Abstract
Telomere length is regulated mostly by proteins directly associated with telomeres. However, genome-wide analysis of Saccharomyces cerevisiae mutants has revealed that deletion of Hpr1p, a component of the THO complex, also affects telomere length. The THO complex comprises four protein subunits, namely, Tho2p, Hpr1p, Mft1p, and Thp2p. These subunits interplay between transcription elongation and co-transcriptional assembly of export-competent mRNPs. Here we found that the deletion of tho2 or hpr1 caused telomere lengthening by ∼50–100 bps, whereas that of mft1 or thp2 did not affect telomere length. Since the THO complex functions in transcription elongation, we analyzed the expression of telomere-associated proteins in mutants depleted of complex components. We found that both the mRNA and protein levels of RIF1 were decreased in tho2 and hpr1 cells. RIF1 encodes a 1917-amino acid polypeptide that is involved in regulating telomere length and the formation of telomeric heterochromatin. Hpr1p and Tho2p appeared to affect telomeres through Rif1p, as increased Rif1p levels suppressed the telomere lengthening in tho2 and hpr1 cells. Moreover, yeast cells carrying rif1 tho2 or rif1 hpr1 double mutations showed telomere lengths and telomere silencing effects similar to those observed in the rif1 mutant. Thus, we conclude that mutations of components of the THO complex affect telomere functions by reducing the expression of a telomere-associated protein, Rif1p.
Introduction
Telomeres are the structure at the ends of eukaryotic linear chromosomes [1], [2]. They are essential for the maintenance of chromosome integrity, and protect natural DNA ends from being recognized as double-strand breaks. In most organisms, the telomeric DNA is composed of short, tandemly repeated sequences with a strand rich in guanine residues (G-strand) running 5′ to 3′ toward the end of the telomere. For example, the telomeric sequences in the brewer's yeast Saccharomyces cerevisiae are ∼250–300 base pair-long TG1–3/C1–3A repeats. Telomeres are maintained by telomerase in most eukaryotes [3]. Telomerase is a ribonucleoprotein containing a catalytic protein component TERT (telomerase reverse transcriptase) and an associated RNA moiety, TER, which serves as the template to extend telomeric DNA sequences. In S. cerevisiae, these two components are encoded by EST2 [4], [5] and TLC1 [6], respectively.
Telomere length homeostasis involves the coordination of telomere lengthening and shortening processes. Although the complete mechanism of telomere length regulation remains to be elucidated, controlling telomerase activity appears to be a major part of the process [7]. For example, the binding of telomerase Tlc1 RNA by yeast Ku proteins contributes to telomere length regulation [8]. The mutation of YKU70 or YKU80 causes shortening of telomere length [9]–[14]. It has also been reported that Pif1p helicase negatively regulates telomerase by removing telomerase from the telomeric DNA [15], [16]. As a consequence, mutation of PIF1 has been shown to lengthen telomeres [17]. These telomere-associated proteins affect telomere length by directly regulating the telomerase activity [7]. Regulation of telomerase could also be achieved through posttranslational modification of telomere-associated proteins. For example, phosphorylation of the telomere binding protein Cdc13p by DNA damage response kinases Tel1p/Mec1p is required for recruiting telomerase onto telomeres for telomere replication [18]. Short telomeres have been observed in cells missing either TEL1 or MEC1 [19]–[21].
Telomere length homeostasis can also be regulated by the repressor/activator protein 1, Rap1p. in S. cerevisiae [22]. It has been reported that several rap1 temperature-sensitive mutants display phenotypes consistent with the role of Rap1p as a negative regulator of telomere length [23]–[25]. Rap1p binds to a loosely defined recognition site within the double-stranded TG1–3 telomeric DNA tracts to affect telomere functions [23]–[25]. Regulation of telomere length by Rap1p appears to be mediated by Rif1p and Rif2p, which bind to the protein-interaction domain at the C-terminus of Rap1p. Deletion of the nonessential RIF1 and RIF2 genes results in extensive telomere elongation [26], [27]. Studies of Kluyveromyces lactis have further confirmed the role of Rap1p in regulating telomere length [28]–[30].
In addition to affecting telomerase and telomere-associated proteins, genes affecting telomere length have also been identified as being involved in diverse cellular functions [31]–[33]. Among them, deletion of HPR1 has been shown to increase telomere length by ∼50–150 bp. HPR1 is a component of the THO complex (a suppressor of the Transcriptional defects of Hpr1 mutants by Overexpression) that is involved in transcriptionelongation and the export of transcribed mRNAs [34], [35]. The THO complex is a conserved nuclear complex that is formed by four protein subunits: Hpr1p, Tho2p (Rlr1p), Mft1p, and Thp2p in S. cerevisiae. Mutations in THO components cause defects in transcription-dependent hyper-recombination and mRNA export [36]–[39]. Moreover, there is evidence that DNA∶RNA hybrids are formed co-transcriptionally, resulting in impaired transcription elongation of GC-rich and long DNA sequences and defective transcription-associated recombination [40]. However, it is unclear how mutations of THO components cause changes in telomere length. Here we found that mutations of THO components affect telomeres by reducing the expression of Rif1p. Our results reveal a novel mechanism by which telomere functions could be affected indirectly through perturbing the expression of a telomere-associated component.
