Abstract
Hematopoietic stem cells (HSCs) are responsible for sustaining hematopoietic homeostasis and regeneration after injury for the entire lifespan of an organism. Maintenance of genomic stability is crucial for the preservation of HSCs, which depends on their efficient repair of DNA damage, particularly DNA double strand breaks (DSBs). Because of the paucity of HSCs and lack of sensitive assays, directly measuring the ability of HSCs to repair DSBs has been difficult. Therefore, we developed a sensitive and quantitative cell free in vitro non-homologous end joining (NHEJ) assay using linearized plasmids as the substrates and quantitative polymerase chain reaction (qPCR) technique. This assay can sensitively detect DSB repair via NHEJ in less than 1 µg 293T cell nuclear proteins or nuclear extracts from about 5,000 to 10,000 human BM CD34+ hematopoietic cells. Using this assay, we confirmed that human bone marrow HSCs (CD34+CD38− cells) are less proficient in the repair of DSBs by NHEJ than HPCs (CD34+CD38+ cells). In contrast, mouse quiescent HSCs (Pyronin-Ylow LKS+ cells) and cycling HSCs (Pyronin-Yhi LKS+ cells) repaired the damage more efficiently than HPCs (LKS− cells). The difference in the abilities of human and mouse HSCs and HPCs to repair DSBs through NHEJ is likely attributed to their differential expression of key NHEJ DNA damage repair genes such as LIG4. These findings suggest that the qPCR-based cell free in vitro NHEJ assay can be used to sensitively measure the ability of human and mouse HSCs to repair DSBs.
Introduction
Hematopoietic stem cells (HSCs) are responsible for sustaining hematopoietic homeostasis and regeneration after injury for the entire lifespan of an organism [1]. Maintenance of genomic stability has been shown to be crucial for the preservation of HSCs, because mice that are deficient in the expression of various DNA damage repair genes exhibit premature exhaustion of HSCs [2], [3]. In addition, HSCs are long living cells that represent ideal cellular targets for acquisition of multiple mutations, leading to cell transformation and leukemia/lymphoma. Therefore, deficiency in the repair of DNA damage by HSCs not only leads to premature HSC exhaustion and bone marrow (BM) failure but also causes cancer and leukemia predisposition as shown in several animal models [1]. For example, mutational inactivation of the “ataxia telangiectasia mutated (ATM)” gene in mice causes progressive BM failure and a high incidence of lymphoma [4], [5]. Similarly, it was reported that mice with a Rad50 hypomorphic mutation (Rad50S/S mice) exhibited progressive HSC failure and cancer predisposition [6]. In addition, HSCs from LIG4 (LIG4Y288C) hypomorphic mutated and KU80 −/− mice exhibited a dramatic decrease in their ability to self-renew and reconstitute hematopoiesis after transplantation [7], [8].
DNA double-strand breaks (DSBs) are considered as the most detrimental genotoxic DNA lesions, which can arise endogenously from an increase in ROS production and collapse of replication forks or exogenously from exposure to various DNA damaging agents, including ionizing radiation (IR) [9]. Failure to rejoin DSBs can lead to gene deletion and even cell death, whereas the misrepair of DSBs may give rise to chromosome translocation and cell transformation. As such, cells have evolved two major repair pathways to repair DSBs: non-homologous end joining (NHEJ) [10] and homologous recombination (HR) [11]. NHEJ repairs DSBs by re-ligating two DNA broken ends, which is often imprecise and thus error prone because the DNA ends frequently require modification before they can be rejoined, leading to deletions or insertions at the break site [10]. In contrast, HR uses sister chromatids as templates to faithfully repair the damage [11]. Since sister chromatids are only available when cells are in the late S and G2 phases, only proliferating cells are able to repair DSBs through this high fidelity repair pathway.
However, whether HSCs can efficiently repair DSBs remain controversial. For example, Milyavsk et al. recently showed that HSCs from human cord blood repaired ionizing radiation (IR)-induced DSBs much slower than hematopoietic progenitor cells (HPCs) [12]. In contrast, at the same time Mohrin et al. reported that quiescent HSCs from adult mouse BM repaired IR-induced DSBs more efficiently than proliferating HPCs primarily via the NHEJ pathway [13]. Several possibilities have been raised to explan the difference in the abilities of human and mouse HSCs to repair DSBs [14], [15]. However, the lacking of an assay that can be used to directly and sensitively measure NHEJ activities in the rare population of stem cells such as HSCs has been a big challenge for studying the difference of human and mouse HSC NHEJ activities. Therefore, we developed a qPCR-based cell free in vitro NHEJ assay to meet the need. We found that this assay could sensitively detect DSB repair via NHEJ in less than 1 µg 293T cell nuclear proteins or nuclear extracts from about 5,000 to 10,000 human BM CD34+ hematopoietic cells. Using this assay, we confirmed that human HSCs (CD34+CD38− cells) are less proficient in the repair of DSBs by NHEJ than HPCs (CD34+CD38+ cells). In contrast, mouse quiescent HSCs (Pyronin-Ylow lin−c-kit+sca1+ cells or PYlowLKS+ cells) and cycling HSCs (Pyronin-Yhi lin−c-kit+sca1+ cells or PYhiLKS+ cells) repaired the damage more efficiently than cycling HPCs (lin−c-kit+sca1− cells or LKS− cells). The difference in the abilities of human and mouse HSCs and HPCs to repair DSBs through NHEJ is likely attributed to their differential expression of key DNA damage repair genes such as LIG4. These findings suggest that the qPCR-based cell free in vitro NHEJ assay can be used to sensitively measure the ability of HSCs to repair DSBs. This assay can also be applied to study DSB repair in other populations of rare tissue stem cells. In addition, it could help us to gain more insights into the mechanisms by which HSCs and tissue stem cells maintain their genomic stability.
