Background: Yeast free oligosaccharides (fOS) are cleaved from glycoproteins during ER-associated degradation of proteins (ERAD), but factors underlying their control remain to be explored.
Results: Only one-half of fOS are regulated by known ERAD processes, and fOS generation and demannosylation are growth-dependent.
Conclusion: Additional complexity of ERAD processes is revealed.
Significance: This is the first demonstration of growth-dependent fOS regulation.
Keywords: Endoplasmic Reticulum (ER), Glycoprotein, Oligosaccharide, Protein Degradation, Yeast, Yeast Metabolism, Endoplasmic Reticulum-associated Degradation, MNL1, MNS1, YOS9
Abstract
In Saccharomyces cerevisiae, proteins with misfolded lumenal, membrane, and cytoplasmic domains are cleared from the endoplasmic reticulum (ER) by ER-associated degradation (ERAD)-L, -M, and -C, respectively. ERAD-L is N-glycan-dependent and is characterized by ER mannosidase (Mns1p) and ER mannosidase-like protein (Mnl1p), which generate Man7GlcNAc2 (d1) N-glycans with non-reducing α1,6-mannosyl residues. Glycoproteins bearing this motif bind Yos9p and are dislocated into the cytoplasm and then deglycosylated by peptide N-glycanase (Png1p) to yield free oligosaccharides (fOS). Here, we examined yeast fOS metabolism as a function of cell growth in order to obtain quantitative and mechanistic insights into ERAD. We demonstrate that both Png1p-dependent generation of Man7–10GlcNAc2 fOS and vacuolar α-mannosidase (Ams1p)-dependent fOS demannosylation to yield Man1GlcNAc2 are strikingly up-regulated during post-diauxic growth which occurs when the culture medium is depleted of glucose. Gene deletions in the ams1Δ background revealed that, as anticipated, Mns1p and Mnl1p are required for efficient generation of the Man7GlcNAc2 (d1) fOS, but for the first time, we demonstrate that small amounts of this fOS are generated in an Mnl1p-independent, Mns1p-dependent pathway and that a Man8GlcNAc2 fOS that is known to bind Yos9p is generated in an Mnl1p-dependent, Mns1p-independent manner. This latter observation adds mechanistic insight into a recently described Mnl1p-dependent, Mns1p-independent ERAD pathway. Finally, we show that 50% of fOS generation is independent of ERAD-L, and because our data indicate that ERAD-M and ERAD-C contribute little to fOS levels, other important processes underlie fOS generation in S. cerevisiae.
Introduction
In yeast and mammalian cells, N-glycans play important roles during ER2-associated degradation (ERAD) of glycoproteins (1, 2). In the case of the disposal of glycoproteins whose misfolded domains are in the lumen of the ER (ERAD-L), prolonged folding reactions favor N-glycan demannosylation leading to the generation of a glycan signal for degradation. In the yeast Saccharomyces cerevisiae, initial experiments demonstrated that ER mannosidase I (Mns1p), through the generation of a specific Man8GlcNAc2 N-glycan (M8(d1,d3); see Fig. 1A), and the Mns1p homolog, Mnl1p (previously known as Htm1p), were required for ERAD of certain substrates (3, 4), and because the latter protein was thought to have no hydrolytic activity, it was proposed to act as a lectin that could target ERAD substrates carrying M8(d1,d3)-N-glycans to the degradation machinery (3). More recently, the combined actions of Mns1p and Mnl1p are thought to generate a Man7GlcNAc2 N-glycan whose structure possesses a terminal non-reducing α1,6-linked mannose residue (M7(d1); see Fig. 1A) (5, 6). As indicated in Fig. 1A, some misfolded glycoproteins are degraded in an Mns1p-independent, Mnl1p-dependent manner (7), and in this case, it is thought that Mnl1p promotes removal of the d3 mannose residue from Man9GlcNAc2 to yield M8(d1,d2). However, this structure, also possessing an exposed α1,6-linked mannose residue (6, 7), has not been formally identified. N-Glycans without the d3 mannose residue bind to the lectin Yos9p in vitro (5, 8, 9) and, through this interaction, are thought to target misfolded glycoproteins to a ubiquitin ligase complex (10, 11). These glycoproteins dislocate into the cytoplasm, where polyubiquitinylation (12), peptide N-glycanase (Png1p)-mediated deglycosylation (13, 14), and proteasomal degradation occur. Many components of the degradation machinery required for disposal of ERAD-L substrates are common with those required for the degradation of ER membrane proteins with misfolded membrane domains (ERAD-M) and ER membrane proteins with misfolded cytoplasmic domains (ERAD-C) (15). The E2 ubiquitin-conjugating enzyme Ubc7p and its partner Cue1p (16, 17) are common to all ERAD pathways. By contrast, whereas Doa10p is specific to the ERAD-C E3 ubiquitin ligase complex, the ER-associated degradation components, Der1p and Hrd1p, are common to both ERAD-L and -M E3 ubiquitin ligase complexes (15). Finally, Yos9p and Hrd3p are thought to be specific for ERAD-L and associate with Hrd1p (15, 18). Presently, there is little knowledge of either the relative quantitative importance of the different ERAD pathways or of the contribution of ERAD to global yeast protein catabolism.
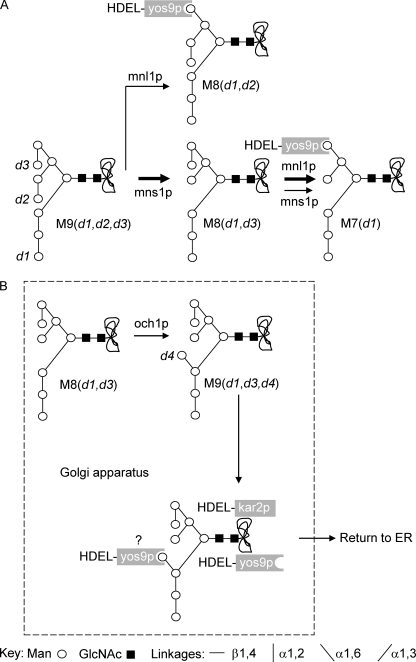
FIGURE 1.
ERAD pathways in S. cerevisiae. A, the best characterized ERAD-L pathway (heavy arrows) involves ER mannosidase I (Mns1p), which cleaves mannose residue d2 from Man9GlcNAc2 to generate Man8GlcNAc2 M8(d1,d3), whose conversion to Man7GlcNAc2 M7(d1) requires ER mannosidase-like protein (Mnl1p) (6). M7(d1) possesses a terminal non-reducing α1,6-linked mannose residue that promotes binding to the lectin Yos9p (5). Another Mns1p-independent, Mnl1p-dependent pathway has been described, and in this case, it is thought, but not proven, that Yos9p-interacting M8(d1,d2) is generated (7). Finally, in vitro data suggest a potential Mnl1p-independent, Mns1p-dependent pathway for the generation of the M7(d1) structure (41). B, in some cases, ERAD processes are thought to involve recovery of misfolded glycoproteins from the Golgi apparatus (47). This is confirmed by the identification of fOS bearing the d4 mannose residue transferred by the Och1p mannosyltransferase that resides in the Golgi apparatus (19, 21). It is thought that chaperones bearing the HDEL ER retention signal, such as Kar2p or Yos9p, could be involved in this process (47). Yos9p is thought to have a lectin-independent role in ERAD (8, 9, 46) as well as its lectin-dependent role, and presently it is not known whether or not the α1,6-linked d4 mannose residue confers Yos9p binding to oligosaccharides.
Png1p-mediated deglycosylation of glycoproteins yields free oligosaccharides (fOS), and an examination of the quantities and structures of these compounds could potentially aid in the understanding of the mechanisms and relative importance of different yeast glycoprotein degradation pathways. Metabolic radiolabeling studies demonstrated that 70–80% of S. cerevisiae [3H]fOS are generated by Png1p (19, 20) and that Ams1p, the vacuolar mannosidase, degrades these compounds (19). In the ams1Δ strain, although the major [3H]fOS is Mns1p-generated [3H]Man8GlcNAc2(d1,d3), small amounts of a [3H]Man7GlcNAc2-like structure, not detected on 3H-glycoproteins, are generated (19). Subsequent quantitative and structural data on fOS at steady state in ams1Δ cells confirmed the preponderance of the M8(d1,d3) fOS and identified the Man7GlcNAc2 structure as M7(d1) (21). Furthermore, in the ams1Δmns1Δ deletion mutant, whereas [3H]Man9GlcNAc2 was found to be the predominant fOS, a [3H]Man8GlcNAc2 compound different from M8(d1,d3) was identified (19). It is presently unknown whether or not this compound corresponds to the M8(d1,d2), Yos9p-interacting structure thought to be required for the Mns1p-independent, Mnl1p-dependent ERAD-L pathway described above (Fig. 1) (6, 7). Therefore, the ensemble of these data is compatible with the idea that only a fraction of fOS manifest ERAD-L-associated trimming (Fig. 1A). It would appear that over one-half of Png1p-generated fOS have structures not attributable to ERAD-L, which points to important roles for ERAD-M and/or ERAD-C or other yeast glycoprotein catabolic processes. In some cases, ERAD involves retrieval of misfolded glycoproteins from the Golgi apparatus, and this is confirmed by the identification of fOS whose structures indicate modification by mannosyltransferases, such as Och1p (see Fig. 1B), known to reside in this compartment (19–21). Presently, it is not known whether or not the α1,6-linked mannose residue (termed the d4 mannose) transferred by Och1p can confer Yos9p binding activity to oligosaccharides (see Fig. 1B).
