Abstract
Plasmodium parasites undergo a clinically silent and obligatory developmental phase in the host’s liver cells before they are able to infect erythrocytes and cause malaria symptoms. To overcome the scarcity of compounds targeting the liver stage of malaria, we screened a library of 1037 existing drugs for their ability to inhibit Plasmodium hepatic development. Decoquinate emerged as the strongest inhibitor of Plasmodium liver stages, both in vitro and in vivo. Furthermore, decoquinate kills the parasite’s replicative blood stages and is active against developing gametocytes, the forms responsible for transmission. The drug acts by selectively and specifically inhibiting the parasite’s mitochondrial bc1 complex, with little cross-resistance with the antimalarial drug atovaquone. Oral administration of a single dose of decoquinate effectively prevents the appearance of disease, warranting its exploitation as a potent antimalarial compound.
The search for active compounds among drugs originally developed and clinically tested for the treatment of other diseases is a particularly advantageous approach in the case of historically neglected diseases such as malaria. Although a number of reports have recently emerged where libraries of existing drugs were screened in the search for inhibitors of the malaria parasite Plasmodium falciparum [1–6], these screens have focused solely on the intraerythrocytic life cycle of Plasmodium infection, when clinical symptoms occur. However, the liver stage of Plasmodium infection is an obligatory step in the maturation and replication of mosquito-delivered parasites toward generating the erythrocyte-infective forms that cause malaria symptoms [7]. To target the hepatic stage is therefore highly desirable in the context of malaria eradication, not only because its asymptomatic nature makes it ideally suited for prophylactic intervention [8], but also because the liver can serve as a reservoir for Plasmodium vivax and Plasmodium ovale hypnozoites, dormant parasite forms that may lead to relapses long after the initial blood infection has been eliminated [9]. In sharp contrast with drugs that kill blood-stage parasites, only a very limited number of drugs are available against liver forms. Among these, primaquine is the only clinically approved drug known to eliminate liver forms of Plasmodium, including hypnozoites, and to kill gametocytes, the sexual forms responsible for transmission of the parasite’s life cycle inside the mosquito vector [10]. Despite primaquine’s potentially lethal side effects that severely limit its use, except for a study from the 1980s [11], systematic efforts toward the identification of novel chemical safe entities efficacious against Plasmodium liver stages have not been reported [12]. Here, we describe the first screen targeted at Plasmodium liver stages and the identification of decoquinate, a compound that is shown to inhibit multiple phases of the parasite’s life cycle. Further investigation revealed decoquinate’s mode of action and established its potential as a potent and selective antimalarial compound.
METHODS
Ethics Statement
All procedures involving animal models complied with European and US regulations.
Cells, Parasites, and Mice
Huh7 cells, a human hepatoma cell line, were cultured as described in the Supplementary Methods.
Two transgenic parasite lines were used in this study: a green fluorescent protein (GFP)–expressing Plasmodium berghei line (parasite line 259cl2) [13], as well as a fusion GFP- and firefly luciferase–expressing P. berghei line (parasite line 676m1cl1) (PbGFP-Luccon) [14]. Sporozoites from both lines were freshly obtained through the disruption of salivary glands of infected female Anopheles stephensi mosquitoes.
Male C57Bl/6 mice aged 6–8 weeks and weighing 20–24 g were purchased from Charles River and housed in the pathogen-free facilities of the Instituto de Medicina Molecular (Lisbon, Portugal).
Drug Library Screen for Activity Against Malaria Liver Stages
Huh7 cells (1800 cells/well) were seeded in 30 μL of complete Roswell Park Memorial Institute medium in 384-well collagen I coated plates (Greiner Bio-one) and incubated at 37°C with 5% CO2. Forty-eight hours after seeding, prediluted drugs (5 μL) were added to wells to achieve a final concentration of 10 μM. After 1 hour, cells were infected with GFP-expressing P. berghei sporozoites (2000 sporozoites/well) freshly obtained through disruption of salivary glands of infected female A. stephensi mosquitoes. Twenty-four hours after infection, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) and permeabilized with 0.1% Triton X-100 in PBS, and cell nuclei were stained with Hoechst-33324 (Invitrogen).
Luminescence Measurement of In Vitro Plasmodium Infection
Inhibition of liver-stage infection was assessed by measuring the luminescence intensity of Huh-7 cells infected with a firefly luciferase–expressing P. berghei line, PbGFP-Luccon, as previously reported [14].
Fluorescence-Activated Cell Sorting (FACS) Analysis
FACS analysis of cells infected with GFP-expressing parasites allows differentiation between the effect of a drug on hepatocyte invasion and on intrahepatocyte parasite development. FACS analysis at 2 and 48 hours after sporozoite addition was performed to determine the percentage of parasite-containing cells and parasite-GFP intensity within infected cells. Cell samples for FACS analysis were processed as previously described [15].