Materials and Methods
Reagents and cells
pDsRed2ER plasmid was ordered from Clontech (Mountain View, CA). BglII and SmaI restriction enzymes were purchased from New England Biolabs (Ipswich, MA). Alexa Fluor® 647-conjugated anti-human CD34 antibody, PE-conjugated anti-human CD38 antibody, Alexa Fluor® 488-conjugated anti-mouse Sca-1, APC-conjugated anti-c-Kit were purchased from BioLegend (San Diego, CA). 293T cells were ordered from ATCC (Manassas, VA). Human BM CD34+ hematopoietic cells were purchased from Lonza (Walkersville, MD). Immortalized wild type (WT), DNA-PKcs −/−, Ku70 −/−, and Ku80 −/− mouse fibroblast cell lines were cultured and purified DNA-PKcs from human HeLa cells were prepared as previously described [16], [17].
Animals
Male C57BL/6-Ly-5.2 mice and PRKDC (DNA-PKcs) null SCID mice were purchased from Jackson Lab (Bar Harbor, MA) and housed at University of Arkansas for Medical Sciences (UAMS) AAALAC certified animal facilities. They received food and water ad libitum. All mice were used at approximately 8–12 weeks of age. The Institutional Animal Care and Use Committees of UAMS approved all experimental procedures used in this study (approval number: AUP3057 & 3093).
Preparation of nuclear protein extracts
Cells were seeded into 100 mm tissue culture dishes at a density of 5×106 per dish. They allowed to growth overnight in Dulbecco's modified Eagle's minimum (DMEM) supplemented with 10% fetal bovine serum (Alanta Biologicals, Lawrenceville, GA), 100 U/ml penicillin and 100 µg/ml streptomycin in a humidified incubator (95% air/5% CO2) at 37°C to reach about 80% confluence. After washed once with 10 ml PBS, they were harvested in 1 ml of PBS containing 2% FBS using a rubber scraper. The cells were transferred to a 1.5 ml microcentrifuge tube and centrifuged at 2000× g for 5 min. The cell pellet was resuspended in 300 µl buffer I containing 10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 500 µM PMSF, 1 mM DTT and protease inhibitor mixture (Cat# p-8340, Sigma, St. Louis, MO) and incubated on ice for 15 min. Six µl of 10% Nonidet P-40 were added to the cell lysates and mixed by vortex for 5 sec. Nuclei were isolated by centrifugation of the lysates at 6,000× g for 5 min. The supernatant (containing cytoplasmic proteins) was transferred to another chilled tube and the nuclear pellet was resuspended in 50 µl of buffer II containing 20 mM HEPES, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA and 25% v/v Glycerol. After 40-min incubation, the nuclear extracts were collected by centrifugation at 13,000× g for 10 min at 4°C. Nuclear proteins were also extracted from different numbers of human and mouse HSCs and HPCs in a similar manner as described above but with less lysis and extraction buffers. The protein concentration of the extracts was determined by a Bradford assay kit (Bio-Rad, Hercules, CA) according to the manufacturer's instruction. The nuclear extracts were immediately used or stored at −80°C.
Standard cell free in vitro NHEJ activity assay
Linearized pDsRed2ER plasmid substrates with either cohesive or blunt ends were generated by the digestion with BglII or SmaI and purified using Qiagen gel extraction kit (Valencia, CA) after separation on 0.7% agarose gels by electrophoresis to remove residual uncut circular plasmids (Fig. 1A & B). An optimized concentration (150 ng) of the purified linearized pDsRed2ER plasmid substrates was incubated with various concentrations of nuclear protein extracts in the end joining reaction buffer (1 mM ATP, 0.25 mM dNTP, 25 mM Tris acetate, 100 mM potassium acetate, 10 mM magnesium acetate, 1 mM DTT at pH 7.5) in a 30-µL reaction volume for 1 h at 37°C. The reaction mixture was deproteinized with 1 mg/ml proteinase K at 65°C for 30 min and then was separated by electrophoresis on a 0.7% agarose gel for 3 h at 90 V. DNA was detected using a gel imaging system (Alpha Innotech Corp. Santa Clara, CA) after ethidium bromide staining.