Attempting to correlate fOS generation with ERAD processes is further complicated by the fact that whereas the latter processes are studied uniquely in rapidly dividing, exponential cultures, Png1p and Ams1p are up-regulated during post-diauxic growth after cells have depleted glucose from the growth medium. Peptide N-glycanase activity is difficult to detect in exponentially growing yeast and increases during post-diauxic growth (22), and Png1p abundance is higher in stationary cells than in exponentially growing cells (23). Likewise, Ams1p-dependent p-nitrophenyl-Man hydrolyzing activity is low in exponential yeast cultures and increases during post-diauxic growth (24). During metabolic radiolabeling studies, [3H]fOS demannosylation parallels the increase in expression of Ams1p-dependent p-nitrophenyl-mannose hydrolysis activity (19). However, more recent studies on the steady state levels of fOS in S. cerevisiae demonstrate little increase in either the quantity or demannosylation of fOS even in stationary cells (21).
Here, in order to better understand the mechanisms underlying fOS metabolism in S. cerevisiae, fOS derived from exponential, post-diauxic, and stationary phase wild type cells and various deletion mutants were quantitated and characterized by HPLC, glycosidase digestion, and mass spectrometry. For the first time, we show that Ams1p-dependent fOS demannosylation increases during post-diauxic growth and that the trisaccharide Man1GlcNAc2 is the limit digest product of fOS demannosylation in stationary yeast cultures. The trisaccharide level in stationary cells was largely Png1p-dependent, did not depend upon vacuolar acidification, could be maximally reduced by only one-half in cells deficient in ERAD-L and/or ERAD-M, and was unaffected in ERAD-C-deficient cells. During post-diauxic growth, fOS levels increase, and when fOS generation was examined in ams1Δ cells, a 7-fold increase in M7(d1) was noted during this period, whereas other fOS increased to a lesser extent. We show for the first time that although Mnl1p and Mns1p are required for efficient generation of the fOS M7(d1), Mnl1p promotes the appearance of the fOS M8(d1,d2) in the absence of Mns1p, and Mns1p can promote the formation of the M7(d1) fOS in the absence of Mnl1p. Accordingly, our data demonstrate that, in yeast, metabolism of Png1p-dependent fOS is regulated by complex growth-dependent processes. Our data confirm the N-glycan trimming pathways presented in Fig. 1A. Other data indicate Yos9p-dependent and Mns1p- and Mnl1p-independent fOS generation. Although a fOS possessing the Och1p-dependent d4 mannose residue as its sole terminal non-reducing α1,6-mannosyl motif was found to be highly regulated by Yos9p, our data cannot distinguish between lectin-dependent and lectin-independent roles for Yos9p during ERAD.
EXPERIMENTAL PROCEDURES
Reagents
The yeast parental strain BY4742 and the single deletion mutants were obtained from Euroscarf (Frankfurt, Germany). Yeast extract and bacto peptone were purchased from Invitrogen. Yeast minimal SD base medium and minimal SD agar base were purchased from Ozyme (Saint-Quentin-en-Yvelines, France). AG 50-X2 (H+ form) and AG 1-X2 (acetate form) came from Bio-Rad. Charcoal was from BDH Prolabo. Biogel P2, 2-aminopyridine (AP), endo-β-N-acetylglucosaminidase, acid-washed glass beads, amyloglucosidase from Rhizopus sp., jack bean α-mannosidase, Helix pomatia β-mannosidase, and protease (Pronase E) from Streptomyces griseus were purchased from Sigma-Aldrich. Aspergillus saitoi α1,2-mannosidase was purchased from Europa Bioproducts Ltd., Ely, UK).
Yeast Strains, Transformation, and Culture
S. cerevisiae parental strains and deletion mutants used in this study are listed in Table 1. All yeast strains were grown in YPD medium (1% yeast extract, 2% Bacto peptone, 2% glucose). Cell cultures were performed in a rotary shaker (250 rpm) at 30 °C with a minimum flask volume/medium volume ratio of 4:1. The evolution of culture growth was monitored by reading the turbidometry of cell suspensions at 600 nm (A600), and overnight preculture in YPG medium was used to inoculate yeast cultures at A600 = 0.1.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Origin |
|---|---|---|
| BY4742 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0 | Euroscarf |
| Y13369 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; gls2::kanMX4 | Euroscarf |
| Y13883 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; vma1::kanMX4 | Euroscarf |
| Y12156 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; png1::kanMX4 | Euroscarf |
| Y16930 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; mns1::kanMX4 | Euroscarf |
| Y12898 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; mnl1::kanMX4 | Euroscarf |
| Y12668 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; mnl2::kanMX; | Euroscarf |
| Y14523 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; ams1::kanMX4 | Euroscarf |
| Y14895 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; dcw1::kanMX4 | Euroscarf |
| Y10824 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; dfg5::kanMX4 | Euroscarf |
| Y14395 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; gls1::kanMX4 | Euroscarf |
| Y13993 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; yos9::kanMX4 | Euroscarf |
| Y11778 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; alg6::kanMX4 | Euroscarf |
| Y14406 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; och1::kanMX4 | Euroscarf |
| Y14156 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; hrd3::kanMX4 | Euroscarf |
| Y13341 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; der1::kanMX4 | Euroscarf |
| Y17299 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; doa10::kanMX4 | Euroscarf |
| Y10850 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; cue1::kanMX; | Euroscarf |
| Y10597 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; ubc7::kanMX4 | Euroscarf |
| Y11907 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; ire1::kanMX4 | Euroscarf |
| BA4 | MATα; ams1::HIST3MX6; leu2Δ0; lys2Δ0; ura3Δ0 | Ref. 29 |
| Y43 | MATα; his3Δ1; yos9::LEU2; lys2Δ0; ura3Δ0 | This study |
| PA1 | MATα; ams1::HIST3MX6; leu2Δ0; lys2Δ0; ura3Δ0; png1::kanMX4 | Ref. 29 |
| MA4 | MATα; ams1::HIST3MX6; leu2Δ0; lys2Δ0; ura3Δ0; mns1::kanMX4 | This study |
| BA4C1 | MATα; ams1::HIST3MX6; leu2Δ0; lys2Δ0; mnl1::URA3 | This study |
| MA4C3 | MATα; ams1::HIST3MX6; leu2Δ0; lys2Δ0; mnl1::URA3; mns1::kanMX4 | This study |
| AY48 | MATα; ams1::HIST3MX6; yos9::LEU2; lys2Δ0; ura3Δ0; | This study |
| AYM58 | MATα; ams1::HIST3MX6; yos9::LEU2; lys2Δ0; ura3Δ0; mns1::kanMX4 | This study |
| AYH53 | MATα; ams1::HIST3MX6; yos9::LEU2; lys2Δ0; mnl1::URA3 | This study |
The yeast genetic background used for making deletion mutants was BY4742 (MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0). As indicated in Table 1, the single, double, or triple deletion mutants were created following a PCR-based targeted gene replacement strategy as described previously (25). Yeast cells were then transformed using lithium acetate, and transformants were isolated after growth on solid selective medium. The disruption of the target gene was confirmed by PCR analysis with oligonucleotides flanking the region of insertion of the auxotrophic marker. For each deletion mutant, at least two independent clones were generated and analyzed. Culture of transformants was performed in liquid selective medium for at least 20 generations before seeding in YPG medium. Exponentially growing yeast were metabolically radiolabeled with [2-3H]mannose as described previously (19).
Extraction of fOS
Yeast cells were extracted as described previously (19). Briefly, 150–400 A600 units of cells (volume of cells recovered (ml) × A600) were harvested, washed twice with ice-cold PBS, and transferred into 50-ml polypropylene tubes. The cell pellets were frozen and thawed before adding one volume of glass beads and one volume of 100 mm Tris-HCl (pH 7.4) containing 4 mm MgCl2. In some experiments, 5 × 10−3 cpm of [2-3H]Man8GlcNAc2 was added to cell pellets as an internal standard. This radioactive compound was purified by HPLC after liberation, by mild acid hydrolysis, from dolichol-linked oligosaccharide obtained from Epstein-Barr virus (EBV)-transformed lymphoblastoid cells that were metabolically radiolabeled with [2-3H]mannose as described previously (26). Cells were disrupted using six 30-s bursts on a Vortex mixer at 4 °C. MeOH and CHCl3 were added to yield a 3:2:1 mixture of CHCl3, MeOH, and buffer, respectively. The mixtures were vigorously shaken at room temperature. After centrifugation, the lower (CHCl3) and upper (methanolic) phases were removed and kept. The interphase proteins were re-extracted with the same volume of solvents, and the newly obtained phases were removed and pooled with the previous phases. Neutral fOS were recovered from the upper phase as described previously (19). Briefly, this phase was dried and dissolved in water, and after desalting on AG-1/AG-50 columns, the unbound neutral material was loaded onto charcoal columns. After washing these columns with water, the neutral fOS were eluted with 30% ethanol. For mass spectrometry, 5 liters of YPD were seeded at 0.1 A600/ml with either ams1Δ or ams1Δmns1Δ cells. After 24-h growth, the cells were harvested and extracted as described above in 10 batches.