In Vivo Drug-Efficacy Studies
All in vivo protocols were approved by the Animal Care Committee of the Instituto de Medicina Molecular and were performed according to the regulations of the European guidelines 86/609/EEG. Drug suspensions in soybean oil (200 μL/mouse) or in an equivalent amount of vehicle were administered orally by gavage. Mice were infected by intravenous injection of 1 × 104 firefly luciferase–expressing P. berghei sporozoites. Alternatively, mice were infected by exposure to the bite of infected female Anopheles stephensi mosquitoes. Mosquitoes (20 per mouse) were allowed to feed for 30 minutes on mice anesthetized by intraperitoneal injection of 180 μL of mixture consisting of 80 mg/kg of ketamine and 10 mg/kg xylazine dissolved in PBS.
Parasite load in the livers was determined 44–46 hours after infection by real-time in vivo imaging with the in vivo IVIS Lumina Imaging System [14]. Infection was allowed to proceed to the blood stage and was monitored by analysis of Giemsa-stained blood smears of tail blood collected between days 3 and 15 after infection. A compound was considered effective in the liver if, by day 15 after infection, no parasite could be detected in the blood smears.
Drug-Susceptibility Testing Against Asexual and Sexual Stages of 164/GFP Parasites
The drug-susceptibility assay was conducted as previously described [6]. Briefly, highly synchronized, young, ring-stage parasites in 110 μL complete medium (4% hematocrit) were plated into 96-well plates (Microtest Tissue Culture Plates, Becton Dickinson) alongside uninfected red blood cells as a negative control. Sexual commitment was induced by a drop in hematocrit 24 hours after seeding the parasites. Serial dilutions of compounds were added at that point or 24 hours later, after reinvasion of parasites. Corresponding dilutions of chloroform were tested, as well, to account for solvent-specific effects. On the day of analysis, cells were stained with Hoechst, and cytometry data were collected as described in detail elsewhere [6]. These data were processed with Quanta software and were analyzed for half-maximal inhibitory concentrations (IC50), using GraphPad Prism.
Ubiquinol-Cytochrome bc1 Oxidoreductase Activity
Ubiquinol-cytochrome c oxidoreductase activity was measured as the antimycin-sensitive decylubiquinol-cytochrome c oxidoreductase level in isolated mitochondria, as detailed in the Supplementary Methods.
Growth-Inhibition Assays for Evaluation of bc1 Inhibition
The sensitivity of P. falciparum–infected erythrocytes to various drugs was determined using a [3H]hypoxanthine incorporation method with an inoculum size of 0.5% parasitemia and 2% hematocrit. Plates were incubated at 37°C in 5% CO2, 5% O2, and 95% N2. After 24 hours of incubation, [3H]hypoxanthine was added and plates were incubated for an additional 24 hours. After that period, plates were harvested on a glass fiber filter, using a TOMTEC Cell harvester 96. Filters were dried and melted on scintillator sheets, and the incorporated radioactivity was quantified by use of a Wallac Microbeta Trilux (Model 1450 LS- Perkin Elmer). The Dd2 cell line containing yeast dihydroorotate dehydrogenase (DHODH) and its parental strain were cultured in the absence or presence of proguanil (1 μM). The ScURA1 gene from Saccharomyces cerevisiae was amplified from genomic DNA and cloned into the pLN-14 vector. The P. falciparum Dd2attB strain was transfected by electroporation, and stable transfectants were selected with blasticidin. Both the Dd2attB_yeastDHODH strain and its parental strain, Dd2attB, were used in these assays. Nonlinear regression analysis was used to fit the normalized results of the dose-response curves, and IC50 data were determined using the Grafit5 software package (Grafit program; Erithacus Software, Horley, Surrey, United Kingdom).
Molecular Modeling Studies
The molecular modeling studies were carried out using the crystal structure of cytochrome bc1 from S. cerevisiae (PDB 3CX5) [16] as a template for generating the Y279S mutation. This was achieved using the Mutate Residue functionality within MOE (CCG MOE), which is an implementation of the method presented by Bower et al [17]. Mutant side-chain conformations are determined from a systematic rotamer search, resulting in acceptable side-chain structures that are based on the local environment. Docking was performed with the GOLD 5.01 (Genetic Optimization for Ligand Docking) package, which searches for the best ligand interaction pose, using a genetic algorithm. Docked ligands were ranked with GoldScore [18], which is included in the software, and were defined by the following components: protein-ligand hydrogen bond energy, protein-ligand van der Waals energy, ligand internal van der Waals energy, and ligand torsional strain energy. This fitness function has been optimized to predict the ligand-binding position and conformation of the ligands. Docking was run with standard settings and 1000 genetic algorithm operations. Atovaquone and decoquinate were docked into the oxidation site in the targeted cytochrome. Visualization of best-fit docking poses was performed with PyMOL after energy-minimization of the binding pocket and docked ligand with MOE (MMFF94x force field).