Results
Mutations of THO complex components HPR1 and THO2 affect telomere length
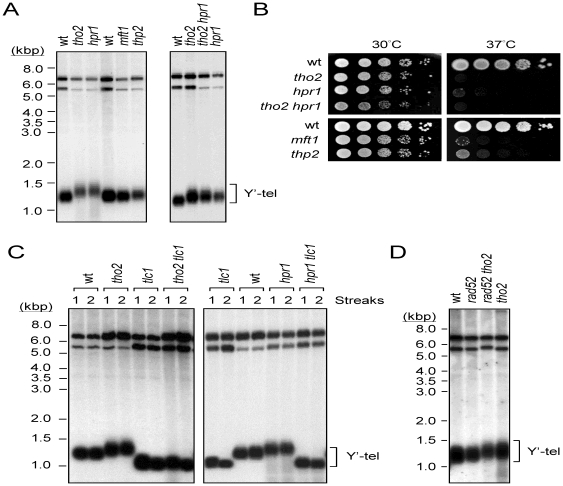
In a genome-wide screen, mutation of HPR1 was found to cause a ∼50–150 bp increase in telomere length [31]. Since Hpr1p is a component of the THO complex, we first tested whether the other three THO components also affect telomere length. In S. cerevisiae, the middle repetitive sequences known as the Y′ elements are found in the subtelomeric regions of most chromosomes [41]. In wild-type cells, XhoI digestion produces a ∼1.3 kbp fragment from the ends of Y′-bearing chromosomes that contains ∼870 bp of Y′ and the terminal tract of ∼350–430 bp of TG1–3/C1–3A DNA. However, the lengths of Y′-bearing telomeres in both hpr1 and tho2 cells appeared ∼75 bp longer than those in wild-type cells (Fig. 1A). This telomere length phenotype was not progressive, as increased cell divisions did not further affect its length (data not shown). Interestingly, mutations of the other two THO complex components, MFT1 and THP2, did not affect telomere length (Fig. 1A), although it has been reported that mutations of these two genes cause slightly shorter telomeres [31]. The reason for the apparent discrepancy is unclear. Telomere lengths of yeast cells harboring both hpr1 and tho2 mutations were also analyzed. As shown in Figure 1A (right panel), hpr1 tho2 double mutations did not further increase the telomere length. Thus, HPR1 and THO2 might affect telomere length through the same pathway. As a control, the temperature sensitivity of the resulting strains were analyzed since mutations of THO components also exhibit severe growth defects at 37°C [42], [43]. Indeed, all of the THO mutant cells showed the expected temperature-sensitive phenotype (Fig. 1B).
Figure 1. THO complex components Hpr1p and Tho2p affect telomere length.
(A) Telomere lengthening in hpr1 and tho2 strains. Yeast DNA was isolated from the YPH499 yeast strain with indicated mutations, digested with XhoI, separated on a 1% agarose gel and analyzed by Southern blotting. The blots were hybridized with probes prepared using the 600-bp sequence from the Y′ element 3′ end. The position of the Y′ telomere is indicated. (B) Temperature-sensitive growth of THO complex mutants. Aliquots of ten-fold serial dilutions of yeast cells were spotted on YEPD plates and incubated at 30 or 37°C for three days. (C) Telomerase is required for the telomere lengthening effects in hpr1 and tho2 cells. Yeast cells carrying the indicated mutations were freshly sporulated and grown on YEPD plates at 30°C after 1 or 2 restreaks. Total yeast DNA was then isolated and analyzed by Southern blotting. (D) Recombination is not required for telomere lengthening in tho2 cells. Total yeast DNA from the indicated strains was isolated and analyzed by Southern blotting.
To determine if the telomere lengthening in hpr1 and tho2 cells required the telomerase activity, deletion of telomerase RNA component TLC1 was introduced into hpr1 and tho2 cells, respectively. Yeast cells losing telomerase RNA undergo progressive telomere shortening which eventually leads to cell death [6]. As shown in Figure 1C, telomere lengths in hpr1 tlc1 and tho2 tlc1 cells were similar to those in tlc1 cells. The results indicate that the long telomeres observed in hpr1 and tho2 cells required functional telomerase. The effect of recombination on telomere lengthening of hpr1 and tho2 cells was also analyzed. A rad52 mutation was introduced into tho2 cells and telomere length was examined. As shown in Figure 1D, rad52 did not affect telomere length in either wild-type or tho2 cells. The results suggest that telomere lengthening observed in THO complex mutants is not due to telomere recombination.