Figure 1. Linearized pDsRed2ER plasmids with cohesive ends are better substrates for the cell free in vitro NHEJ assay than those with blunt ends.
A. A diagram to illustrate the strategy for how to prepare linearized pDsRed2ER plasmid substrates with cohesive and blunt ends by enzymatic digestion with Bgl II and Sma I, respectively. B. Confirmation of the purity of the linearized pDsRed2ER plasmid substrates by agarose gel electrophoresis. A representative agarose gel electrophoresis image is shown. C. Representative agarose gel electrophoresis images to illustrate the formation of various NHEJ products after the Bgl II (the upper panel)- and Sma I (the lower panel)-digested linearized pDsRed2ER plasmid substrates were incubated with increasing concentrations (0.25 µg to 8.0 µg) of 293T cell nuclear protein extracts. D. A representative agarose gel electrophoresis image to illustrate the formation of ligated NHEJ products after the Sma I-digested linearized pDsRed2ER plasmid substrates were incubated with higher concentrations (15 µg to 120.0 µg) of 293T cell nuclear protein extracts.
qPCR-based cell free in vitro NHEJ activity assay
Linearized pDsRed2ER plasmid substrates (150 ng) were incubated with nuclear protein extracts as described above. The resulting ligated DNAs were diluted with 10 mM pH 8.5 Tris buffer (1∶10,000). One µL of the diluted DNAs was amplified using three pairs of primers, e.g. Ds-F1 and Ds-R2, Ds-F1 and Ds-R3, and Ds-F2 and Ds-R2 designed for the head-to-tail (H-T), head-to-head (H-H), and tail-to-tail (T-T) ligated DNAs, respectively, by qPCR as described below (Fig. 2B). Amplifications from Ds-F1 and Ds-R1 primers were used as an internal control. All primers' sequences were listed in Table 1. For each sample, SYBR Green real-time qPCR was conducted in duplicate or triplicate in 96-well plates using an ABI StepOne plus real time PCR System (Applied Biosystems, Foster City, CA). The 20 µL PCR reaction was prepared as follow: 10 µL of 2× SYBR Green PCR master mix, 0.4 µL of 10 µM of appropriate forward and reverse primers, 7.6 µL RNase-free water, and 2 µL DNA template. A negative control (digested plasmid DNA without nuclear protein) and a positive control (undigested plasmid DNA) were also included for each qPCR reaction. The qPCR was performed with 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. A dissociation melting curve was generated using thermal conditions from 60°C to 95°C. An arbitrary unit was calculated by the comparative CT method according to the CT values of the internal control as described previously [18].
Figure 2. The formation of the NHEJ products depends on DNA-PKcs, Ku70 and Ku80.
A–B. Nuclear protein extracts from 293T cells and DNA-PKcs null SCID mouse BM cells (SCID) (A) and those from immortalized wild type (WT), DNA-PKcs −/−, Ku70 −/−, and Ku80 −/− mouse fibroblast cell lines (B) were incubated with the Bgl II-digested linearized pDsRed2ER plasmid substrates. C. Nuclear protein extracts from immortalized wild type (WT) or DNA-PKcs −/− were complemented with 0.24 ng DNA-PKcs purified from HeLa cells and then incubated with the Bgl II-digested linearized pDsRed2ER plasmid substrates. The formation of various NHEJ products was analyzed by agarose gel electrophoresis. A representative agarose gel electrophoresis image is shown. CTL means no nuclear proteins added to the assay.
Table 1. Sequences for the primer used in the in vitro qPCR NHEJ assay.
| Bgl II generated linear plasmids | Forward sequences | Reverse sequences |
| Ds-F1 | 5-TCTCGAGCTCAAGCTTCGAATTCTGCAGTC-3 | |
| Ds-F2 | 5-GGCGGCCACTACCTGGTGGAGTTCAAGTC-3 | |
| Ds-F3 | 5-TGTACGATCTCAGGAACAGGTGGTGGCG-3 | |
| Ds-R1 | 5-TACAGCTCGTCCTTCTTGTACTAGCTCAG-3 | |
| Ds-R2 | 5-TGGCTGATTATGATCAGTTATCTAGATC-3 | |
| Ds-R3 | 5-GAGGGCCGCCACCACCTGTTCCTGAGATC-3 |
Western blot analysis
Total cell lysates and cytoplasmic and nuclear protein extracts were separated on a 12% SDS–PAGE and transferred onto a nitrocellulose membrane. Immunoblotting was conducted using 1∶1000 anti-Lamin A/C primary antibody (Cat#2032, cell signaling, Danvers, MA) and 1∶10,000 goat anti-rabbit secondary antibody (Jackson ImmunoResearch Labs., West Grove, PA). Immunocomplexes were detected using Amersham ECL Western Blotting Detection Reagents (GE Healthcare, Piscataway, NJ) and exposed to ECL Plus film. After detection, the membrane was stripped and reprobed with anti-β-actin antibody (Cat# SC-1616, Santa Cruz Biotechnology, Santa Cruz, CA) to control for cytoplasmic protein contamination.