RESULTS
Chemical Screen Identifies Decoquinate as a Potent Inhibitor of Plasmodium Liver Infection
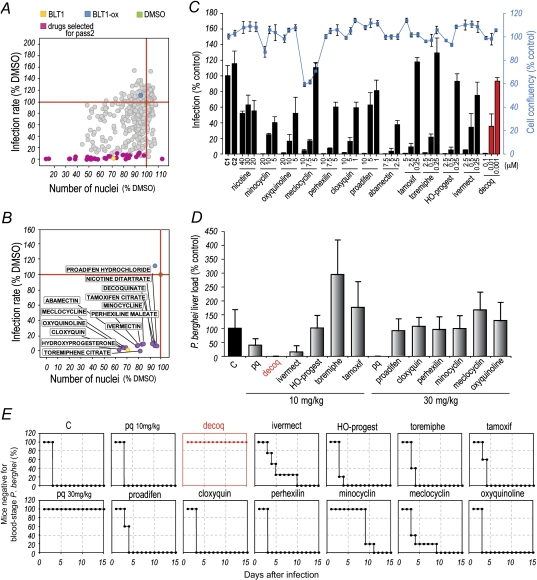
We employed an in vitro infection model combining P. berghei parasites and the Huh7 human hepatoma cell line [15] to screen a commercially available library composed of 1037 compounds that have reached clinical stages (eg, clinical trials) in the United States (Supplementary Table 1) for their activity against Plasmodium liver stages. The high-throughput screen was carried out in a 384-well format combined with an automated microscopy high content readout (Supplementary Figure 1). The effect of each drug on the course of infection was assessed with the help of customized image analysis algorithms that allow for automatic quantification of both cell numbers and amount of intracellular parasites [19, 20]. The library was screened 3 independent times, using a fixed concentration of 10 μM of each compound. This process identified 116 compounds that decreased infection by >50% relative to the control. It is interesting to note that these compounds are distributed across a wide range of drug classes, suggesting a multiplicity of possible targets (Supplementary Figure 2). Among these compounds, the 41 producing a reduction in infection >90% were selected to undergo a second screening pass (Figure 1A and Supplementary Table 1), where their dose-response behavior was evaluated. On the basis of that response, and after exclusion of cytotoxic drugs, compounds with previously reported antiplasmodial activity, and topical drugs, 13 compounds were chosen to undergo a final round of in vitro screening (Figure 1A and 1B and Supplementary Table 1). In this assay, Huh7 cells and luciferase-expressing P. berghei parasites were used as previously described [14]. Each compound was assayed at 3 different concentrations, and their effects on infection load and cell confluence were evaluated by luminescence and fluorescence measurements, respectively. With the exception of nicotine, for which no dose-response was observed, all of the drugs led to a dose-dependent reduction in the P. berghei infection rate among Huh7 cells, albeit within different ranges of compound concentrations (Figure 1C). These results defined a final list of 12 compounds with proven in vitro activity against Plasmodium liver stages, which were selected for in vivo evaluation (Supplementary Table 1).
Figure 1.
Screen of compound library. A, Results of the first screening round of 1037 compounds, highlighting the compounds leading to >90% decrease in infection. Blocker of lipid transport 1 (BLT-1) and its oxidized version (BLT-ox), previously used in the context of liver-stage Plasmodium infection [20], were used as positive and negative controls, respectively. B, Thirteen compounds selected after the second screening round. The plots in A and B depict the effect of the drugs on Huh7 cell proliferation (x-axis) and infection by Plasmodium berghei sporozoites (y-axis), measured 24 h after parasite addition. Each circle represents a compound. Green, yellow, and blue dots represent the dimethyl sulfoxide, positive control, and negative control, respectively, as above. The horizontal and vertical red lines indicate 100% infection and 100% confluency, respectively. C, Luminescence-based measurement of dose-dependent effects of selected compounds. Bars represent infection loads, and the red line represents cell confluency. Error bars represent SDs from 3 independent measurements. D, Luminescence-based measurements of liver parasite loads in mice following oral administration of selected compounds. Compounds were administered in the indicated amounts 24 h before, concomitantly with, and 24 h after intravenous injection of 10 000 luciferase-expressing P. berghei sporozoites. Error bars represent SDs (n = 5). E, Blood-stage patency of infection following oral administration of the same drugs as in D. Decoquinate is highlighted in red in both D and E.
The ability of the selected compounds to inhibit infection in vivo was evaluated in an established rodent model of malaria. Mice received 3 treatments with 10 or 30 mg/kg of each compound, administered orally 24 hours before, concomitantly with, and 24 hours after intravenous injection of 10 000 luciferase-expressing P. berghei sporozoites. Liver parasite burdens were determined by luminescence 44 hours after infection (Figure 1D), and blood parasitemias were monitored daily for 15 days (Figure 1E). Most notably, decoquinate consistently emerged from our studies as the most potent inhibitor of Plasmodium infection in vivo, completely abrogating liver parasitemia and fully preventing the appearance of parasites in the blood at 10 mg/kg. Decoquinate’s IC50 against liver stages in vitro was estimated at 2.6 nM, approximately 3000-fold lower than that of primaquine (Table 1 and Supplementary Figure 3A). Flow cytometry analysis of Huh7 cells infected with GFP-expressing P. berghei sporozoites further demonstrated that decoquinate acts by inhibiting Plasmodium’s intracellular replication, rather than its ability to invade hepatic cells (Supplementary Figure 3B).