Analytical Procedures
For analytical HPLC, fOS mixtures were derivatized with AP as described previously (27), and derivatized fOS were separated from excess reagents on columns of Biogel P2 (28). fOS-AP mixtures were resolved by HPLC using an amine-bonded silica column (LiChrospher Amino 5 μm, 250 × 4.6 mm, Sulpelco Inc.) and detected with a Jasco FP-2020 Plus fluorescence detector (excitation wavelength 310 nm, emission wavelength 380 nm) as described previously (29). The isomers of Man8GlcNAc2 were separated using reverse phase HPLC as their Man8GlcNAc-AP derivatives as described previously (28). A. saitoi α1,2-mannosidase incubations contained 0.5 milliunits of enzyme in a total volume of 20 μl of 100 mm acetate buffer, pH 5.0. Digestion was carried out for 2 days at 37 °C. H. pomatia β-mannosidase digestions were carried out for 16 h at 25 °C in 35 μl of 100 mm acetate buffer, pH 5.0, containing 30 milliunits of enzyme. Oligosaccharides were also treated with 0.4 units of jack bean α-mannosidase in 20 μl of 100 mm acetate buffer, pH 5.0, at 37 °C for 16 h. MALDI-TOF mass spectra were recorded in positive ion mode with a Shimadzu-Biotech AXIMA TOF2 mass spectrometer (Shimadzu Biotech, Manchester, UK). Samples were prepared by spotting 0.5 μl of an aqueous solution of the glycans onto the target plate together with 0.5 μl of matrix solution (2,5-dihydroxybenzoic acid (10 mg) in water (0.5 μl) and acetonitrile (0.5 μl)). The mixture was allowed to dry under ambient conditions, followed by recrystallization from ethanol. Negative ion electrospray spectra and collision-induced dissociation were obtained with a Waters Q-TOF (version 1) mass spectrometer (Waters MS Technologies, Manchester, UK) using Proxeon nanoelectrospray capillaries (Proxeon Biosystems (now part of Thermo-Fisher Scientific), Odense, Denmark) for sample introduction. Samples were dissolved in water/MeOH (1:1, v/v) containing 0.5 mm ammonium nitrate (the ammonium nitrate was to form stable [M + NO3]− ions to assist fragmentation). The mass spectrometer ion source was maintained at 120 °C, the voltage on the capillary was 1.1 kV, and fragmentation was performed by argon collisions in the collision cell, which was operated at a voltage appropriate to the glycan mass. Spectral acquisition and processing was performed with MassLynx version 4.0.
RESULTS
Png1p-dependent fOS Are Regulated during Post-diauxic Growth
First, we examined yeast fOS steady state levels during yeast growth because both Png1p and Ams1p, the enzymes responsible for the production and processing of fOS, respectively, are known to be regulated in a growth-dependent manner. Under standard laboratory conditions, a wild type yeast population doubles about every 90 min (exponential culture) until the glucose required for fermentation is consumed (30). At this time (diauxic shift), there is an inflection in the growth curve, and thereafter, slow post-diauxic growth occurs, during which time yeast metabolizes non-fermentable carbon sources. When there is no further increase in cell number, the yeast culture is said to have reached the stationary phase, and under standard laboratory conditions, this occurs after about 6 days (30). Peptide N-glycanase activity is difficult to detect in exponentially growing yeast and increases during post-diauxic growth (22). Likewise, during metabolic radiolabeling studies, [3H]fOS demannosylation parallels the increase in expression of Ams1p-dependent p-nitrophenyl-mannose hydrolysis activity (19) that is known to be growth-dependent (24). By contrast, more recent studies on the steady state levels of fOS in S. cerevisiae demonstrate little demannosylation of fOS, even in stationary cells (21). In order to explore these issues, fOS generation and processing were examined during the different growth phases of S. cerevisiae.
In exponentially growing (5-h cultures) wild type yeast, several pools of oligosaccharide were detected. One pool comprised structures (Fig. 2A, top, peaks a–e) that were sensitive to an amyloglucosidase preparation from Rhizopus mold3 and occurred in png1Δ (Fig. 2A, top, red trace) and ams1Δ (Fig. 2A, top, green trace) deletion mutants as well as in the wild type strain (Fig. 2A, top, blue trace). Although amyloglucosidase cleaves α1,3-, α1,4-, and α1,6-glucose residues, it is possible that peaks a–e do not correspond to oligomers of α-linked glucose but rather correspond to β-glucans previously described to occur in yeast (Glc3–15 (21)) because the amyloglucosidase preparation used in our studies is probably contaminated with β-glucanase (31). Another oligosaccharide pool was noted in both wild type and ams1Δ cells but occurred in strikingly reduced amounts in the in png1Δ deletion mutant (Fig. 2A, top). The retention times of the predominant Png1p-dependent oligosaccharides correspond to [3H]Man9–7GlcNAc2 structures described previously in metabolically radiolabeled S. cerevisiae (19). A third oligosaccharide pool comprises Png1p-independent structures with retention times similar to those of the Png1p-dependent pool and whose components are distributed similarly but not identically to those of Png1p-dependent compounds (data not shown). When oligosaccharide levels were examined in wild type yeast after 6 days of culture (Fig. 2A, bottom, blue trace), Man9–7GlcNAc2 structures were much reduced, and structure f comigrating with standard Man1GlcNAc2 predominated. Upon digestion of f with H. pomatia β-mannosidase, a compound that co-eluted with standard di-N-acetylchitobiose was obtained (Fig. 2B), substantiating that structure f is Man1GlcNAc2. In the 6-day culture of png1Δ cells, Man1GlcNAc2 occurs at ∼6%, the level seen in wild type cells (Fig. 2A, bottom, compare blue and red traces), whereas this compound was not detected in 6-day cultures of ams1Δ cells (Fig. 2A, bottom, compare blue and green traces), indicating that Png1p-independent fOS are also demannosylated by Ams1p. Compound g, also observed in wild type cells after 6 days of culture (Fig. 2A, bottom, blue trace), migrates similarly to a Man5GlcNAc2 structure but was found to be greatly reduced in alg6Δ cells (Fig. 2C) that lack the capacity to glucosylate the dolichol-linked oligosaccharide that serves as donor for protein N-glycosylation in the ER. Taking into account that 6-day cultures of ER glucosidase 1-deficient gls1Δ and ER glucosidase 2-deficient gls2Δ cells reveal accumulations of Glc3Man4GlcNAc2 and Glc2Man4GlcNAc2, respectively (data not shown), compound g probably corresponds to Glc1Man4GlcNAc2. In order to understand whether or not the Man1GlcNAc2 and Glc1Man4GlcNAc2 compounds are generated in vivo or ex vivo during fOS work up of stationary cells, [3H]Man8GlcNAc2(d1,d3) was added to pellets of stationary cells before homogenization and extraction. As demonstrated in Fig. 2D, the tritiated compound remained largely intact throughout the fOS preparation procedures, indicating that the Man1GlcNAc2 and Glc1Man4GlcNAc2 compounds are generated physiologically. The trisaccharide Man1GlcNAc2 appeared to be generated from larger fOS because (i) it was not detected in the ams1Δ deletion mutant (Fig. 2A, bottom, green trace), and the area under the peak corresponding to Man1GlcNAc2 derived from 6-day cultures of wild type cells was similar to the summed areas under the peaks corresponding to the Man9–7GlcNAc2 structures derived from 6-day cultures of ams1Δ cells (Fig. 2E, left); (ii) similarly (Fig. 2E, right), Man1GlcNAc2 occurred in strikingly reduced amounts in gls1Δ and gls2Δ cells, and the quantities of Glc3Man4–8GlcNAc2 and Glc2Man4–8GlcNAc2 compounds, respectively, that occur in these 6-day cultures were similar to the amount of Man1GlcNAc2 occurring in control wild type cells.
FIGURE 2.
Identification of a Png1p-dependent fOS pool that is catabolized by Ams1p. A, the indicated yeast strains were cultivated for either 5 h (top) or 6 days (bottom), and 250 A600 units of cells were harvested and extracted to yield oligosaccharides, as described under “Experimental Procedures.” Oligosaccharide-AP derivatives were resolved by HPLC and detected by on-line fluorimetry. The peaks labeled a–e are described under “Results.” Peaks corresponding to [3H]Man9–7GlcNAc2 oligosaccharides (M9–7) previously reported to occur in metabolically radiolabeled wild type S. cerevisiae are also indicated. B, material eluting under peak f recovered from stationary wild type cells (A, bottom) was analyzed by HPLC after either mock digestion or digestion with H. pomatia β1,4-mannosidase. The arrows indicate the elution times of standard di-N-acetylchitobiose (GN2) and Manβ1,4GlcNAc2 (M1GN2). C, fOS derived from 6-day cultures of wild type and alg6Δ cells were analyzed by HPLC as described above. Only the regions of the chromatographs containing peak g are shown. D, [2-3H]Man8GlcNAc2 was subjected to the homogenization and fOS extraction procedures in either the absence (−cells) or presence (+cells) of yeast cells. After derivatization with AP, fOS were subjected to HPLC and detected by both on-line fluorimetry and scintillation counting. E, the indicated yeast strains were cultivated for 6 days (left) or 5 days (right), and fOS were analyzed as described above. The peak areas associated with Man1GlcNAc2 (M1GN2) and the summed areas of peaks corresponding to Glc0–3Man4–9GlcNAc2 (G0–3M4–9GN2) compounds have been plotted. AU, arbitrary units.
Previously, we showed that processing of Png1p-dependent [3H]Man9–7GlcNAc2 fOS by the vacuolar mannosidase Ams1p occurred normally in the vma1Δ deletion mutant that is deficient in vacuolar acidification, indicating that fOS processing occurs in the cytosol (20), where Ams1p is synthesized and known to be active (32). In agreement with these data, it has been shown that at least the initial steps of fOS demannosylation are accelerated in atg19Δ cells in which cytoplasm-to-vacuole targeting of Ams1p is blocked (21). Data reported in the Fig. 2E (right) now show that the appearance of Man1GlcNAc2 is little affected in vma1Δ cells, indicating that removal of all α-linked mannose residues from fOS may occur in the cytoplasm.