Flow cytometry and cell sorting
Human BM CD34+ hematopoietic cells were stained with Alexa Fluor® 647-conjugated anti-CD34 and PE-conjugated anti-CD38 antibodies. Dead cells were excluded by staining with Propidium Iodide (PI). Human CD34+CD38− HSCs and CD34+CD38+ HPCs were sorted using an Aria II cell sorter. To isolate mouse HPCs (LKS− cells), cycling HSCs (PYhiLKS+ cells) and quiescent HSCs (PYlowLKS+ cells), mouse BM Lin− cells were prepared as previously described [18]. They were blocked with anti-CD16/32 antibody and then stained with Alexa Fluor® 488-conjugated anti–Sca1 antibody and APC-conjugated anti–c-Kit antibody followed by incubation with PY (1 µg/ml) for 15 min at 37°C in HBSS medium containing 10% FCS, 10 mM HEPES, 1 mg/ml glucose, and 50 µg/ml Verapamil. HPCs, PYhiLKS+ and PYlowLKS+ cells were sorted immediately using an Aria II cell sorter [19].
Quantitative reverse transcriptase PCR (qRT-PCR)
Total RNA was isolated using the Qiagen RNeasy Mini Kit (Valencia, CA) according to the manufacturers' instructions. RNA yield and quality were determined by measuring absorbencies at 260 nm and 280 nm, respectively. First-strand cDNA was synthesized in a final volume of 20 µl using the Superscript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). Quantitative PCR (qPCR) analyses were performed using a SYBR Green mix on an ABI StepOne Plus Real-Time PCR System (Applied Biosystems, Foster City, CA). Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and mouse hypoxanthinephophoribosyltransferase (HPRT) transcripts were used as housekeeping internal references for human and mouse mRNA, respectively. The expression of various DSB repair genes were calculated by the comparative CT method as described previously [18]. The sequences for all the primers used in the qRT-PCR assays are listed in Table 2.
Table 2. Primer sequences used for real time RT-PCR.
| Genes | Forward sequences | Reverse sequences |
| human ATM | 5-GATCTTGTGCCTTGGCTACAGATTG-3 | 5-ATGTCGCTGTTGGGGTAGAAGCTGAG-3 |
| human DNA-PKcs | 5-GGTGAACTTAAGACCCAGATGACATC-3 | 5-TCCCTTGAAGTCTGGGGATCTTCTTC-3 |
| human KU70 | 5-GTGATCTCCGAGATACAGGCATCTTC-3 | 5-TCCTCAAAGTGAACCCTGAGGTCCTC-3 |
| human KU80 | 5-TTCTCAACAGGCTGACTTCCTGGATG-3 | 5-TGAGGTCAGTGAATATTTCAATATGC-3 |
| human Ligase4 | 5-TCTTCAACTTATAACTCAGAGTTCAG-3 | 5-TGTAGTGACATTATGCAACTCAGCAG-3 |
| human XRCC4 | 5- CCTGAAAGATGTCTCATTCAGACTTG-3 | 5- TGAACATCATTCCAATCTCTCAGAAG-3 |
| human GAPDH | 5-CGGAGTCAACGGATTTGGTCGTAT-3 | 5-AGCCTTCTCCATGGTGGTGAAGAC-3 |
| mouse ATM | 5-TGTGTATACATACCAAGCATACGC-3 | 5-CTGGATGTACTCCGTGATCCAATG-3 |
| mouse DNA-PKcs | 5-GAATTGCATTAGTGCTGTGGTGCAC-3 | 5-GTACGTAGGTACTTTCCACCTGC-3 |
| mouse KU70 | 5- AAGATTTGGACAACCCAGGCGCTAAG -3 | 5-AGCTGGACGTCGCTGAAGAGGTTGGC-3 |
| mouse KU80 | 5-GAGGACACTATTCAAGGGTACCG-3 | 5-GCAACAGCTGCCGCCTCATCATC-3 |
| mouse Ligase4 | 5-GAGTTGCACAACGTCACCACAGATC-3 | 5-CATGTCCTTCTCCACACGCTCCAC-3 |
| mouse XRCC4 | 5-AGTCCAGCAGCTGGAGGAGAGTAC-3 | 5-TCATCCTTATGAAGAGTTTCTGCAGC-3 |
| mouse HPRT | 5-AGCAGTACAGCCCCAAAATGGTTA-3 | 5-TCAAGGGCATATCCAACAACAAAC-3 |
Statistical analysis
The data were analyzed by analysis of variance (ANOVA). In the event that ANOVA justified post hoc comparisons between group means, these were conducted using the Student-Newman-Keuls test for multiple comparisons. For experiments in which only single experimental and control groups were used, group differences were examined by unpaired Student t test. Differences were considered significant at P<0.05. All of these analyses were done using GraphPad Prism from GraphPad Software (San Diego, CA).