Table 1.
Effect of Decoquinate on Liver and Blood Stages of Plasmodium Infection, as Represented by Its Half-Maximal Inhibitory Concentration (IC50)
| Blood Stages (IC50) |
|||
| Compound | Liver Stages (IC50) | Asexual | Sexual |
| Decoquinate | 2.6 ± 0.7 | 10 ± 8 | 36 ± 30 |
| Primaquine | 7500 | 11 000 ± 5000 [6] | No effect [6] |
| Artemisinin | NA | 62 ± 14 | 92 ± 31 |
| Atovaquone | NA | 1.8 ± 0.2 [6] | Slight reduction [6] |
IC50 data are mean nM ± SD. Published IC50 data given for comparison were taken from [6].
Abbreviation: NA, not available.
Decoquinate Acts on Asexual and Presexual Plasmodium Blood Forms
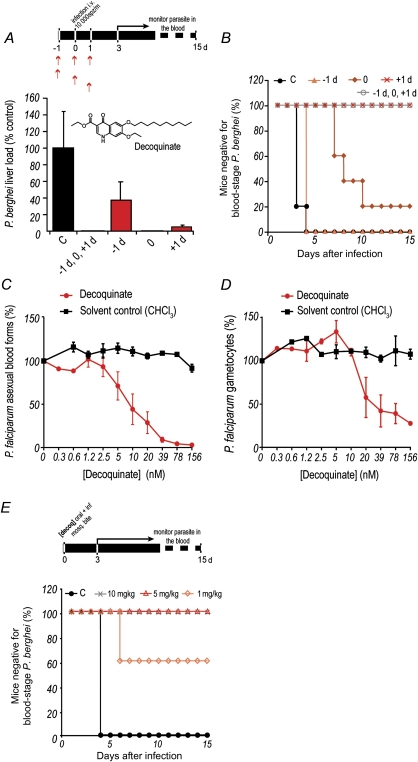
We then investigated the degree of protection conferred by different decoquinate administration schedules relative to parasite inoculation. To this end, the drug was administered orally to mice at 10 mg/kg, at different times relative to intravenous injection of 10000 luciferase-expressing sporozoites (Figure 2A). Our results showed that administration of decoquinate in any of these schedules significantly decreased parasite liver load, compared with untreated controls (Figure 2A). Interestingly, liver parasite load decreased to below detectable levels in mice treated immediately prior to infection, but they eventually developed blood parasitemia, although significantly delayed (P = .00159), while mice treated 24 hours after infection remained negative for blood-stage parasites (Figure 2B). This suggests that decoquinate acts not only on Plasmodium liver stages but also on the blood forms of the parasite. To confirm this, the effect of decoquinate on in vitro cultures of P. falciparum strain 164/GFP blood stages was evaluated using methods described elsewhere [6] (Figure 2C and Table 1). Indeed, decoquinate displayed a marked effect on asexual blood stages of Plasmodium, with an IC50 of the same order of magnitude of that of artemisinin, an effective blood schizonticidal antimalarial, and approximately 1000-fold lower than that of primaquine. Crucially, our data also showed that decoquinate is active against Plasmodium gametocytes, the presexual blood forms of the parasite responsible for transmission to the invertebrate host, with an IC50 of 36 nM (Figure 2D and Table 1).
Figure 2.
Inhibition of Plasmodium by decoquinate. A, B, Oral administration of 10 mg/kg of decoquinate at various times relative to intravenous injection of 10 000 luciferase-expressing Plasmodium berghei sporozoites. Luminescence-based measurement of liver parasite loads are shown in A, and blood-stage patency of infection is shown in B. The inset depicts the structure of the decoquinate molecule. Results shown are representative from 3 independent experiments. Error bars represent SDs (n = 5). C, D, Half-maximal inhibitory concentration curves representing the effect of decoquinate on parasite (C) and gametocyte (D) load. Each assay represents at least 3 biological replicates with 2 technical replicates per plate. The graphs show mean values of the experiments, whereas the error bars represent the standard error of the mean. E, Blood-stage patency of infection in mice following oral administration of a single dose of decoquinate immediately prior to P. berghei sporozoite delivery by mosquito bite (n = 5). Abbreviation: C, control.
Finally, we sought to establish the minimal decoquinate dose that would provide complete protection against infection transmitted to mice by mosquito bite, the most physiologically relevant transmission route. A single decoquinate dose of 10, 5, or 1 mg/kg was administered orally prior to sporozoite injection by P. berghei–infected mosquitoes, followed by daily monitoring of blood parasitemia. Results showed that whereas 1 mg/kg of decoquinate could already confer partial protection, a single 5 mg/kg dose of the compound completely prevented the appearance of blood-stage parasites (Figure 2E).