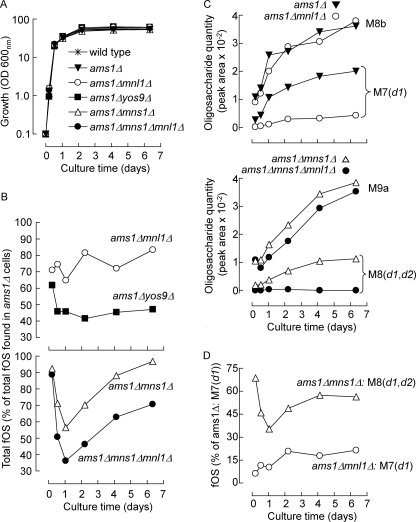
Data in Fig. 2A show that steady state levels of fOS are higher in 6-day cultured ams1Δ cells than in their 5-h cultured counterparts, and those in the right-hand panel of Fig. 3A demonstrate that, in ams1Δ cells, this rise in fOS levels occurs mainly during post-diauxic growth. In a parallel culture of wild type cells, large fOS (Man7–9GlcNAc2) decline at a time when the level of Man1GlcNAc2 begins to increase (Fig. 3A, left). Importantly, after 6 days of culture, the Man1GlcNAc2 level in wild type cells appears to be attaining the same level as that of M7–9 fOS in the ams1Δ strain (compare M1 and M7–9 curves in Fig. 3A, right and left, respectively). As demonstrated in a separate experiment using ams1Δ cells (Fig. 3B, top left), it was noted that, whereas HPLC peaks M7 and M8 underwent striking increases during the post-diauxic growth phase, M9a and M9b peak sizes fluctuated less. By contrast, the levels of Png1p-independent Man7–9GlcNAc2 compounds remain extremely low throughout the growth cycle of ams1Δpng1Δ cells (Fig. 3B, top right). Finally, the levels of compounds a–d (compound e was not quantitated in this experiment; see Fig. 2A, top) declined rapidly during exponential growth of both ams1Δ and ams1Δpng1Δ cells (Fig. 3B, bottom left and right). This is an important point because, for characterization purposes, fOS can be isolated from 24-h cultures with minimum interference from these glucose oligomers (see below).
FIGURE 3.
Changes in steady state levels of fOS as a function of yeast growth. A, wild type and ams1Δ cells were cultivated for the indicated times. Growth was measured as a function of culture time by turbidometry (A600 (OD 600 nm)) and is plotted on a logarithmic scale. The different phases of the yeast growth cycle are indicated by the bars above the growth curves. E, exponential growth. The post-diauxic shift occurs at the first inflection of the growth curve and is indicated by the heavy vertical bar. Post-diauxic growth (Post diauxic) occurs before the true stationary phase (Stat), where there is no net increase in cell number. At the indicated culture time, cellular fOS were isolated from 150 A600 units of cells and resolved by HPLC as shown in Fig. 2A. The area under the peak corresponding to Man1GlcNAc2 (M1) and the summed areas of peaks M7–9 (see Fig. 2A) are shown. B, the ams1Δ and ams1Δpng1Δ deletion mutants were cultivated for 9 days, and growth curves were established as above. Areas under the M7–9 peaks (top) as well as those of peaks a–d (see Fig. 2A, bottom) were calculated and are expressed per 400 A600 units of cells.
ERAD Processes Account for Only One-half of fOS Generation
As summarized in Fig. 4A, we have followed the fate of fOS throughout the different yeast growth phases. For the first time, we show that Ams1p-dependent fOS processing is a feature of post-diauxic growth and that the trisaccharide Man1GlcNAc2 is the major limit digest product of this process. Furthermore, for the first time, we demonstrate structure-dependent increases in fOS during the post-diauxic growth period. Importantly, although abolishing fOS processing, invalidation of the AMS1 gene does not greatly affect the total level of fOS generated during yeast growth. Although Png1p seems to account for greater than 90% of fOS in S. cerevisiae, it is not known whether or not all Png1p-dependent fOS are derived from misfolded glycoproteins. To evaluate this question, the trisaccharide was quantitated in stationary single yeast deletion mutants in which genes involved in different ERAD pathways or in the unfolded protein response are invalidated. Whereas deletion of the ERAD-C-specific gene DOA10 had little if any effect on the level of Man1GlcNAc2, single deletion mutants targeting either ERAD-L alone (hrd3Δ and yos9Δ cells) or ERAD-L + ERAD-M together (der1Δ) manifested similar (∼50%) reductions in trisaccharide levels (Fig. 4, B and C). Deletion of UBC7 and CUE1, genes whose products are involved in ubiquitination of ERAD-L, -C, and -M substrates, provoked smaller (∼25%) inhibitions of the appearance of the trisaccharide (Fig. 4, B and C). Strains deficient in Mns1p and Mnl1p displayed 10–20% reductions in the trisaccharides, whereas another mannosidase-like protein predicted to reside in the Golgi apparatus with unknown function (Mnl2p) appears to have no significant effect on the appearance of the trisaccharide (Fig. 4C). Finally, ire1Δ cells (deficient in the transmembrane protein kinase, Ire1), which are unable to mount an unfolded protein response, manifest the same level of trisaccharide as that found in control cells, indicating that this pathway is not involved in fOS generation under our growth conditions (Fig. 4C).
FIGURE 4.
The steady state level of Man1GlcNAc2, the major fOS identified in stationary wild type cells, is reduced in cells defective in ERAD. A, our interpretation of results shown in Figs. 2 and 3 is summarized. Generation of a mixture of large fOS occurs in both exponential and post-diauxic cultures but is much reduced in stationary cells. Demannosylation of fOS (processing) occurs only in post-diauxic and stationary cultures. By contrast, fOS are not further processed beyond Man1GlcNAc2 under standard growth conditions. B, ERAD-L, -C, and -M pathways are defined in the Introduction. Each requires an E3 ubiquitin ligase complex comprising many components. Only those components whose roles are evaluated in the present report are indicated. The E2 ubiquitin conjugation complex that comprises Cue1p and Ubc7p, among other proteins, appears to be common to all ERAD pathways. Misfolded glycoproteins that are dislocated into the cytoplasm are deglycosylated by the peptide N-glycanase, Png1p, and degraded by the proteasome. C, the indicated deletion mutants were cultivated for 6 days, and Man1GlcNAc2 was quantitated. The trisaccharide levels are expressed as a percentage of that found to occur in wild type cells. Error bars, S.D. (n = 3).
These results indicate that only about one-half of fOS can be attributed to known ERAD processes and that, of the ERAD pathways, ERAD-L is the major source of fOS. Although disruption of the E3 ubiquitin ligase complex involved in ERAD-L strikingly reduces fOS generation, Mns1p and Mnl1p, which, together, are thought to generate the M7(d1) glycoprotein degradation signal, perturb fOS generation to a lesser degree. These data suggest that Mns1p and Mnl1p have overlapping functions. Alternatively, other mechanisms, such as Och1p modification, could give rise to Yos9p-interacting N-glycans. Finally, Yos9p could have a function in glycoprotein degradation that is independent of its lectin activity. In order to look more closely at these questions, we characterized fOS generated in ams1Δ cells and then examined how the different structures are modulated in cells deficient in various gene products involved in ERAD-L.
Structural Determination of fOS in ams1Δ Cells
fOS, prepared from 5 liters of 24-h (post-diauxic) ams1Δ cells, were derivatized with AP, and after HPLC (Fig. 5A), fractions corresponding to peaks M7, M8b, M9a, and M9b were collected and analyzed as described in the legends to Figs. 6–8. MALDI-TOF spectrometry of underivatized material revealed compounds with compositions of Hex7–10HexNAc2, and as shown in Figs. 6–8, the negative ion collision-induced dissociation spectra of the nitrate adducts of Hex7–9HexNAc2 structures were obtained and interpreted as described earlier (33–35). All of the fragmentation spectra contained the same group of ions in the high mass region that arise from fragmentation of the chitobiose core. Ion nomenclature is that devised by Domon and Costello (36).
FIGURE 5.
Structural characterization of Hex7GlcNAc2 oligosaccharides isolated from the ams1Δ deletion mutant. A, oligosaccharides were isolated from 5 liters of 24-h cultures of the ams1Δ deletion mutant, and an aliquot (0.3%) was derivatized with AP and analyzed by HPLC. The indicated M7–9 peaks were collected for further analyses. Peaks labeled a–d (see Figs. 2A and 3B) are described in the text. Inset, an aliquot of the underivatized material was subjected to MALDI-TOF mass spectrometry, as described under “Experimental Procedures.” B, negative ion fragmentation spectrum (m/z 1620). The ions generated during fragmentation are compatible with the compound M7(d1), and this spectrum was identical to that of the reference M7(d1) compound (data not shown). C, the HPLC peak M7 was reanalyzed by HPLC after sham treatment or digestion with either A. satoi α1,2-mannosidase (α1,2M'ase) or jack bean mannosidase (JBM'ase). The compounds indicated by M1 and M5 have the same retention times as standard Man1GlcNAc2 and Man5GlcNAc2, respectively.
FIGURE 6.
Structural determination of Hex8GlcNAc2 oligosaccharides isolated from the ams1Δ deletion mutant. A, negative ion fragmentation spectra (m/z 1782). Inset, examination of the different fragmentation ions indicates the presence of the indicated compounds as described under “Results.” B, the HPLC peak M8b was subjected to HPLC after digestion with α1,2-mannosidase (α1,2-M'ase). The major digestion product co-eluted with standard Man5GlcNAc2 (M5). A minor digestion product eluting as a Man6GlcNAc2 (M6) structure was also identified. C, oligosaccharides were harvested from exponentially growing wild type cells and the och1Δ deletion mutant. After derivatization with AP and separation of oligosaccharide derivatives by HPLC, M9–7 compounds were collected as a pool and digested with the α1,2-mannosidase.
FIGURE 7.