Results
Linearized plasmids with cohesive ends are better substrates for the cell free in vitro NHEJ assay than those with blunt ends
Linearized pDsRed2ER plasmid substrates with cohesive and blunt ends were generated by the digestion with BglII and SmaI, respectively, and then purified as shown in Fig. 1A & B. After purification, they were incubated with different concentrations of 293T cell nuclear protein extracts. The formation of circular and linear dimeric, trimeric, and multimeric NHEJ DNA products was analyzed by gel electrophoresis. It was found that the circular NHEJ DNA products were barely detectable with both substrates under the experimental conditions. The predominant linear NHEJ DNA products detected were dimers, followed by trimers and then multimers, for the Bgl II-digested substrates (upper panel of Fig. 1C). The amounts of these products formed were dose-dependent to the concentrations of nuclear proteins added to the assay. However, no NHEJ DNA products were formed until the SmaI-digested substrates were incubated with more than 15 µg 293T cell nuclear proteins (lower panel of Fig. 1C and 1D). In addition, only the dimeric NHEJ DNA products were produced for the SmaI-digested substrates. These findings suggest that DNAs with cohesive ends can be repaired more efficiently than those with blunt ends in the cell free in vitro NHEJ assay. In addition, as it would be expected that the formation of the NHEJ DNA products requires Ku70, K80 and DNA-PKcs [10], the nuclear proteins from DNA-PKcs null SCID mouse BM cells and these from immortalized DNA-PKcs −/−, Ku70 −/−, and Ku80 −/− mouse fibroblast cells failed to generate any of these NHEJ DNA products after they were incubated with the Bgl II-digested substrates (Fig. 2A and B). Complementation of DNA-PKcs −/− cell nuclear proteins with DNA-PKcs purified from HeLa cells produced NHEJ ligated DNA (Fig. 2C). Therefore, these results confirm that the ligated DNA products were generated through NHEJ.
Detection and quantification of NHEJ DNA products by qPCR
As shown in Fig. 3A, linearized pDsRed2ER plasmid substrates can be ligated by NHEJ in three different orientations, e.g. head-to-tail (H-T or product a), tail-to-tail (T-T or product b), and head-to-head (H-H or product c). Each of them can be specifically detected by the pairs of primers Ds-F1 and Ds-R2, Ds-F1 and Ds-R3, and Ds-F2 and Ds-R2, respectively. In addition, all ligated and non-ligated pDsRed2ER plasmid DNAs can be amplified by the Ds-F1 and Ds-R1 primers, which was included in every qPCR reaction as an internal control for normalization. Data presented in Fig. 3B are an example of the qPCR reactions. An arbitrary unit was calculated for each NHEJ DNA product after normalization with the internal control (Fig. 3C). The data showed that product a was the most abundant NHEJ products in the cell free in vitro NHEJ assay, followed by product b and then product c. In addition, the production of all these products was dose-dependent to the concentrations of the nuclear proteins used in the assay (Fig. 4A–D). Based on these findings, we selected product a for the qPCR assay to analyze NHEJ activities of nuclear proteins in the subsequent experiments and found that this assay is highly sensitive because it could measure NHEJ activities in 1 µg 293T cell nuclear proteins (Fig. 4) and nuclear extracted from as low as a few thousands of 293T cells (Fig. 5).
Figure 3. qPCR analysis of NHEJ activity.
A. A diagram to illustrate the strategy for how to amplify various NHEJ products by qPCR with their respective pairs of primers. H-T (product a), T-T (product b), and H-H (product c) represent different NHEJ products with the orientations of head-to-tail, tail-to-tail, and head-to-head ligations of the Bgl II-digested linearized pDsRed2ER plasmid substrates, respectively. B. An example of qPCR analysis of different NHEJ products after the Bgl II-digested linearized pDsRed2ER plasmid substrates were incubated with 8 µg of 293T cell nuclear protein extracts. Products a (H-T), b (T-T) and c (H-H) were amplified with the primers of Ds-F1 and Ds-R2, Ds-F1 and Ds-R3, and Ds-F2 and DS-R2, respectively. The internal control (IC) for all ligated and non-ligated pDsRed2ER plasmid DNAs was amplified by the primers of Ds-F1 and Ds-R1. C. The arbitrary units for all NHEJ products analyzed by qPCR are presented as mean ± SD (n = 3 independent assays). The arbitrary units were calculated by the comparative CT method (arbitrary units = 2[−ΔCT] while ΔCT = CT-Product−CT-IC). a, p<0.05 vs. product b; b, p<0.05 vs. product c.
Figure 4. Dose-dependent production of NHEJ products.
