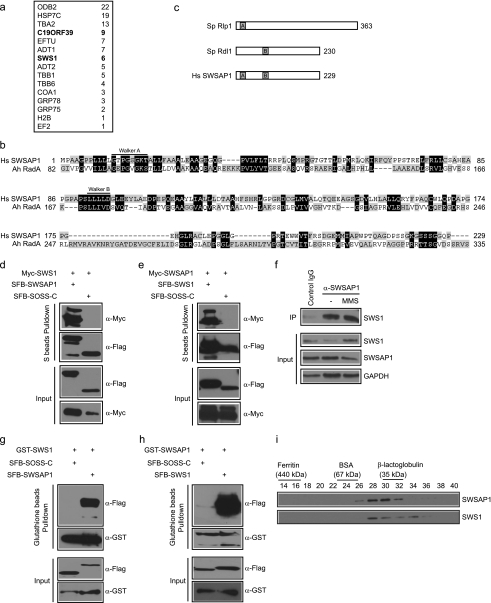
In an attempt to understand how the human Shu complex is assembled in vivo, we performed tandem affinity purification using HEK293T cells stably expressing streptavidin-FLAG-S protein (SFB)-tagged wild-type hSWS1, the homolog of budding yeast Shu2, for the identification of hSWS1-containing protein complexes. We repeatedly found a novel protein C19orf39 as the major hSWS1 binding partner (Fig. 1a). On the basis of its functions described below, we designated this protein as SWSAP1 (hSWS1-associated protein 1).
FIGURE 1.
Identification of SWSAP1 as hSWS1-binding protein. a, 293T cells stably expressing SFB-tagged (S-tag, FLAG epitope tag, and streptavidin-binding peptide tag) hSWS1 was used for tandem affinity purification of protein complexes isolated from chromatin fractions. Tables are summaries of proteins identified by mass spectrometry analysis. Letters in bold indicate the bait proteins. b, alignment of human SWSAP1 with Arcanobacterium hemolyticum RadA. The conserved Walker A and Walker B motifs are indicated. c, schematic representation of fission yeast Rlp1, Rdl1, and human SWSAP1. d and e, the interaction between hSWS1 and SWSAP1 was confirmed by co-immunoprecipitation (IP) experiments. 293T cells were transfected with plasmids encoding Myc-tagged hSWS1 or SWSAP1 together with plasmids encoding SFB-tagged SWSAP1 or hSWS1 as indicated. Cells were collected 24 h after transfection. Precipitation reactions were performed using S beads, and immunoblotting was carried out using antibodies as indicated. f, endogenous hSWS1 and SWSAP1 form a complex in vivo. HeLa cells were mock treated or treated with MMS. Control or anti-SWSAP1 immunoprecipitates were immunoblotted with anti-hSWS1 antibody (top). The expression levels of endogenous proteins were revealed by immunoblotting using anti-hSWS1 and anti-SWSAP1 antibodies (bottom). g and h, hSWS1 and SWSAP1 directly bind to each other. SF9 cells were co-infected with baculoviruses expressing GST-tagged hSWS1 or SWSAP1 together with those expressing SFB-tagged SWSAP1 or hSWS1. Pulldown experiments and immunoblotting were carried out as indicated. i, heterodimeric complex formation was studied by gel filtration chromatography as described under “Experimental Procedures.” Aliquots from peak fractions were analyzed on 12.5% SDS-PAGE and confirmed by Western blot analysis.
Human SWSAP1 contains 229 amino acid residues. However, there was no further characterization of this protein concerning its domain structure, activity, or biological functions. We used full-length of SWSAP1 in PSI BLAST-based database searches and found that SWSAP1 has similarity to RadA, a RecA family protein in archaeal (10, 11) (Fig. 1b). All RecA/RadA-like family members have two NTP binding motifs: Walker A box and Walker B box. Structural analysis of SWSAP1 protein sequence revealed that both the Walker A and B boxes are present in SWSAP1 (Fig. 1c). This is in contrast to the SWS1-Rlp1-Rdl1 complex in S. pombe, where the walker A and B motifs are present in two separate components, Rlp1 (containing a Walker A motif) and Rdl1 (a Walker B motif), respectively (14) (Fig. 1c).