Decoquinate Specifically and Selectively Inhibits Plasmodium Mitochondrial bc1 Complex
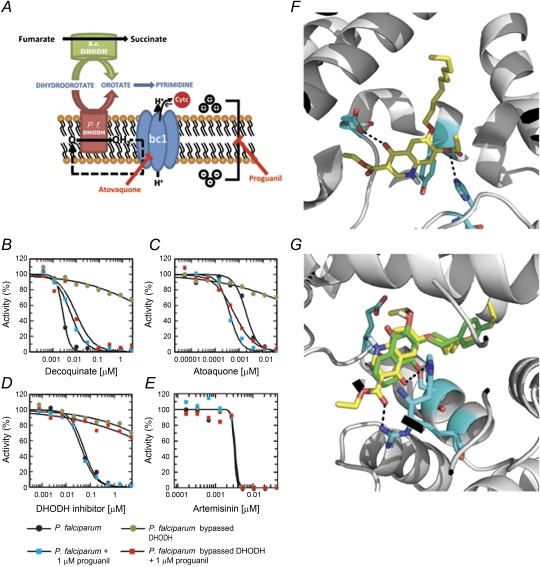
Given the structural similarities with other quinolones known to inhibit Plasmodium’s cytochrome bc1 complex (ubiquinol:cythocrome c oxidoreductase or respiratory complex III; bc1; P. falciparum genes 2655541, 811955) [21], we investigated whether the mechanism of decoquinate’s antimalarial activity could be explained by bc1 inhibition. To address this, we initially assessed decoquinate inhibition of bc1 activity on mitochondria isolated from P. falciparum 3D7A parasites and human HEK293 cells. We found plasmodial bc1 complex to be susceptible to decoquinate, with an IC50 of 0.002 μM and a selectivity index of >5000, when compared with its human counterpart (Table 2). To confirm the antimalarial mode of action of decoquinate by inhibition of bc1 complex, we then carried out a series of growth inhibition experiments (Figure 3). The mitochondrial electron transport chain (ETC) is critical for parasite survival, inhibition of the P. falciparum cytochrome bc1 being the mode of action of the antimalarial drug atovaquone [22] (Figure 3A). ETC is used by Plasmodium as the only way to regenerate mitochondrial coenzyme Q. This is the electron receptor used by DHODH, a mitochondrion-located, membrane-anchored enzyme of the pyrimidine biosynthetic pathway. DHODH is also essential for survival because Plasmodium cannot salvage pyrimidines [23] (Figure 3A). Artemisinin, a known potent antimalarial, kills the parasite by a different mechanism, presumably one involving hemoglobin digestion [24], and can therefore be used as an appropriate control for assays to measure the mode of action of growth inhibition. We showed that P. falciparum growth was effectively inhibited by decoquinate, atovaquone, a specific DHODH inhibitor, and artemisinin (Figure 3B–E and Table 3). These drugs were then assayed on a P. falciparum strain transfected with cytoplasmic S. cerevisiae DHODH (ScDHODH), which bypasses the endogenous Plasmodium counterpart that is required for pyrimidine biosynthesis. ScDHODH uses fumarate instead of mitochondrial coenzyme Q as an electron acceptor, and therefore the transgenic strain is resistant to both bc1 and DHODH inhibitors [23]. This Plasmodium strain remained sensitive to artemisinin but was not killed by decoquinate, atovaquone, or the DHODH inhibitor, suggesting that the latter 3 compounds block pyrimidine biosynthesis (Figure 3B–E and Table 3). Finally, to distinguish bc1 from DHODH inhibition, the assay was performed in the presence of proguanil, which collapses the mitochondrial membrane potential when electron transport is inhibited and renders the ScDHODH-expressing strain sensitive to ETC inhibitors [23]. Addition of proguanil restored inhibition by decoquinate and atovaquone but not by the DHODH inhibitor (Figure 3B–E and Table 3). Overall, these results show that decoquinate’s antimalarial mode of action is the potent, selective, and specific inhibition of P. falciparum mitochondrial bc1 complex.
Table 2.
Sensitivity of Ubiquinol:Cytochrome bc1 Activity to Decoquinate and Atovaquone on Isolated Mitochondria
| IC50 (mean μM ± SD) |
||
| Ubiquinol Cytochrome c Reductase | Decoquinate | Atovaquone |
| P. falciparum | 0.002 ± 0.0005 | 0.0002 ± 0.00003 |
| HEK293 | >10 | 0.327 ± 0.03 |
Abbreviation: P. falciparum, Plasmodium falciparum.
Figure 3.