A. An example of qPCR analysis of product a after the Bgl II-digested linearized pDsRed2ER plasmid substrates were incubated with increasing concentrations of 293T cell nuclear protein extracts. A negative control (digested plasmid DNA without nuclear protein, NEG) and a positive control (undigested plasmid DNA, POS) were also included in the assay. B–D. The arbitrary units for NHEJ product a (B), b (C), c (D) were calculated as described in Figure 3 and presented as mean ± SD (n = 3). **p<0.01, ***p<0.001 vs. 0.25 µg proteins.
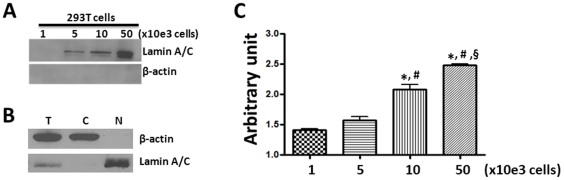
Figure 5. Sensitive detection of NHEJ activity in 293T cell nuclear protein extracts by the qPCR cell free in vitro NHEJ assay.
A. Nuclear protein extracts from 1,000–50,000 293T cells were immunoblotted with an antibody against Lamin A/C. β-actin blot is included as a control to detect cytoplasmic protein contamination. B. The purity of the nuclear protein extracts (N) was further confirmed by Western blot with antibodies against Lamin A/C and β-actin in comparison with total cell lysates (T) and cytoplasmic protein extracts (C). C. qPCR analysis of product a after the Bgl II-digested linearized pDsRed2ER plasmid substrates were incubated with the nuclear proteins extracted from increasing number of 293T cells. The data are presented as mean ± SD of arbitrary units (n = 3 independent assays). The symbols of *, # and § represent p<0.05 vs. nuclear proteins from 1,000, 5,000, and 10,000 cells, respectively.
Human and mouse HSCs and HPCs exhibit different proficiencies in the repair of DSBs
Recently, Milyavsky et al. used the γH2AX foci and neutral comet assays to compare the ability of human cord blood HSCs and HPCs to repair DSBs generated by ionizing radiation (IR) and found that human HPCs repaired DSBs more rapidly than HSCs [12]. However, the mechanisms underlying the difference between human HSCs and HPCs are not known. Therefore, we first examined if we can confirm the finding using our newly developed qPCR-based cell free in vitro NHEJ activity assay. As shown in Fig. 6A, this assay is very sensitive as it could measure the NHEJ activities in nuclear proteins extracted from a few thousands of human BM CD34+ hematopoietic cells. Nuclear protein extracts from the same number of human BM HPCs (CD34+CD38+ cells) generated significantly more NHEJ DNA products than those from human BM HSCs (CD34+CD38− cells) in the cell-free in vitro NHEJ assay (Fig. 6B & C), confirming that human HPCs indeed have a higher level of NHEJ activity than HSCs. This difference can be attributed at least in part to their differential expression of LIG4, as qRT-PCR analysis revealed that human HPCs expressed significantly higher levels of ligase 4 mRNA than HSCs (Fig. 6D)
Figure 6. Nuclear protein extracts from human BM HPCs have higher NHEJ activity than those from HSCs.
A. qPCR analysis of the NHEJ activity in nuclear proteins extracted from different numbers of human BM CD34+ cells. The data are presented as mean ± SD of arbitrary units (n = 3). The symbols of * and # represent p<0.05 vs. nuclear proteins from 1,000 and 5,000 cells, respectively. B. A representative gating strategy to sort human HSCs (CD34+CD38− cells) and HPCs (CD34+CD38+ cells) from human BM CD34+ cells. C. NHEJ activity in nuclear proteins extracted from the same number (6,600) of human HSCs and HPCs. The data are presented as mean ± SD of arbitrary units (n = 3 independent experiments). D. Analysis of the expression of ATM, DNA-PKcs, KU70, KU80, Ligase 4, and XRCC4 mRNA in human BM HSCs and HPCs by qRT-PCR. The data are presented as ratios between HPCs and HSCs (n = 3 independent experiments). **p<0.01 and ***p<0.001 vs. HSCs.
In contrast, Mohrin et al. recently reported that mouse BM HSCs, particularly those in quiescence, repaired IR-induced DSBs more efficiently via the NHEJ pathway than mouse BM HPCs using the γH2AX foci and alkaline comet assays [13]. These findings raise an intrigue question whether there is a species difference between human and mouse HSCs and HPCs in their ability to repair DSBs [14], [15]. Therefore, we also compared the NHEJ activities of nuclear protein extracts from mouse BM HPCs (LKS− cells), cycling HSCs (PYhiLSK+ cells) and quiescent HSCs (PYlowLSK+ cells) using our qPCR cell free in vitro NHEJ assay (Fig. 7A & B). As shown in Fig. 7B, it was found that the nuclear protein extracts from both cycling and quiescent mouse BM HSCs had a significantly higher level of NHEJ activity than those from mouse BM HPCs. This finding is in agreement with the observation that both cycling and quiescent mouse BM HSCs expressed significantly great levels of KU70, KU80, ligase 4 and XRCC4 mRNA than HPCs (Fig. 7C). In addition, using IR-induced DSB foci assays Mohrin et al. showed that quiescent mouse BM HSCs exhibited a higher NHEJ activity than the HSCs that were in cell cycle and proliferating after they were activated by cytokines or mobilized by cyclophosphamide/granulocyte colony-stimulating factor treatment [13]. This finding is in agreement with our observation that the nuclear protein extracts from quiescent mouse BM HSCs had about 2-fold greater NHEJ activity than those from cycling mouse BM HSCs (Fig. 7C). These results demonstrate that our qPCR cell free in vitro NHEJ assay can be used to sensitively and accurately measure NHEJ activity in both human and mouse HSCs.