The components of Shu complex in yeast have been shown to form a stable complex (12–17). To verify that human SWS1 and SWSAP1 also physically interact with each other, we first performed co-immunoprecipitation experiments using epitope-tagged hSWS1 and SWSAP1. We found that SFB-tagged SWSAP1 interacted strongly with Myc-tagged hSWS1 but not with the SOSS-C control protein (19), and the reverse experiment confirmed this result (Fig. 1, d and e). Interestingly, we found that the expression of tagged hSWS1 or SWSAP1 was greatly enhanced when they were co-expressed in cells (Fig. 1, d and e), which indicates that SWSAP1 and hSWS1 might form a stable complex in vivo and are mutually interdependent for their stability. To determine whether endogenous hSWS1 and SWSAP1 interact, HeLa cell lysates were prepared and subjected to immunoprecipitation with either a control or anti-SWSAP1 antibodies. As expected, endogenous hSWS1 was immunoprecipitated by anti-SWSAP1 antibody, but not by control antibody (Fig. 1f). This interaction does not change before or after treatment of cells with MMS or CPT (Fig. 1f and data not shown), indicating that the interaction between SWSAP1 and hSWS1 is not DNA damage-dependent.
Moreover, we expressed SFB- and GST-tagged SWSAP1 and hSWS1 in insect cells. Pull down assays demonstrated that SWSAP1 and hSWS1 bind directly to each other (Fig. 1, g and h). To further confirm that SWSAP1 and hSWS1 exist in the same complex, insect cells were co-infected with baculovirus expressing His-tagged SWSAP1 and non-tagged hSWS1, and complex formation was studied by FPLC. As shown in Fig. 1I and supplemental Fig. 1Sa, SWSAP1 and hSWS1 co-eluted as a heterodimeric complex with a molecular mass of ∼40 kDa. Together, these data support that human SWSAP1 and hSWS1 form a stable complex.
hSWS1 and SWSAP1 Are Interdependent for Their Stability
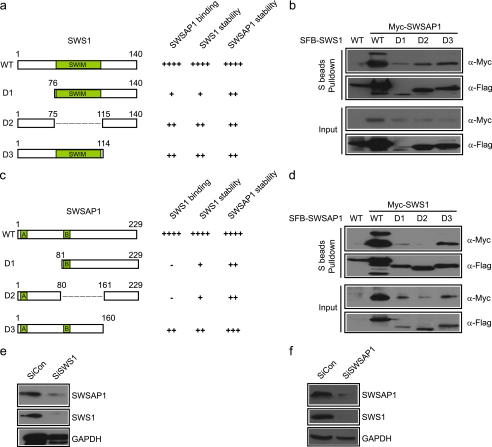
To further define the binding between hSWS1 and SWSAP1, we generated a series of truncation or internal deletion mutants of hSWS1 and SWSAP1. As shown in Fig. 2 (a–d), we found that, although full-length hSWS1 and SWSAP1 associated with each other, any deletion mutants would decrease this interaction. Interestingly, analysis of full-length or deletion mutants of hSWS1 or SWSAP1 revealed a general correlation between the expression level of a mutant with its ability to bind to its partner (Fig. 2, a–d).
FIGURE 2.
hSWS1 and SWSAP1 are interdependent for their stability. a–c, schematic representation of human SWS1 and SWSAP1 and their deletion mutants used in this study. b–d, mapping of the corresponding regions required for hSWS1-SWSAP1 interaction. Precipitation reactions were performed using S beads, and immunoblotting was carried out using antibodies as indicated. e and f, depletion of hSWS1 or SWSAP1 leads to the loss of both proteins in the cell. HeLa cells were transfected with control siRNA or siRNAs specifically targeting hSWS1 or SWSAP1. Lysates were analyzed by immunoblotting using antibodies recognizing hSWS1, SWSAP1, or GAPDH.