Characterization of Hex9GlcNAc2 oligosaccharides isolated from the ams1Δ deletion mutant. A, negative ion fragmentation spectra (m/z 1944). Inset, examination of the different fragmentation ions indicates the presence of the indicated compounds as described under “Results.” B, HPLC peak M9a was reanalyzed by HPLC after digestion with α1,2-mannosidase (α1,2M'ase). As well as the Man5GlcNAc2 (M5) digestion product, a product, h, which appeared to be one hexose unit smaller than the starting material was also observed. C, product h was analyzed by HPLC after digestion with jack bean α-mannosidase (JBM'ase). A component behaving as Man1GlcNAc2 (M1) was not detected. D, oligosaccharides were harvested from exponentially growing wild type cells and the alg6Δ deletion mutant. After derivatization with AP and separation of oligosaccharide derivatives by HPLC, M9–7 compounds were quantitated, the -fold change (alg6Δ/wild type) in the steady state level of each oligosaccharide was computed, and the value was imposed on a logarithmic scale. E, HPLC peak M9b was subjected to HPLC after digestion with α1,2-mannosidase (α1,2M'ase).
FIGURE 8.
Mns1p and Mnl1p regulate fOS structure. A, ams1Δ, ams1Δmnl1Δ (left), ams1Δmns1Δ, and ams1Δmns1Δmnl1Δ (right) cells were cultivated for 1 day. 170 A600 units of cells were harvested, and fOS were analyzed by HPLC. Left, inset, the indicated yeast strains were metabolically radiolabeled with [2-3H]mannose for 30 min, and the resulting [2-3H]fOS were resolved by TLC and visualized by fluorography. The migration positions of standard Man9–7GlcNAc2 (M9–7) oligosaccharides are indicated to the left of the chromatographs. B, fOS were prepared from ams1Δmns1Δ cells as described in Fig. 3B and subjected to mass spectrometry. Alternatively, an aliquot of fOS material was subjected to HPLC. The negative ion fragmentation spectrum (m/z 1782) is shown. Inset, the M8 components derived from the ams1Δmns1Δ cells were collected and digested with endo-β-N-acetylglucosaminidase before being rederivatized with AP. The resulting AP derivatives were resolved using reversed phase HPLC (green trace). The same sample was co-chromatographed (red trace) with a mixture of the three Man8GlcNAc-AP isomers (blue trace). The elution positions of the three isomers of Man8GlcNAc-AP are indicated. C, N-glycans derived from the yeast cultures described in the left and right panels of A were examined by HPLC after release of oligosaccharides from Pronase-digested glycoproteins with endo-β-N-acetylglucosaminidase. Left, inset, [2-3H]N-glycans derived from the metabolically radiolabeled cells described in A were prepared and examined by TLC. The migration positions of standard Man9–7GlcNAc (M9–7) oligosaccharides are indicated to the left of the chromatographs.
Hex7GlcNAc2 from ams1Δ cells (m/z 1620)
In the spectrum (Fig. 5B), the ions at m/z 1396 and 1193 are the 2,4A6 and 2,4A5 cross-ring fragments, respectively (inset). m/z 1336 is from the B5 cleavage (between the two GlcNAc rings). The D, [D-18]−, O,3A3, B2β, and C2β ions at m/z 647, 629, 575, 485, and 503, respectively, are characteristic of a 6-antenna containing three mannose residues (inset). Therefore, the additional mannose residue must be on the 3-antenna. This spectrum is identical to that of reference M7(d1). Material eluting under the M7 peak after HPLC was subjected to jack bean (α1,2-, α1,3-, and α1,6-) mannosidase and A. saitoi α1,2-mannosidase digestion. Consistent with the structure being M7(d1), it can be seen that, whereas the former enzyme removed six residues of mannose from the starting material, the latter enzyme removed only two mannose residues (Fig. 5C).
Hex8GlcNAc2 from ams1Δ cells (m/z 1782)
In the spectrum (Fig. 6A), the D, [D-18]−, O,3A3 B3β, and C3β ions are 162 mass units higher than in the spectrum of Man7GlcNAc2 shown in Fig. 4A. Therefore, there is an additional mannose residue on the 6-antenna. The high relative abundance of the ion at m/z 485 (ion D′) indicates that this mannose residue is located on the 6-branch of the 6-antenna. This spectrum is identical to that of the reference M8(d1,d3) (Fig. 5A, inset, left-hand structure). Minor ions at m/z 503 (C2β) and 575 (O,3A2) suggest the presence of a compound with three mannose residues in the 6-antenna (other diagnostic ions are coincident with major fragments from the above compound). The 3-antenna of this second compound must, therefore, contain four hexose residues. Possible structures of this chain are indicated and include a structure bearing either an Alg6p glucosyltransferase-mediated glucose residue (37) or an mnn1p mannosyltransferase-mediated mannose residue in α1,3-linkage (38, 39) to the outermost mannose of the 3-antenna (Fig. 6A, inset, right-hand structures) and a structure whose innermost mannose of the 3-antenna contains an Och1p mannosyltransferase-mediated α1,6-linked mannose residue (40) (Fig. 6A, inset, middle structure). A. satoi α1,2-mannosidase digestion of material in HPLC fraction M8b yields two major limit digest products (Fig. 6B); Man5GlcNAc2 (M5) is the expected limit digest product of M8(d1,d3), whereas Man6GlcNAc2 (M6) would be the expected product generated from the Och1p-mediated structure. When total fOS derived from the exponentially growing och1Δ deletion mutant were digested with A. satoi α1,2-mannosidase, the M6 digestion product was detected in strikingly reduced amounts (Fig. 6C). By contrast, the structure bearing either a glucose or a mannose residue in α1,3-linkage to the outermost mannose of the 3-antenna (Fig. 6A, inset, right-hand structures) resists α1,2-mannosidase digestion. However, only trace amounts of a mannosidase-resistant component were noted, and this structure appeared to correspond, in part, to the trailing edge of the undigested material (Fig. 6B).
Hex9GlcNAc2 from ams1Δ cells (m/z 1782)
In the spectrum shown in Fig. 7A, the D, [D-18]−, O,3A3, B3β, and C3β ions are at m/z 809, 791, 737, 665, and 647, respectively, as in the Man8GlcNAc2 spectrum (Fig. 6A). The ions in the region of m/z 467–809 are virtually identical to those in the spectrum of Man8GlcNAc2, indicating a similar substitution. Therefore, the additional hexose residue must be in the 3-antenna. The ion at m/z 545 is characteristic of oligosaccharides with either a glucose or a mannose residue attached to the terminal mannose residue (Fig. 7A, inset, left-hand structures). This ion is of relatively low abundance, suggesting that it derives from a minor isomer and that at least one other isomer is present. The most likely substitution is a mannose 1,6-linked to the first mannose of this chain. This conclusion is supported by the increased relative abundance of the ion at m/z 383, corresponding to loss of the terminal two mannose residues in a 1,3A3 cleavage. Formation of this ion only leaves positions 4 and 6 available for the 9th mannose residue (position 3 is eliminated in the fragmentation), position 6 being favored. This compound appears to be the Och1p-modified M8(d1,d3,d4) structure shown (Fig. 7A, inset, right-hand structure). Additionally, the D, [D-18]−, and O,3A3 ions, 162 units higher than those of the predominant structure, indicate the presence of M9(d1,d2,d3) (Fig. 7A, inset, middle structure). Indeed, the HPLC peak M9a has the same retention time as this compound. A. satoi α1,2-mannosidase digestion of material recovered under peak M9a yielded two major limit digest products (Fig. 7B); M5 is the expected limit digest product of mammalian-type Man9GlcNAc2, whereas the elution time of the digest product labeled h is consistent with the excision of a single mannose residue from the starting material. Compound h was digested with jack bean mannosidase, and a structure one hexose larger than M5 was generated (Fig. 7C), consistent with the 3-antenna of the starting material possessing a non-reducing glucose residue (Glc1Man8GlcNAc2). The presence of a glucosylated compound eluting under the M9a peak was confirmed by the fact that, when the fOS population derived from exponentially growing ER glucosyltransferase I-deficient alg6Δ cells is compared with that derived from wild type cells, the area of the M9a peak was reduced by ∼75% in the alg6Δ deletion mutant (Fig. 7D). Finally, A. satoi α1,2-mannosidase digestion of material associated with HPLC peak M9b yields predominantly an M6 structure, consistent with M9b comprising mainly the Och1p-modified M9(d1,d3,d4) structure (Fig. 7A, inset, right-hand structure).
A summary of the structural characterization of fOS derived from ams1Δ cells is shown in Table 2, where it can be seen that the predominant structures are M8(d1,d3) and M7(d1) and their Och1p-modified counterparts M9(d1,d3,d4) and M8(d1,d4). In fact, these four compounds accounted for 96% of all of the fOS that were analyzed.
TABLE 2.
Distribution of major free oligosaccharides in post-diauxic ams1Δ and ams1 Δyos9Δ deletion mutants
a Oligosaccharides were prepared from 24-h (post-diauxic) cultures of the indicated yeast strains, derivatized with AP, and resolved by HPLC, and compounds eluting under the indicated peaks were collected. AU, arbitrary units.
b The symbols are as described for Fig 1, and glucose residues are indicated by open squares.
c Although we have demonstrated that the compound eluting as HPLC peak M9b possesses the d4 α1,6-mannosyl moiety, the isomeric configuration of the α1,6 antenna remains uncertain, with the possibility that either mannose d2 or d3 is missing. However, because the size of the M9b peak does not change when MNL1 is deleted in ams1Δ cells (Fig 8A, left), it has been assumed that mannose d2 has been removed by Mns1p. In agreement with this, peak M9b is absent from the HPLC profile of material obtained from ams1Δmns1Δ cells (Fig 8A, right).