Figure 7. Nuclear protein extracts from mouse BM HSCs have higher NHEJ activity than those from HPCs.
A. A representative gating strategy to sort HPCs (LKS− cells) and cycling (PYhiLKS+ cells) and quiescent HSCs (PYlowLKS+ cells) from mouse BM lineage negative cells (Lin− cells). B. NHEJ activity in nuclear proteins extracted from the same number (20,000) of mouse HPCs and cycling and quiescent HSCs. The data are presented as mean ± SD of arbitrary units (n = 3). C. Analysis of the expression of ATM, DNA-PKcs, KU70, KU80, Ligase 4, and XRCC4 mRNA in mouse BM HPCs and cycling and quiescent HSCs by qRT-PCR. The data are presented as ratios between HPCs and HSCs (n = 3 independent assays). The symbols of * and # represent p<0.01 vs. HPCs and PYhiLKS+ cells, respectively.
Discussion
Currently, the γH2AX foci assay is the most widely used method to analyze DSBs and DSB repair [20]. It is several orders of magnitude more sensitive than the neutral comet assay and pulsed-field gel electrophoresis (PFGE) [20]. Therefore, the γH2AX foci assay has been widely used to analyze DSB repair in HSCs [12], [13]. However, the γH2AX foci assay is an indirect assay for DSBs, because the appearance and disappearance of γH2AX foci depend on the phosphorylation and dephosphorylation cascades that sense DSBs and mediate DSB repair and lag behind the occurrence of the actual damage and repair [20]. In addition, the formation and resolution of γH2AX foci can be affected by many other factors [20]. Therefore, validation of the results from the γH2AX foci assay by a method that can direct measure DSBs is always warranted. However, this cannot be accomplished by the use of the neutral comet assay and pulsed-field gel electrophoresis (PFGE) for HSCs, because they can only reliably detect tens or hundreds of DSBs which rarely occur in HSCs under a physiological condition or after exposure to stress without killing the cells instantly [20]. A cell free in vitro NHEJ assay has the potential to fill the gap by directly measuring the ability of HSCs to repair DSBs. This is because the majority of adult HSCs are quiescent and quiescent HSCs primarily repair DSBs through the NHEJ pathway due to a lacking of sister chromatids as the substrates for HR [13].
Unfortunately, the conventional cell free in vitro NHEJ assay is insensitive and requires tens µg nuclear proteins or nuclear extracts from a large number of cells for the assay. This is very difficult to achieve for HSCs and other tissue stem cells considering the scarcity of these cells in the body. As an example, Sotiropoulou et al. recently reported to use 40 µg nuclear proteins from CD34+ hair follicle bulge stem cells and CD34− non-stem cells to perform the conventional cell free in vitro NHEJ assay in order to confirm that the stem cells have a higher DSB repair activity than CD34− non-stem cells [21]. It is likely that to obtain such a large quantify of nuclear proteins requires isolation of hundreds of thousands or even millions of CD34+ hair follicle bulge stem cells from a large number of mice. Therefore, developing a sensitive and quantitative cell free in vitro assay that can be used to measure NHEJ activity in a few thousands of cells is highly desirable for studying DSB repair function of HSCs.
Here, we report the development of a qPCR-based cell free in vitro NHEJ assay using linearized plasmids as the substrates and qPCR technique. We showed that the linearized pDsRed2ER plasmids with cohesive ends generated by BglII were better substrates for the cell free in vitro NHEJ assay than those with blunt ends produced by SmaI. This assay is highly specific for NEHJ, because the formation of the ligated DNA products in the cell free in vitro NHEJ assay depends on DNA-PKcs, Ku70 and Ku80, as the nuclear proteins from DNA-PKcs −/−, Ku70 −/− and Ku80 −/− cells failed to generate any of these NHEJ DNA products after they were incubated with the Bgl II-digested substrates. The NHEJ activity of the nuclear proteins extracted from DNA-PKcs −/− cells was restored after complementation of the nuclear extracts with DNA-PKcs proteins purified from HeLa cells. Interestingly, the most of the NHEJ DNA products generated in this assay were linear and had a head-to-tail (H-T) orientation. Therefore, we could acurately measure NHEJ activity in cell nuclear extracts by quantifying the abundance of the H-T products (product a) using qPCR. We showed that we could measure NHEJ activity in less than 1 µg 293T cell nuclear proteins with this qPCR-based cell free in vitro NHEJ assay. In addition, we could quantify NHEJ activity in nuclear extracts from a few thousands of human BM CD34+ hematopoietic cells using this assay. Therefore, this qPCR-based NHEJ assay is much more sensitive than the conventional cell free in vitro NHEJ assay.