The stability of one component in a multiprotein complex often depends on the presence of other components so that when one protein is depleted, the others become unstable and show reduced expression levels (19). Indeed, reduction of hSWS1 expression by siRNA led to a dramatic decrease of SWSAP1 protein level in the cell (Fig. 2e). Conversely, depletion of SWSAP1 by siRNA also resulted in a reduction of hSWS1 expression (Fig. 2f). Together, these results demonstrate that the maintenance of hSWS1 protein level depends on SWSAP1 and vice versa.
SWSAP1 Is a DNA-stimulated ATPase and Preferentially Binds Single-stranded DNA
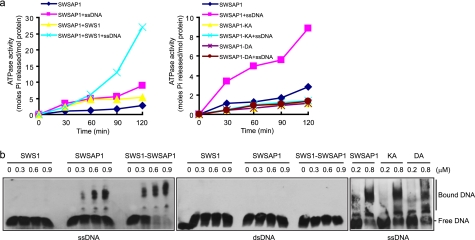
Like RecA/RadA-like family members, SWSAP1 contains Walker A/Walker B motifs and is, therefore, predicted to be a nucleotide-binding protein that can hydrolyze ATP. Indeed, SWSAP1 or hSWS1·SWSAP1 complex was able to hydrolyze ATP in vitro. Interestingly, the ATPase activity was significant increased in the presence of single-stranded DNA (ssDNA) compared with double-stranded DNA (dsDNA) (Fig. 3a and data not shown). As expected, site-directed mutagenesis of SWSAP1 at highly conserved residue Lys-18 or Asp-96 in the Walker A and B motifs, respectively, led to severe defects in ssDNA-stimulated ATPase activity (Fig. 3a and supplemental Fig. 1Sb).
FIGURE 3.
SWSAP1 is a DNA-stimulated ATPase and preferentially binds Single-stranded DNA. a, time course for ATPase activity of SWSAP1 and hSWS1·SWSAP1 complex. SWSAP1, hSWS1·SWSAP1 complex, or SWSAP1 ATPase-inactivating mutants was incubated with or without ssDNA as described under “Experimental Procedures.” b, increasing concentrations of hSWS1, SWSAP1, hSWS1·SWSAP1 complex and SWSAP1 ATPase-inactivating mutants were incubated with 5′-biotin-labeled ssDNA or dsDNA as described under “Experimental Procedures.” Protein·DNA complexes and unbound DNA are indicated on the right.
Next we sought to test whether SWSAP1, like members of the RecA/RadA-like family, has a capacity to bind DNA. As shown in Fig. 3b, SWSAP1 or the hSWS1·SWSAP1 complex, but not hSWS1 alone, was found to preferentially bind to ssDNA compared with dsDNA. Moreover, DNA binding by SWSAP1 or SWS1·SWSAP1 complex was not dependent on the presence of ATP (data not shown). Accordingly, ATPase-inactivating mutants of SWSAP1 retained the ability to bind ssDNA (Fig. 3b and supplemental Fig. 1Sc).
SWSAP1 Interacts with RAD51 and RAD51 Paralogs and Is Involved in Homologous Recombination
Cells deficient in hSWS1 displayed increased MMS sensitivity and reduced RAD51 foci formation (14). We thus examined whether the loss of SWSAP1 would result in similar defects in DNA damage repair. As shown in Fig. 4a, SWSAP1-depleted cells were more sensitive to MMS than control cells. Moreover, we performed a gene conversion assay to examine HR efficiency using the DR-GFP reporter system. HR repair efficiency was reduced by ∼2-fold in SWSAP1-depleted cells (Fig. 4, b and c). The recombination protein RAD51 is the key component of the homologous recombination repair machinery, and the formation of Rad51 foci can be used as another indicator of HR repair. In agreement with the results from gene conversion assay, DNA damage-induced RAD51 foci formation was also impaired in SWSAP1-depleted cells (Fig. 4, d and e). Significantly, depletion of SWSAP1 leads to decreased homologous recombination frequencies, reduced RAD51 foci formation ability, and increased sensitivity to genotoxic stress to levels similar to those achieved by depleting human SWS1 (Fig. 4, a–f). Moreover, depleting hSWS1 and SWSAP1 together did not decrease homologous recombination efficiency further than was achieved by hSWS1 or SWSAP1 depletion alone (Fig. 4, a–f). These findings support that hSWS1 and SWSAP1 promote homologous recombination via a common pathway. Flow cytometry analyses showed that cell cycle distribution was not affected by suppression of hSWS1 and/or SWSAP1 expression (data not shown), ruling out the possibility that the phenotypes observed in hSWS1- and/or SWSAP1-depleted cells may be due to any change in cell cycle distribution.