Examination of the Role of Yos9p, Mns1p, and Mnl1p in fOS Generation
Next, a series of double and triple deletion mutants were generated in order to explore the role of Yos9p, Mns1p, and Mnl1p in fOS generation during the different growth phases. First, the effects of these deletions on the quantities and distribution of fOS in 1-day cultures were explored.
The Role of Yos9p
A comparison of the steady state levels of fOS in ams1Δ and ams1Δyos9Δ deletion mutants is shown in Table 2. Deletion of Yos9p causes variable reductions in the steady state levels of the different fOS; whereas M7(d1) and the Och1p-modified structures M8(d1,d4) and M9(d1,d3,d4) are reduced by ≥70%, M9(d1,d2,d3) is reduced by only 21% (Table 2). In fact, fOS structures whose presence is most dependent upon Yos9p either are missing mannose residue d3 or possess the d4 mannose residue, and consequently all bear a terminal non-reducing α1,6-mannosyl residue.
The Role of Mnl1p
A striking reduction in the steady state level of M7(d1) in the ams1Δmnl1Δ deletion mutant compared with parental ams1Δ cells is apparent (Fig. 8A, left). The small reduction in size of HPLC peak M8b is probably due to reduction in M8(d1,d4). HPLC peaks M9a and M9b are unaffected by the absence of Mnl1p, indicating that this protein does not appear to be involved in their generation.
The Role of Mns1p
Deletion of Mns1p provokes the concomitant disappearance of HPLC peak M8b and appearance of peak M8a (Fig. 8A, right). The fragmentation spectrum of Hex8GlcNAc2 (m/z 1782) from ams1Δmns1Δ cells is of a mixture of compounds (Fig. 8B). The D, [D-18]−, O,3A3β, and C3β ions at m/z 809, 791, 737, and 665, respectively, show four mannose residues in the 6-antenna leaving three for the 3-antenna, consistent with either M8(d1,d3) or M8(d1,d2). However, there are two points about the spectrum that are not consistent with M8(d1,d3). First, the ratio of the ions m/z 647/665 is the reverse of that derived from the spectrum of M8(d1,d3) found in ams1Δ cells (m/z 665 is much more abundant than m/z 647). Second, in the ion mobility spectra, both positive and negative ion, this compound shows a different mobility to that of a reference M8(d1,d3) sample.4 Taking into account the above two points and that the chromatographic properties of this compound are different from those of M8(d1,d3) during HPLC and TLC (19) as well as the fact that the distribution of residues between the 3- and 6-antennae is as in M8(d1,d3), it is deduced that this compound is M8(d1,d2). A further weak set of ions corresponding to m/z 971, 953, 899, and 827 is consistent with the presence of a further mannose in the 6-antenna. This leaves only two mannose residues for the 3-antenna and indicates the presence of M8(d2,d3) shown on the right-hand side of the inset in Fig. 8B. The data shown in the inset to Fig. 8B confirm the presence of M8(d1,d2) in the ams1Δmns1Δ deletion mutant, but M8(d2,d3), if present, was below the limit of detection using this technique.
In order to better determine the individual roles of Mns1p and Mnl1p in fOS production, the ams1Δmns1Δmnl1Δ triple deletion mutant was generated. Man7GlcNAc2 was not detected in this strain (Fig. 8A, right), indicating that Mns1p is required for production of this compound in the ams1Δmnl1Δ deletion mutant. Furthermore, as indicated by the data shown in Fig. 8A (right), the absence of M8(d1,d2) in the triple mutant indicates that Mnl1p is involved in the generation of this compound in the ams1Δmns1Δ strain. The ensemble of these results is compatible with the scheme for ERAD-L as outlined in Fig. 1A, where the major route for the degradation of misfolded glycoproteins is shown to involve both Mns1p and Mnl1p (heavy arrows). An Mns1p-independent, Mnl1p-dependent ERAD pathway has also been demonstrated (7), and in agreement with this, we demonstrate for the first time the presence of Mns1p-independent Mnl1p-dependent M8(d1,d2) fOS in ams1Δmns1Δ cells. As indicated in Fig. 1A, this structure is known to have high affinity with the lectin Yos9p. Finally, although it is known that the catalytic subunit of Mns1p is capable of generating the M7(d1) structure in vitro (41), our results demonstrate for the first time that, in vivo, this structure can be generated in an Mnl1p-independent, Mns1p-dependent manner.
Png1p-dependent fOS Are Derived from Glycoproteins Whose N-Glycans Are Processed Differently from N-Glycans Present on the Bulk of Glycoproteins at Steady State
The above data show that substantial quantities of fOS continue to be liberated in yeast strains deficient in proteins required for ERAD-L and, in ams1Δmns1Δmnl1Δ cells, contain 9–10 hexose residues (Fig. 8A, right). To understand the origin of these fOS, we asked whether or not this fOS profile simply reflects the steady state distribution of N-glycans in the same cells. Comparison of the HPLC profiles shown in Fig. 8, A (fOS) and C (N-glycans), demonstrates that, whatever the yeast strain considered, compared with N-glycans, fOS are relatively enriched in ER-type structures or the same ER structures that have been modified by Och1p. On the other hand, N-glycan structures are enriched in larger structures that display more complex modification by mannosyltransferases of the Golgi apparatus. These data demonstrate that fOS generation in ERAD-L-deficient cells remains selective for ER-type or Och1p-modified ER-type structures and suggest the presence of an another ERAD process.
M7 and M8 N-glycan structures were noted in ams1Δ and ams1Δmns1Δ deletion mutants, respectively, but, by contrast to their fOS counterparts, these M7 and M8 N-glycans persist in the absence of Mnl1p and indicate that yeast N-glycan processing may be more complex than previously thought. Apart from AMS1, MNS1, and MNL1, the yeast genome data base (available on the World Wide Web) indicates the presence of at least three other genes (MNL2, DFG5, and DCW1) that encode mannosidase-like proteins. However, N-linked M7 was detected in the mnl2Δ, dfg5Δ, and dcw1Δ deletion mutants as well as in wild type cells (data not shown). To further understand the origin of the M7 N-glycan, exponentially growing ams1Δ cells were metabolically radiolabeled for 30 min. Under these conditions, N-glycan structures smaller than [3H]Man8GlcNAc2 are not detected (Fig. 8C, left, inset) despite the appearance of an Mnl1p-dependent fOS behaving as [3H]Man7GlcNAc2 (Fig. 8A, left, inset). These metabolic radiolabeling data are consistent with the notion that once a [3H]N-glycan is trimmed to [3H]M7(d1), the glycoprotein is rapidly degraded. Whatever the origin of truncated N-glycans observed at steady state in the ams1Δ and ams1Δmns1Δ deletion mutants, they are not radiolabeled under our metabolic radiolabeling conditions and do not appear to give rise to fOS.
Reductions in fOS Steady State Levels in Mns1p-, Mnl1p-, and Yos9p-deficient Cells Reveal Different Dependences on Growth
Although data shown in Fig. 4C demonstrate little difference in the steady state levels of the trisaccharide Man1GlcNAc2 in stationary wild type, mnl1Δ, and mns1Δ cells, those reported in Fig. 8A indicate that the different strains reveal striking differences in total fOS levels, after a 24-h culture. In order to investigate this further, fOS were monitored during exponential and post-diauxic growth. As shown in Fig. 9A, the ams1Δmns1Δ, ams1Δmnl1Δ, ams1Δyos9Δ, and ams1Δmns1Δmnl1Δ deletion mutants grow similarly to both wild type and ams1Δ cells. Whereas total fOS levels in ams1Δyos9Δ cells remain at about 40–50% of those in the parental ams1Δ strain throughout post-diauxic growth, those in ams1Δmnl1Δ cells increased slowly from 70% to 80% of parental strain values throughout the post-diauxic period (Fig. 9B, top). By contrast, total fOS levels in ams1Δmns1Δ cells reveal striking biphasic changes (Fig. 9B, bottom). In exponential ams1Δmns1Δ cells, total fOS are 90% of those of parental cells, but at the post-diauxic shift, fOS levels do not increase, and at this time the ams1Δmns1Δ cell fOS level is only 55% that of the parental strain. However, during post-diauxic growth of ams1Δmns1Δ cells, the total fOS level slowly returns to that of parental cells. Data shown in Fig. 9B demonstrate that a similar phenomenon occurs in the ams1Δmns1Δmnl1Δ deletion mutant and that the inhibition of fOS seen in ams1Δmnl1Δ cells is additive to that seen in ams1Δmns1Δ cells. Accordingly, the reductions in appearance of fOS observed when ERAD-L is blocked at the level of N-glycan signal generation are distinct from that when Yos9p, the lectin required for decoding these signals, is absent. These observations suggest but do not prove that Yos9p may have a role in ERAD that is independent of Mns1p- and Mnl1p-generated N-glycan signals. Additionally, it can be seen that the quantities of the M7(d1) and M8(d1,d2) structures that correspond to the major Yos9p-interacting structures in ams1Δmnl1Δ and ams1Δmns1Δ cells, respectively, only achieve 60 and 20%, respectively, of the M7(d1) level noted in parental ams1Δ cells (Fig. 9, C and D), despite the fact that, in these two cell lines, total fOS levels reach 80 and 95% of those of parental cells (Fig. 9B). Accordingly, total fOS inhibition appears not be correlated with loss of these two Yos9p-interacting structures.
FIGURE 9.