Antiplasmodial mode of action of decoquinate. A, Generation of P. falciparum mitochondrial membrane potential by bc1 complex–mediated and dihydroorotate dehydrogenase (DHODH)–mediated reactions. Plasmodium falciparum (P. f.) DHODH is the key enzyme in the reduction of coenzyme Q (from Q to QH2). QH2 is reoxidized by the bc1 complex with subsequent electron transfer to cytochrome c, resulting in proton translocation and generation of electropotential across the inner membrane. Inhibitors of the bc1 complex prevent this mode of electropotential generation, as well as reoxidation of QH2. In this scenario, pyrimidine synthesis can be secured by Saccharomyces cerevisiae DHODH, which bypasses plasmodial DHODH by using fumarate as the electron acceptor, while a proguanil-sensitive alternate path can provide the necessary membrane potential. Thus, in the presence of atovaquone or other electron transport inhibitors, the generation of electropotential becomes hypersensitive to proguanil (adapted from [18]). B–E, P. falciparum in vitro growth curves and genetic rescue with yeast DHODH. Addition of proguanil restores sensitivity of the P. falciparum line with S. cerevisiae DHODH to decoquinate and atovaquone but not to a specific DHODH inhibitor. Artemisinin is used as a bc1 complex–independent and DHODH-independent control for growth inhibition. F, Docking pose of decoquinate for the wild-type bc1 complex. A hydrogen bond with H181 and a carbonyl-carbonyl interaction with E272 are established between protein and ligand. G, Docking poses of ligands for the mutant cytochrome bc1 model. Decoquinate is represented in yellow and atovaquone in green. Decoquinate is within hydrogen bond distance of H181 and R283, whereas atovaquone loses the ability to interact with E272 and H181.
Table 3.
Growth Inhibition of Parental and Transgenic Plasmodium falciparum Strains in the Absence or Presence of Proguanil
| IC50 (mean μM ± SD) |
|||||
| Parasite Strain | Proguanil (1 μM) | Decoquinate | DHODH Inhibitor | Atovaquone | Artemisinin |
| Dd2attB | Absent | 0.003 ± 0.0001 | 0.046 ± 0.006 | 0.002 ± 0.00002 | 0.004 ± 0.0008 |
| Dd2attB | Present | 0.006 ± 0.0004 | 0.048 ± 0.002 | 0.0004 ± 2 × 10 –5 | 0.004 ± 0.0008 |
| Dd2attB_yeastDHODH | Absent | >2.5 | >5 | >0.035 | 0.004 ± 0.0008 |
| Dd2attB_yeastDHODH | Present | 0.011 ± 0.001 | >5 | 0.001 ± 7.5 × 10−5 | 0.003 ± 4 × 10−5 |
Decoquinate Presents Little Cross-Resistance With Atovaquone
Resistance to atovaquone, presently the only drug in clinical use that targets bc1, emerged rapidly [25]. This raised the need for alternative bc1 inhibitors with little cross-resistance with atovaquone-resistant strains, as is the case of quinolones [21]. The fact that, unlike atovaquone [6], decoquinate shows potent gametocidal activity indicated that the 2 drugs interact differently with their bc1 complex target. To further predict whether decoquinate presents cross-resistance with atovaquone, we carried out a molecular docking study that used S. cerevisiae bc1 complex structure (PDB 3CX5) [16] as a surrogate for the P. falciparum protein. This approach has been used effectively to study the binding mode of atovaquone, given the high sequence identity at the Qo site of cytochrome b [26]. Decoquinate presented a similar docking pose to 4(1H)-pyridones in the wild-type protein [27], which is consistent with its IC50 (Figure 3F). The Y279S mutation (yeast numbering) corresponds to one of the most frequent mutations in P. falciparum and results in resistance to atovaquone [26]. Molecular-docking runs on a mutated model generated from 3CX5 showed that Y279 is critical for atovaquone binding, as mutation to S279 (Figure 3G) leads to loss of interaction with E272 and H181 [27, 28]. Introduction of the Y279S mutation led to inversion in the docking pose of decoquinate. However, the protein-inhibitor complex is still stabilized by hydrogen bonds with H181 and R283 and by hydrophobic interactions with residues from the binding pocket (Figure 3G). Therefore, decoquinate is expected to inhibit the Y279S mutant of cytochrome bc1 effectively and is therefore expected to present little cross-resistance with atovaquone. This prediction was confirmed in a very recent report that showed that decoquinate indeed possesses limited cross-resistance against 5 atovaquone-resistant P. falciparum lines [29].