FIGURE 4.
SWSAP1 interacts with RAD51 and RAD51 paralogs and is involved in homologous recombination repair. a, cells with hSWS1 or SWSAP1 down-regulation display increased MMS but not CPT sensitivity. Cell survival assays were performed as described under “Experimental Procedures.” Data are presented as means ± S.D. (error bars) from three different experiments. b, schematic representation of HR assay. The DR-GFP construct consists of direct repeats of two mutated GFP genes, SceGFP and the truncated iGFP. When a single double strand DNA break generated by I-Sce1 is repaired via gene conversion with iGFP, expression of GFP is restored and can be measured by FACS analysis. c, decreasing hSWS1 or SWSAP1 expression impairs HR repair. U2OS DR-GFP cells were electroporated with pCBASce plasmids (see “Experimental Procedures” for details). The percentage of GFP-positive cells was determined by flow cytometry 48 h after electroporation. The data were normalized to those obtained from cells transfected with control siRNA (set as 1.0). Means ± S.E. (error bars) shown are obtained from three independent experiments. d and e, down-regulation of hSWS1 or SWSAP1 impairs MMS-induced RAD51 foci formation. Immunostaining was performed 6 h after MMS treatment using the indicated antibodies. Representative RAD51 foci are shown in d. Data were presented as means ± S.D. (error bars) from three different experiments, more than 100 cells were counted in each experiment (e). f, knockdown efficiency was confirmed by immunoblotting. g, SWSAP1 interacts with RAD51 and several RAD51 paralogs, but not with XRCC2 in vivo. 293T cells were transfected with plasmids encoding SFB-tagged SWSAP1 together with plasmids encoding Myc-tagged RAD51 or RAD51 paralogs. Cells were collected 24 h after transfection. Precipitation reactions were performed using S beads, and immunoblotting was carried out using antibodies as indicated. h, SWSAP1 interacts with RAD51 and several RAD51 paralogs, but not with XRCC2 in vitro. SF9 cells were co-infected with baculoviruses expressing GST-tagged RAD51 or RAD51 paralogs together with those expressing SFB-tagged SWSAP1. Pulldown experiments and immunoblotting were carried out as indicated. i, the ATPase activity of SWSAP1 is required for restoring cellular resistance to MMS. HeLa-derivative cell lines stably expressing siRNA-resistant HA-FLAG-tagged WT (SWSAP1SiR-WT), K18A mutant (SWSAP1SiR-KA), and D96A mutant (SWSAP1SiR-DA) of SWSAP1 were generated. Cell survival assays were performed as described under “Experimental Procedures.” Data are presented as means ± S.D. (error bars) from three different experiments. j, the ATPase activity of SWSAP1 is required for its function in promoting RAD51 foci formation. Immunostaining was performed 6 h after MMS treatment using indicated antibodies. Data are presented as means ± S.D. (error bars) from three different experiments, more than 100 cells were counted in each experiment. k, SWSAP1 expression was confirmed by immunoblotting with the use of FLAG antibody, and extracts were prepared from cells transfected with SWSAP1 siRNA#2.