The roles of Yos9p and N-glycan trimming during fOS generation. A, the growth of the different deletion mutants was measured at the indicated times. B, 170 A600 (OD 600nm) units of cells were harvested, and after separation and quantitation of fOS, the peak areas corresponding to Man7–9GlcNAc2 fOS were summed. The quantities of fOS, expressed as a percentage of those recovered from the parental ams1Δ deletion mutant, are plotted as a function of growth time. C, using data derived from the same experiment, the areas of the HPLC peaks (M8b and M9a; see Fig. 8A) or of the peaks known to correspond to M7(d1) and M8(d1,d2) are plotted as a function of growth time for the indicated deletion mutants. D, using the same data, the amounts of M7(d1) and M8(d1,d2) recovered from the ams1Δmnl1Δ and ams1Δmns1Δ cells, respectively, expressed as a percentage of the M7(d1) occurring in parental ams1Δ cells, are plotted as a function of culture time.
DISCUSSION
Growth-dependent fOS Demannosylation
In the present study, we identify, for the first time, the trisaccharide Man1GlcNAc2 as the limit catabolic product of fOS in stationary phase cells. Under standard growth conditions, this structure can only be detected during post-diauxic growth and was found to be stable in stationary cells. These results are strikingly different from those obtained by Hirayama et al. (21) which demonstrate very limited fOS demannosylation in stationary cells. Apart from the possibility that Hirayama et al. (21) may define stationary cells differently from the generally accepted method and that their stationary cells are in fact undergoing early post-diauxic growth, another explanation is possible. The extraction procedure employed by Hirayama et al. (21) is reported to allow examination of only cytosolic oligosaccharides. Accordingly, by analogy to that occurring in mammalian cells where cytosolic fOS are partially trimmed in the cytosol by the Ams1p homolog Man2c1p prior to being translocated into lysosomes (42), highly demannosylated yeast fOS may be transferred to the vacuole. Indeed, Ams1p is synthesized in the cytoplasm before being translocated into the vacuole by the cytoplasm-to-vacuole targeting pathway and is known to be active in both compartments (32). However, first, our results demonstrate that vacuolar acidification is not required for fOS metabolism, and, second, the yeast vacuole is fragile, and it remains to be demonstrated that this organelle is not ruptured during the homogenization procedure described by Hirayama et al. (21), which is performed with glass beads in hypotonic buffer. Subcellular localization of fOS in yeast will require experiments in which the intactness of subcellular organelles is monitored. Thus, the origin of the differences between our data and those of Hirayama et al. (21) remains to be determined. However, our data on fOS structures occurring at steady state during the different yeast growth phases are in agreement with previous metabolic radiolabeling studies in which we show that, after 24-h chase incubations, the major Png1p-dependent [3H]fOS appear to contain only 3–4 mannose residues (19, 20). If the role of Ams1p is solely to demannosylate fOS (and other glycoconjugates) in order to recycle mannose into biosynthetic pathways, it makes sense that this process is only activated during post-diauxic growth when external glucose (which can be converted into mannose intracellularly) becomes limiting.
The ultimate fate of the trisaccharide Man1GlcNAc2 remains puzzling. Examination of the S. cerevisiae genome data base suggests that this organism does not possess a β-mannosidase with close homology to mammalian lysosomal β-mannosidases. Although an absence of β-mannosidase activity may explain the pile-up of Man1GlcNAc2 in yeast, our preliminary observations indicate that the level of this structure declines when stationary cells are reincubated in fresh growth medium.3 Whatever the fate of this compound, we show that its steady state level in post-diauxic/stationary cells can be modulated in certain ERAD-defective yeast deletion mutants. Because the trisaccharide can be easily quantitated using as little as 500 μl of stationary cell culture, screening single gene deletion mutants for this compound will be possible. Such an approach may reveal novel gene products involved in glycoprotein quality control and degradation.
Growth-dependent Increases in fOS Steady State Levels
The Man7–9GlcNAc2 fOS species that we characterized comprise >90% of total fOS recovered from post-diauxic ams1Δ cells. Furthermore, the structures that we identified in post-diauxic ams1Δ cells have been found to occur in exponentially growing ams1Δ cells although in different amounts (21). In fact, we noted that the levels of different fOS change in a structure-dependent manner during post-diauxic growth. In exponentially growing ams1Δ cells, fOS levels remain rather constant, and in some experiments, we noted a slight decline in total fOS during exponential/late exponential growth (Fig. 3B, top left). It is possible that even if fOS generation remains constant during rapid growth, cell division and expansion of daughter cells outstrips fOS generation so that there is net reduction in fOS levels per cell. Later, when ams1Δ cell growth slows down, fOS steady state levels stabilize and then rise in the face of ongoing fOS generation. Peptide N-glycanase activity increases 7-fold at the diauxic shift (22). It may be hypothesized that increased Png1p expression during post-diauxic growth (23) reflects an increased need to deal with misfolded glycoproteins. Accordingly, the 7-fold increase in the Man7GlcNAc2 fOS may be due to an increase in flux of misfolded glycoproteins through the ERAD-L pathway. Further experiments will be required in order to determine if changes in Png1p expression underlie increased fOS levels. Again, results reported here contrast with those of Hirayama et al. (21) in that the small increase in fOS in stationary cells with respect to exponential cells that they note is similar to the fOS increase that we see during early post-diauxic growth. As mentioned above, it is possible that either Hirayama et al. (21) have a definition of stationary cells different from the generally accepted one, or perhaps the larger increases in fOS that are noted by us may be due to generation of fOS in a compartment other than the cytoplasm.
Is Mnl1p an α-Mannosidase?
Data reported here demonstrate that both Mns1p and Mnl1p are required for efficient generation of M7(d1), and Mns1p but not Mnl1p is essential for this to occur. Although the yeast Mns1p catalytic subunit can generate M7(d1) from Man9GlcNAc2 in vitro (41), purified Mnl1p has not yet been demonstrated to have enzymic activity in vitro. Presently, the only confirmed mannosidases in S. cerevisiae are Ams1p and Mns1p. The S. cerevisiae genome database indicates that, in addition to Mnl1p, three other proteins (Mnl2p, Dfg5p, and Dcw1p) have mannosidase-like sequences. In fact, our data (Fig. 8B) suggest that mannose trimming of N-glycans in yeast appears to be more complex than previously thought. The presence of Man7GlcNAc2 and Man8GlcNAc2 N-glycans in ams1Δmnl1Δ and ams1Δmns1Δmnl1Δ cells, respectively, suggests a third mannosidase and creates further incertitude surrounding the role of Mnl1p. However, the fact that we were able to detect these unusually truncated N-glycans during steady state measurements but not in pulse metabolically radiolabeled cells may indicate that these structures are formed late in the lifetime of glycoproteins and are not involved in ERAD. Furthermore, Mnl2p, Dfg5p, and Dcw1p do not modify either N-glycan or fOS structures under our experimental conditions (data not shown). Accordingly, our results suggest the presence of an as yet uncharacterized mannosidase in S. cerevisiae, but whether or not this activity plays a role in ERAD remains unknown.
The Roles of Mns1p and Mnl1p in fOS Generation
The roles of proteins required for ERAD have generally been investigated by following the rate of degradation of misfolded (glyco)proteins in exponentially growing cells in which ERAD genes have either been invalidated or overexpressed (6, 11, 15). Measuring fOS steady state levels as a function of the yeast culture cycle yields different but nevertheless important information on ERAD processes. Although our fOS data are in agreement with the N-glycan trimming pathways shown in Fig. 1A, the data shown in Table 2 indicate that only 38% (M7(d1) + M7(d1,d4)) of fOS appear to be derived from N-glycans that have lost mannose residues d2 and d3. Although this figure may increase somewhat in stationary cells, a substantial proportion of fOS do not have structures compatible with the demannosylation scheme outlined in Fig. 1A. This could be explained by evidence indicating that glycan trimming of misfolded glycoproteins for degradation during ERAD-L is site- and context-specific (43). Thus, if a glycoprotein has multiple N-glycans, only one need be acted on by Mns1p and Mnl1p to ensure efficient degradation; in this situation, Png1p will generate one M7(d1) fOS and several other structures that need not possess the degradation signal (43). This may be true, but some of our data argue against it. When compared with that noted in ams1Δ cells, there is a striking, sustained inhibition of M7(d1) in ams1Δmnl1Δ cells that is not accompanied by a sustained decrease (or increase) in HPLC peak M8b (Fig. 9C, top), although, when compared with that noted in ams1Δmns1Δ cells, there is a small reduction in the HPLC M9 peak accompanying the inhibition of M8(d1,d2) observed in ams1Δmns1Δmnl1Δ cells (Fig. 9C, bottom). This is probably due to elimination of M9(d1,d2,d4) and not reduction of M9(d1,d2,d3) (data not shown). Thus, either most glycoproteins destined for degradation have a single N-glycan, or the mannosidases involved are not glycan-specific but rather trim all the N-glycans of a target glycoprotein. Alternatively, these data indicate the presence of other, trimming-independent, processes for glycoprotein degradation. Indeed, some of our data point to the possibility that, at least during post-diauxic growth, N-glycan trimming is not essential for fOS generation but accelerates the process. Thus, as data presented in Fig. 9, B and D, demonstrate, deletions of Mns1p and/or Mnl1p lead to sustained and striking inhibitions of fOS missing the d3 mannose residue, whereas total fOS generation, although slowed down, is relatively less affected (Fig. 9B, top and bottom).