DISCUSSION
Decoquinate is a cheap and widely available coccidiostat commonly used in livestock ranging from poultry to mammals [30] and has been recently identified in a screen for blood-stage inhibitors of Plasmodium infection [29]. The data presented here demonstrate that decoquinate is a potent antimalarial, with transmission blocking potential due to the marked effect on various stages of Plasmodium’s life cycle. Its identification stemmed from the first high-throughput, microscopy-based screen performed to identify compounds that inhibit the hepatic stage of plasmodial infection. Besides its ability to inhibit the development of the parasite’s liver stages, decoquinate demonstrated strong activity against Plasmodium blood forms, including gametocytes, which are latent presexual parasite forms responsible for the transmission of Plasmodium to Anopheles mosquitoes. This is in contrast with atovaquone, which has been shown to present activity against Plasmodium liver stages [31] but to be only slightly effective against parasites in the early sexual stage [6]. Importantly, a previous study has concluded that compounds shown to be gametocytocidal also possess radical curative effects against true relapsing malarias [32]. Thus, although the present study does not provide direct evidence for this, decoquinate’s strong effect against latent gametocytes suggests that it might be active against hypnozoites, which are latent parasite forms in the liver. On the other hand, drugs targeting multiple stages of Plasmodium’s life cycle may present added advantages in terms of efficacy. Decoquinate inhibits the bc1 complex, an important, yet underexploited antimalarial target. The rapid emergence of resistance to atovaquone led to its combination with proguanil (Malarone), the cost of which limited its widespread use in resource-poor, disease-endemic areas and highlighted the need for cheaper alternatives that can overcome resistance [21]. Given its little cross-resistance with atovaquone-resistant strains [29], decoquinate may constitute one such alternative. In this context, and at a time when novel drugs against malaria are urgently required, this study paves the way not only for future screening efforts of a similar nature but also for further exploitation of decoquinate as part of an antimalarial strategy.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://www.oxfordjournals.org/our_journals/jid/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments.
F. P. C. acknowledges FCT for postdoctoral grant BPD/64539/2010. K. B. acknowledges a postdoctoral Feodor Lynen fellowship from the Alexander von Humboldt foundation. T. R. was supported by the Chemical Computing Group, which provided a research license to ETH for the use of MOE. M. M. M. is a Howard Hughes Medical Institute International Scholar. M. P. is a holder of a Ciência 2007 position of the Portuguese Ministry of Science.
Financial support.
This work was supported by Fundação para a Ciência e Tecnologia (grant PTDC/SAU-MII/099118/2008 to M P; grants PTDC/SAU-GMG/100313/2008 and HMSP-CT/SAU-ICT/10068/2009 to M M M), by the NGFN Transfer program of the German Ministry of Education and Research, and by the European Union's Framework Programme 7 (grant 242095 to M M M). This work was developed under the scope of EVIMalaR (EC-FP7).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Yuan J, Cheng KC, Johnson RL, et al. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science. 2011;333:724–9. doi: 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chong CR, Chen X, Shi L, Liu JO, Sullivan DJ., Jr A clinical drug library screen identifies astemizole as an antimalarial agent. Nat Chem Biol. 2006;2:415–16. doi: 10.1038/nchembio806. [DOI] [PubMed] [Google Scholar]
- 3.Guiguemde WA, Shelat AA, Bouck D, et al. Chemical genetics of Plasmodium falciparum. Nature. 2010;465:311–15. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamo FJ, Sanz LM, Vidal J, et al. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–10. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 5.Rush MA, Baniecki ML, Mazitschek R, et al. Colorimetric high-throughput screen for detection of heme crystallization inhibitors. Antimicrob Agents Chemother. 2009;53:2564–8. doi: 10.1128/AAC.01466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchholz K, Burke TA, Williamson KC, Wiegand RC, Wirth DF, Marti M. A high-throughput screen targeting malaria transmission stages opens new avenues for drug development. J Infect Dis. 2011;203:1445–53. doi: 10.1093/infdis/jir037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prudencio M, Rodriguez A, Mota MM. The silent path to thousands of merozoites: the Plasmodium liver stage. Nat Rev Microbiol. 2006;4:849–56. doi: 10.1038/nrmicro1529. [DOI] [PubMed] [Google Scholar]
- 8.Mazier D, Renia L, Snounou G. A pre-emptive strike against malaria's stealthy hepatic forms. Nat Rev Drug Discov. 2009;8:854–64. doi: 10.1038/nrd2960. [DOI] [PubMed] [Google Scholar]
- 9.Wells TN, Burrows JN, Baird JK. Targeting the hypnozoite reservoir of Plasmodium vivax: the hidden obstacle to malaria elimination. Trends Parasitol. 2010;26:145–51. doi: 10.1016/j.pt.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Vale N, Moreira R, Gomes P. Primaquine revisited six decades after its discovery. Eur J Med Chem. 2009;44:937–53. doi: 10.1016/j.ejmech.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Davidson DE, Jr, Ager AL, Brown JL, Chapple FE, Whitmire RE, Rossan RN. New tissue schizontocidal antimalarial drugs. Bull World Health Organ. 1981;59:463–79. [PMC free article] [PubMed] [Google Scholar]