DNA repair through homologous recombination requires the recombinase RAD51, and in vertebrates five RAD51 paralogs (22, 23). Since SWSAP1 shares some sequence homology with archaeal RecA family protein RadA, we examined whether SWSAP1 would be involved in HR through a direct interaction with RAD51 and/or its paralogs. Indeed, as shown in Fig. 4 (g and h), SWSAP1 interacts with RAD51 and several RAD51 paralogs, except XRCC2 in vivo and in vitro. Moreover, hSWS1 can also interact with RAD51D and XRCC3 in vitro (supplemental Fig. 2Sa). The observation that double knockdown of SWSAP1 and RAD51C led to increased MMS sensitivity and further reduced RAD51 foci formation when compared with SWSAP1 or RAD51C single knockdown indicates that hSWS1·SWSAP1 and RAD51 paralogs likely represent independent subpathways, which may function together in homologous recombination repair (supplemental Fig. 2S (b–d)).
To explore the physiological relevance of the highly conserved Walker A/Walker B motifs, we knocked down SWSAP1 in HeLa cells using siSWSAP1#2 (SWSAP1-specific small interfering RNA (siRNA) 2) and reintroduced siRNA-resistant full-length SWSAP1, or the ATPase-inactivating mutants (K18A and D96A) of SWSAP1. Clonogenic and immunostaining assays indicated that reconstitution with WT SWSAP1, but not its ATPase-inactivating mutants, restored cell survival and RAD51 foci formation after MMS treatment (Fig. 4, i–k), suggesting that the ATPase activity of SWSAP1 is important for SWSAP1 function in promoting homologous recombination after MMS treatment. Moreover, the phenotypes observed in SWSAP1 ATPase-inactivating mutant-reconstituted cells are not due to a failure in forming hSWS1·SWSAP1 protein complex (supplemental Fig. 3S).
DISCUSSION
In this study, we adopted the TAP approach to isolate human SWS1-containing protein complex and identified a novel component, SWSAP1, in this complex. Despite the low sequence similarity between SWSAP1 and the respective yeast proteins, we found that human SWS1 and SWSAP1 form a tight complex, similar to that of fission yeast Sws1, Rlp1, and Rdl1 or budding yeast Csm3, Psy3, Shu1, and Shu2, respectively (12–17). The integral nature of this complex is emphasized by the mutual interdependence of these two proteins for their stability and by the fact that depletion of hSWS1, SWSAP1, or together always produce similar phenotypes. These observations strongly support that hSWS1 and SWSAP1 form and act as a complex in vivo.
It is worthy to mention that, although an early study implied that XRCC2·RAD51D might be two of the components in human Shu complex (14), several lines of evidence argue against this possibility: 1) XRCC2, RAD51D, or other RAD51 paralogs were not detected in the mass spectrometry analysis of hSWS1-containing complexes; 2) hSWS1 was not detected in the mass spectrometry analysis of complexes containing various members of RAD51 paralogs (data not shown); 3) unlike that of hSWS1 and SWSAP1, the interdependence between hSWS1 and RAD51 or RAD51 paralogs was not observed (Fig. 4f). Because SWSAP1 shares some sequence similarity with archaeal RadA, we favor the idea that the human SWS1·SWSAP1 complex represents another so-called “RAD51 paralog.” The existence of several RAD51-like protein complexes, like hSWS1·SWSAP1, RAD51B/C/D·XRCC2, and RAD51C·XRCC3, underscores the importance of controlling homologous recombination repair in mammalian cells.
Although depletion of hSWS1 and/or SWSAP1 lead to increased MMS sensitivity and reduced HR efficiency, it should be noted that, unlike RAD51-depleted cells, the hSWS1- or SWSAP1-depleted cells do not exhibit appreciable sensitivity to CPT (Fig. 4a). Thus, it is very likely that this human Shu complex functions as a non-essential HRR accessory factor, which affects the efficiency and/or the timing of HRR. These findings are consistent with our hypothesis mentioned above that the human Shu complex may represent another “RAD51 paralog,” which may only be required for the repair of specific types of DNA lesions.
In summary, we have identified human Shu complex, which consists of hSWS1 and SWSAP1, that is required for efficient homologous recombination repair in mammalian cells. The importance of homologous recombination repair is well recognized, because defective HR repair is associated with genomic instability and cancer prone syndromes. The potential roles of human Shu complex in the maintenance of genomic stability and tumor suppression should be explored in the near future.