A Potential Role for Och1p in fOS Generation
Affinity studies and x-ray crystallography data suggest that the α1,6-antenna of N-glycans binds to Yos9p, as illustrated in Fig. 1A. However, because structural data showing that the human Yos9p ortholog (OS-9) binds M7(d1) and M8(d1,d2) with similar affinity (8, 9), the crucial event for the recognition of N-glycans by Yos9p involves removal of mannose-d3. The levels of M7(d1), M8(d1,d4), and M8(d1,d3,d4) fOS are most reduced in ams1Δyos9Δ cells (Table 2). The appearance of the latter structure in this fOS pool is difficult to reconcile with the idea that Yos9p interacts only with glycans missing mannose residue d3, and it is tempting to hypothesize that the Och1p modification could itself promote interaction of a misfolded glycoprotein with Yos9p after return to the ER (Fig. 1B). This idea is reinforced by data presented in Fig. 9B showing that total fOS levels remain lower in ams1Δyos9Δ cells than in ams1Δmns1Δmnl1Δ cells. One interpretation of this observation is that glycan signals other than those generated by Mns1p and/or Mnl1p are involved. However, other facts weaken this hypothesis. In general, it may not be surprising that such a high fraction of the fOS that are highly regulated by Yos9p have undergone Och1p modification because Yos9p itself possesses a C-terminal HDEL ER retention signal (44), suggesting that Yos9p-misfolded glycoprotein complexes are potentially retrieved from the Golgi apparatus after modification by Och1p. Similarly, the yeast ER luminal binding protein BiP homolog, Kar2p, possesses a C-terminal HDEL ER retention signal (45), indicating that Kar2p-misfolded glycoprotein complexes could also be retrieved from the Golgi apparatus after modification by Och1p. Furthermore, recent reports show that Yos9p also has a lectin-independent role in ERAD (8, 9, 46). In vitro binding studies will be required in order to evaluate whether or not the Och1p modification could enhance oligosaccharide binding to Yos9p.
In conclusion, information derived from the structural and quantitative analysis of fOS generated in deletion mutants has allowed us to gain insight into ERAD pathways as a function of cell growth. The utility of this approach is attested to by the fact that we have been able to correlate the appearance of certain fOS with known ERAD pathways. However, our data point to more complex origins for fOS. The observation that Man1GlcNAc2 is the final product of fOS catabolism in stationary cells suggests that screening commercially available single deletion mutants for modified trisaccharide levels will allow identification of other pathways for fOS generation.
This work was supported by the Mizutani Foundation, the GIS-Institut des Maladies Rares/INSERM-funded French CDG Research Network, EUROGLYCANET (LSHM-CT-2005-512131), La Fondation pour la Recherche Médicale, and institutional funding from INSERM.
I. Chantret and S. E. H. Moore, unpublished data.
D. J. Harvey, unpublished data.
- ER
- endoplasmic reticulum
- ERAD
- endoplasmic reticulum-associated degradation
- fOS
- free oligosaccharide(s)
- Glc or G
- glucose
- GlcNAc or GN
- N-acetylglucosamine
- Hex
- hexose
- Man or M
- mannose
- AP
- 2-aminopyridine.
REFERENCES
- 1. Aebi M., Bernasconi R., Clerc S., Molinari M. (2010) Trends Biochem. Sci. 35, 74–82 [DOI] [PubMed] [Google Scholar]
- 2. Molinari M. (2007) Nat. Chem. Biol. 3, 313–320 [DOI] [PubMed] [Google Scholar]
- 3. Jakob C. A., Bodmer D., Spirig U., Battig P., Marcil A., Dignard D., Bergeron J. J., Thomas D. Y., Aebi M. (2001) EMBO Rep. 2, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jakob C. A., Burda P., Roth J., Aebi M. (1998) J. Cell Biol. 142, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quan E. M., Kamiya Y., Kamiya D., Denic V., Weibezahn J., Kato K., Weissman J. S. (2008) Mol. Cell 32, 870–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clerc S., Hirsch C., Oggier D. M., Deprez P., Jakob C., Sommer T., Aebi M. (2009) J. Cell Biol. 184, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hosomi A., Tanabe K., Hirayama H., Kim I., Rao H., Suzuki T. (2010) J. Biol. Chem. 285, 24324–24334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hosokawa N., Kamiya Y., Kamiya D., Kato K., Nagata K. (2009) J. Biol. Chem. 284, 17061–17068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mikami K., Yamaguchi D., Tateno H., Hu D., Qin S. Y., Kawasaki N., Yamada M., Matsumoto N., Hirabayashi J., Ito Y., Yamamoto K. (2010) Glycobiology 20, 310–321 [DOI] [PubMed] [Google Scholar]
- 10. Denic V., Quan E. M., Weissman J. S. (2006) Cell 126, 349–359 [DOI] [PubMed] [Google Scholar]
- 11. Gauss R., Jarosch E., Sommer T., Hirsch C. (2006) Nat. Cell Biol. 8, 849–854 [DOI] [PubMed] [Google Scholar]
- 12. Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009) Nature 458, 453–460 [DOI] [PubMed] [Google Scholar]
- 13. Suzuki T., Park H., Hollingsworth N. M., Sternglanz R., Lennarz W. J. (2000) J. Cell Biol. 149, 1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suzuki T., Park H., Kwofie M. A., Lennarz W. J. (2001) J. Biol. Chem. 276, 21601–21607 [DOI] [PubMed] [Google Scholar]
- 15. Carvalho P., Goder V., Rapoport T. A. (2006) Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 16. Bazirgan O. A., Hampton R. Y. (2008) J. Biol. Chem. 283, 12797–12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kostova Z., Mariano J., Scholz S., Koenig C., Weissman A. M. (2009) J. Cell Sci. 122, 1374–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie W., Ng D. T. (2010) Semin. Cell Dev. Biol. 21, 533–539 [DOI] [PubMed] [Google Scholar]
- 19. Chantret I., Frénoy J. P., Moore S. E. (2003) Biochem. J. 373, 901–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chantret I., Moore S. E. (2008) Glycobiology 18, 210–224 [DOI] [PubMed] [Google Scholar]
- 21. Hirayama H., Seino J., Kitajima T., Jigami Y., Suzuki T. (2010) J. Biol. Chem. 285, 12390–12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suzuki T., Park H., Kitajima K., Lennarz W. J. (1998) J. Biol. Chem. 273, 21526–21530 [DOI] [PubMed] [Google Scholar]
- 23. Davidson G. S., Joe R. M., Roy S., Meirelles O., Allen C. P., Wilson M. R., Tapia P. H., Manzanilla E. E., Dodson A. E., Chakraborty S., Carter M., Young S., Edwards B., Sklar L., Werner-Washburne M. (2011) Mol. Biol. Cell 22, 988–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Opheim D. J. (1978) Biochim. Biophys. Acta 524, 121–130 [DOI] [PubMed] [Google Scholar]
- 25. Petracek M. E., Longtine M. S. (2002) Methods Enzymol. 350, 445–469 [DOI] [PubMed] [Google Scholar]
- 26. Peric D., Durrant-Arico C., Delenda C., Dupré T., De Lonlay P., de Baulny H. O., Pelatan C., Bader-Meunier B., Danos O., Chantret I., Moore S. E. (2010) PLoS One 5, e11675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hase S., Ibuki T., Ikenaka T. (1984) J. Biochem. 95, 197–203 [DOI] [PubMed] [Google Scholar]
- 28. Moore S. E., Spiro R. G. (1994) J. Biol. Chem. 269, 12715–12721 [PubMed] [Google Scholar]
- 29. Chantret I., Fasseu M., Zaoui K., Le Bizec C., Yayé H. S., Dupré T., Moore S. E. (2010) PLoS One 5, e11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gray J. V., Petsko G. A., Johnston G. C., Ringe D., Singer R. A., Werner-Washburne M. (2004) Microbiol Mol. Biol. Rev 68, 187–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kennedy L. D., Court C. B. (1983) Anal. Biochem. 128, 465–467 [DOI] [PubMed] [Google Scholar]
- 32. Hutchins M. U., Klionsky D. J. (2001) J. Biol. Chem. 276, 20491–20498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harvey D. J. (2005) J. Am. Soc. Mass Spectrom. 16, 631–646 [DOI] [PubMed] [Google Scholar]
- 34. Harvey D. J. (2005) J. Am. Soc. Mass Spectrom. 16, 622–630 [DOI] [PubMed] [Google Scholar]
- 35. Harvey D. J., Royle L., Radcliffe C. M., Rudd P. M., Dwek R. A. (2008) Anal. Biochem. 376, 44–60 [DOI] [PubMed] [Google Scholar]
- 36. Domon B., Costello C. E. (1988) Glycoconj. J. 5, 397–409 [Google Scholar]
- 37. Reiss G., te Heesen S., Zimmerman J., Robbins P. W., Aebi M. (1996) Glycobiology 6, 493–498 [DOI] [PubMed] [Google Scholar]
- 38. Wiggins C. A., Munro S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7945–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yip C. L., Welch S. K., Klebl F., Gilbert T., Seidel P., Grant F. J., O'Hara P. J., MacKay V. L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 2723–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakanishi-Shindo Y., Nakayama K., Tanaka A., Toda Y., Jigami Y. (1993) J. Biol. Chem. 268, 26338–26345 [PubMed] [Google Scholar]
- 41. Herscovics A., Romero P. A., Tremblay L. O. (2002) Glycobiology 12, 14G-15G [PubMed] [Google Scholar]
- 42. Moore S. E. (1999) Trends Cell Biol. 9, 441–446 [DOI] [PubMed] [Google Scholar]
- 43. Spear E. D., Ng D. T. (2005) J. Cell Biol. 169, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Friedmann E., Salzberg Y., Weinberger A., Shaltiel S., Gerst J. E. (2002) J. Biol. Chem. 277, 35274–35281 [DOI] [PubMed] [Google Scholar]
- 45. Normington K., Kohno K., Kozutsumi Y., Gething M. J., Sambrook J. (1989) Cell 57, 1223–1236 [DOI] [PubMed] [Google Scholar]
- 46. Bhamidipati A., Denic V., Quan E. M., Weissman J. S. (2005) Mol. Cell 19, 741–751 [DOI] [PubMed] [Google Scholar]
- 47. Kawaguchi S., Hsu C. L., Ng D. T. (2010) PLoS One 5, e15532 [DOI] [PMC free article] [PubMed] [Google Scholar]