- 12.Prudencio M, Mota MM, Mendes AM. A toolbox to study liver stage malaria. Trends Parasitol. 2011;27:565–574. doi: 10.1016/j.pt.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Franke-Fayard B, Trueman H, Ramesar J, et al. A Plasmodium berghei reference line that constitutively expresses GFP at a high level throughout the complete life cycle. Mol Biochem Parasitol. 2004;137:23–33. doi: 10.1016/j.molbiopara.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Ploemen IH, Prudencio M, Douradinha BG, et al. Visualisation and quantitative analysis of the rodent malaria liver stage by real time imaging. PLoS One. 2009;4:e7881. doi: 10.1371/journal.pone.0007881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prudencio M, Rodrigues CD, Ataide R, Mota MM. Dissecting in vitro host cell infection by Plasmodium sporozoites using flow cytometry. Cell Microbiol. 2008;10:218–24. doi: 10.1111/j.1462-5822.2007.01032.x. [DOI] [PubMed] [Google Scholar]
- 16.Solmaz SR, Hunte C. Structure of complex III with bound cytochrome c in reduced state and definition of a minimal core interface for electron transfer. J Biol Chem. 2008;283:17542–9. doi: 10.1074/jbc.M710126200. [DOI] [PubMed] [Google Scholar]
- 17.Bower MJ, Cohen FE, Dunbrack RL., Jr Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J Mol Biol. 1997;267:1268–82. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- 18.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–48. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 19.Prudencio M, Rodrigues CD, Hannus M, et al. Kinome-wide RNAi screen implicates at least 5 host hepatocyte kinases in Plasmodium sporozoite infection. PLoS Pathog. 2008;4:e1000201. doi: 10.1371/journal.ppat.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodrigues CD, Hannus M, Prudencio M, et al. Host scavenger receptor SR-BI plays a dual role in the establishment of malaria parasite liver infection. Cell Host Microbe. 2008;4:271–82. doi: 10.1016/j.chom.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 21.Barton V, Fisher N, Biagini GA, Ward SA, O'Neill PM. Inhibiting Plasmodium cytochrome bc1: a complex issue. Curr Opin Chem Biol. 2010;14:440–6. doi: 10.1016/j.cbpa.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Fry M, Pudney M. Site of action of the antimalarial hydroxynaphthoquinone, 2-[trans-4-(4′-chlorophenyl) cyclohexyl]-3-hydroxy-1,4-naphthoquinone (566C80) Biochem Pharmacol. 1992;43:1545–53. doi: 10.1016/0006-2952(92)90213-3. [DOI] [PubMed] [Google Scholar]
- 23.Painter HJ, Morrisey JM, Mather MW, Vaidya AB. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature. 2007;446:88–91. doi: 10.1038/nature05572. [DOI] [PubMed] [Google Scholar]
- 24.Klonis N, Crespo-Ortiz MP, Bottova I, et al. Artemisinin activity against Plasmodium falciparum requires hemoglobin uptake and digestion. Proc Natl Acad Sci U S A. 2011;108:11405–10. doi: 10.1073/pnas.1104063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Looareesuwan S, Viravan C, Webster HK, Kyle DE, Hutchinson DB, Canfield CJ. Clinical studies of atovaquone, alone or in combination with other antimalarial drugs, for treatment of acute uncomplicated malaria in Thailand. Am J Trop Med Hyg. 1996;54:62–6. doi: 10.4269/ajtmh.1996.54.62. [DOI] [PubMed] [Google Scholar]
- 26.Kessl JJ, Ha KH, Merritt AK, et al. Cytochrome b mutations that modify the ubiquinol-binding pocket of the cytochrome bc1 complex and confer anti-malarial drug resistance in Saccharomyces cerevisiae. J Biol Chem. 2005;280:17142–8. doi: 10.1074/jbc.M500388200. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigues T, Guedes RC, dos Santos DJ, et al. Design, synthesis and structure-activity relationships of (1H-pyridin-4-ylidene)amines as potential antimalarials. Bioorg Med Chem Lett. 2009;19:3476–80. doi: 10.1016/j.bmcl.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Kessl JJ, Lange BB, Merbitz-Zahradnik T, et al. Molecular basis for atovaquone binding to the cytochrome bc1 complex. J Biol Chem. 2003;278:31312–18. doi: 10.1074/jbc.M304042200. [DOI] [PubMed] [Google Scholar]
- 29.Nam TG, McNamara CW, Bopp S, et al. A chemical genomic analysis of decoquinate, a Plasmodium falciparum cytochrome b inhibitor. ACS Chem Biol. 2011 doi: 10.1021/cb200105d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorne JL, Fernandez-Cruz ML, Bertelsen U, et al. Risk assessment of coccidostatics during feed cross-contamination: animal and human health aspects. Toxicol Appl Pharmacol. 2011 doi: 10.1016/j.taap.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Davies CS, Pudney M, Nicholas JC, Sinden RE. The novel hydroxynaphthoquinone 566C80 inhibits the development of liver stages of Plasmodium berghei cultured in vitro. Parasitology. 1993;106:1–6. doi: 10.1017/s0031182000074746. [DOI] [PubMed] [Google Scholar]
- 32.Gwadz RW, Koontz LC, Miller LH, Davidson DE., Jr Plasmodium gallinaceum: avian screen for drugs with radical curative properties. Exp Parasitol. 1983;55:188–96. doi: 10.1016/0014-4894(83)90013-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.