Acknowledgments
We thank all our colleagues in the Huang laboratory for insightful discussions and Dr. Shen Ye for technical assistance.
This work was supported in part by the National Natural Science Foundation of China (Grant 31071243), the Natural Science Foundation of Zhejiang Province (Grant R2110569), and the China's Fundamental Research Funds for the Central Universities (to J. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1, S2, and S3.
- HRR
- homologous recombination repair
- SFB
- streptavidin-FLAG-S protein
- MMS
- methylmethane sulfonate
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA.
REFERENCES
- 1. van Gent D. C., Hoeijmakers J. H., Kanaar R. (2001) Nat. Rev. Genet. 2, 196–206 [DOI] [PubMed] [Google Scholar]
- 2. Branzei D., Foiani M. (2008) Nat. Rev. Mol. Cell Biol. 9, 297–308 [DOI] [PubMed] [Google Scholar]
- 3. Moynahan M. E., Jasin M. (2010) Nat. Rev. Mol. Cell Biol. 11, 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weinstock D. M., Richardson C. A., Elliott B., Jasin M. (2006) DNA Repair (Amst.) 5, 1065–1074 [DOI] [PubMed] [Google Scholar]
- 5. Lukas J., Bartek J. (2004) Cell 118, 666–668 [DOI] [PubMed] [Google Scholar]
- 6. Huang J., Huen M. S., Kim H., Leung C. C., Glover J. N., Yu X., Chen J. (2009) Nat. Cell Biol. 11, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. West S. C. (2003) Nat. Rev. Mol. Cell Biol. 4, 435–445 [DOI] [PubMed] [Google Scholar]
- 8. Thacker J. (2005) Cancer Lett. 219, 125–135 [DOI] [PubMed] [Google Scholar]
- 9. Komori K., Miyata T., DiRuggiero J., Holley-Shanks R., Hayashi I., Cann I. K., Mayanagi K., Shinagawa H., Ishino Y. (2000) J. Biol. Chem. 275, 33782–33790 [DOI] [PubMed] [Google Scholar]
- 10. Sandler S. J., Satin L. H., Samra H. S., Clark A. J. (1996) Nucleic Acids Res. 24, 2125–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seitz E. M., Brockman J. P., Sandler S. J., Clark A. J., Kowalczykowski S. C. (1998) Genes Dev. 12, 1248–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang M. E., Rio A. G., Nicolas A., Kolodner R. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11529–11534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shor E., Weinstein J., Rothstein R. (2005) Genetics 169, 1275–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martín V., Chahwan C., Gao H., Blais V., Wohlschlegel J., Yates J. R., 3rd, McGowan C. H., Russell P. (2006) EMBO J. 25, 2564–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mankouri H. W., Ngo H. P., Hickson I. D. (2007) Mol. Biol. Cell 18, 4062–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bernstein K. A., Reid R. J., Sunjevaric I., Demuth K., Burgess R. C., Rothstein R. (2011) Mol. Biol. Cell 22, 1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ball L. G., Zhang K., Cobb J. A., Boone C., Xiao W. (2009) Mol. Microbiol. 73, 89–102 [DOI] [PubMed] [Google Scholar]
- 18. Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang J., Gong Z., Ghosal G., Chen J. (2009) Mol. Cell 35, 384–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu T., Ghosal G., Yuan J., Chen J., Huang J. (2010) Science 329, 693–696 [DOI] [PubMed] [Google Scholar]
- 21. Weinstock D. M., Nakanishi K., Helgadottir H. R., Jasin M. (2006) Methods Enzymol. 409, 524–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Masson J. Y., Tarsounas M. C., Stasiak A. Z., Stasiak A., Shah R., McIlwraith M. J., Benson F. E., West S. C. (2001) Genes Dev. 15, 3296–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sigurdsson S., Van Komen S., Bussen W., Schild D., Albala J. S., Sung P. (2001) Genes Dev. 15, 3308–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]