More importantly, using this assay, we found that the nuclear protein extracts from human HSCs had a lower level of NHEJ activity than those from HPCs. In contrast, it was found that the nuclear extracts from both mouse cycling (PYhiLKS+ cells) and quiescent HSCs (PYlowLKS+ cells) had a higher level of NHEJ activity than those from cycling HPCs (LKS− cells). Because almost all HPCs and the cycling HSCs isolated from mouse BM by cell sorting are PYhi cells and in the cell cycle [19], the difference between mouse BM HSCs and HPCs cannot be simply ascribed to a difference in their cell cycle status. These findings confirm that human and mouse HSCs and HPCs have different abilities to repair DSBs via NHEJ as shown in the recent studies [12], [13]. The difference in the abilities of human and mouse HSCs and HPCs to repair DSBs through NHEJ is likely attributed to their differential expression of key NHEJ DNA damage repair genes, such as LIG4. This is because it was found that human HSCs expressed less mRNA for ligase 4 than human HPCs, while mouse HSCs expressed higher levels of ligase 4 mRNA than mouse HPCs. In addition, both cycling and quiescent mouse HSCs also expressed higher levels of KU70, KU80 and XRCC4 mRNA than mouse HPCs. The mechanisms underlying the differential expression of these DSB repair genes in human and mouse HSCs and HPCs are unknown at the present. Similarly, the biological significance of these differences has yet to be determined.
In summary, the findings presented in this manuscript suggest that the qPCR-based cell free in vitro NHEJ assay we developed can be used to sensitively measure the ability of HSCs to repair DSBs. This assay can also be applied to study DSB repair in other populations of rare tissue stem cells. In addition, it could help us to gain more insights into the mechanisms by which HSCs and tissue stem cells maintain their genomic stability.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding was provided by the National Institutes of Health (US) R01-CA086688, CA102558, and AI080421; the Winthrop Rockefeller Endowment for Leukemia Research; and the Arkansas Research Alliance Scholarship from the Arkansas Science & Technology Authority. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 2.Kenyon J, Gerson SL. The role of DNA damage repair in aging of adult stem cells. Nucleic Acids Res. 2007;35:7557–7565. doi: 10.1093/nar/gkm1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niedernhofer LJ. DNA repair is crucial for maintaining hematopoietic stem cell function. DNA Repair (Amst) 2008;7:523–529. doi: 10.1016/j.dnarep.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 5.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 6.Bender CF, Sikes ML, Sullivan R, Huye LE, Le Beau MM, et al. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev. 2002;16:2237–2251. doi: 10.1101/gad.1007902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 8.Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 9.Phillips ER, McKinnon PJ. DNA double-strand break repair and development. Oncogene. 2007;26:7799–7808. doi: 10.1038/sj.onc.1210877. [DOI] [PubMed] [Google Scholar]
- 10.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 11.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milyavsky M, Gan OI, Trottier M, Komosa M, Tabach O, et al. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell. 2010;7:186–197. doi: 10.1016/j.stem.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seita J, Rossi DJ, Weissman IL. Differential DNA damage response in stem and progenitor cells. Cell Stem Cell. 2010;7:145–147. doi: 10.1016/j.stem.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Lane AA, Scadden DT. Stem cells and DNA damage: persist or perish? Cell. 2010;142:360–362. doi: 10.1016/j.cell.2010.07.030. [DOI] [PubMed] [Google Scholar]
- 16.Tomimatsu N, Tahimic CG, Otsuki A, Burma S, Fukuhara A, et al. Ku70/80 modulates ATM and ATR signaling pathways in response to DNA double strand breaks. J Biol Chem. 2007;282:10138–10145. doi: 10.1074/jbc.M611880200. [DOI] [PubMed] [Google Scholar]
- 17.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Schulte BA, LaRue AC, Ogawa M, Zhou D. Total body irradiation selectively induces murine hematopoietic stem cell senescence. Blood. 2006;107:358–366. doi: 10.1182/blood-2005-04-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Passegue E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 21.Sotiropoulou PA, Candi A, Mascre G, De CS, Youssef KK, et al. Bcl-2 and accelerated DNA repair mediates resistance of hair follicle bulge stem cells to DNA-damage-induced cell death. Nat Cell Biol. 2010;12:572–582. doi: 10.1038/ncb2059. [DOI] [PubMed] [Google Scholar]