RIF1 expression is reduced in hpr1 and tho2 cells
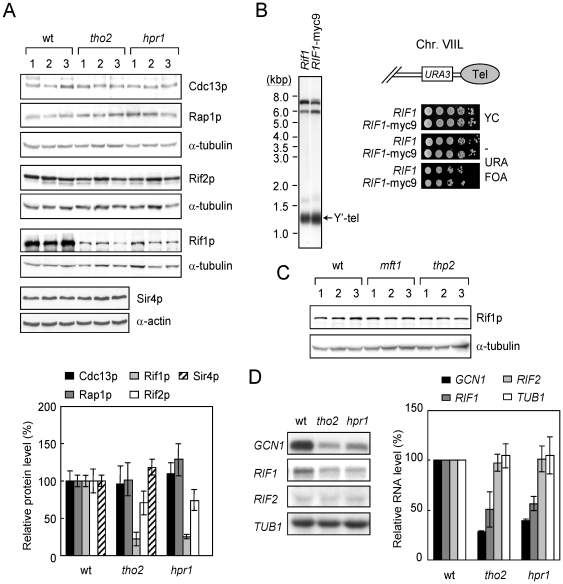
Next, we explored the mechanism by which HPR1 and THO2 affect telomere length. The THO complex is involved in transcription elongation. Hybrids of template DNA and transcribed RNA are formed during transcription in mutants of the THO complex components [40]. As a result, the transcription of long genes is preferentially repressed [39], [44]. We hypothesized that mutations of HPR1 and THO2 might affect telomere length by influencing the expression of genes that encode telomere-associated proteins. We first surveyed the length of telomere-associated proteins (Table 1). Among those we examined, RIF1 is the longest gene with a size of 5,751 bps that encodes a protein with 1,916 amino acid residues. To test if hpr1 and tho2 affect the expression of the long genes, we analyzed the Rif1p protein level in mutant cells. As shown in Figure 2A, Rif1p levels were decreased by ∼4 folds in both hpr1 and tho2 cells. In comparison, SIR4 (4,077 bps), CDC13 (2,775 bps), RAP1 (2,484 bps), and RIF2 (1,188 bps) were also analyzed. The results shown in Figure 2A indicate that hpr1 and tho2 did not affect the expression of SIR4, CDC13, or RAP1. The Rif2p level was only slightly decreased in these two mutants. Since a yeast strain with 9 repeats of Myc attached to the 3′ end of the chromosomal RIF1 gene was used in our analysis, the effect of Myc-tagging on Rif1p was also analyzed. The RIF1-myc9 tagged allele caused similar, if not identical, effects on telomere length and telomere silencing to those observed in cells with the wild-type allele, implying that the tagged Rif1p is expressed and functional (Fig. 2B).
Table 1. The sizes and G+C contents of genes encoding telomere-associated proteins.
| Gene name | ORF size | G+C content |
| RIF1 | 5751 | 36.24 |
| SIR4 | 4077 | 37.23 |
| SIR3 | 2937 | 36.50 |
| CDC13 | 2775 | 43.93 |
| EST2 | 2655 | 33.79 |
| RAP1 | 2484 | 41.51 |
| PIF1 | 2580 | 39.34 |
| EST1 | 2100 | 36.00 |
| YKU80 | 1890 | 39.52 |
| STN1 | 1485 | 36.97 |
| YKU70 | 1809 | 33.78 |
| SIR2 | 1689 | 39.67 |
| TLC1 | 1301 | 35.59 |
| RIF2 | 1188 | 34.68 |
| EST3 | 547 | 38.90 |
| TEN1 | 483 | 35.60 |
| TEL1 | 8364 | 35.80 |
| LacZ | 3069 | 56.21 |
Figure 2. tho2 and hpr1 selectively decrease the expression level of Rif1p.
(A) The Rif1p protein level is decreased in tho2 and hpr1 cells. Total yeast proteins from wild-type, tho2 or hpr1 yeast cells were precipitated with TCA, separated on an 8% SDS-polyacrylamide gel, and analyzed by immunoblotting using antibodies against Rap1p, Cdc13p, α-tubulin, α-actin, Myc (for myc9-tagged Rif1p and Rif2p), or TAP (for TAP-tagged Sir4p) (top panel). The percentages of protein expression relative to the wild-type cells are quantified (bottom panel). The bars were standard deviations determined using data from three different colonies of the indicated yeast strain. (B) The myc9-epitope tagging does not affect the effect of Rif1p on telomere length and telomere silencing. Telomere length of wild-type RIF1 or RIF1-myc9 cells were determined by Southern blotting using the Y′ element probe (left panel). Telomere silencing effects were determined in RIF1 or RIF1-myc9 cells. Yeast cells in 10-fold serial dilutions were spotted on YC, YC lacking uracil, or plates containing 5-FOA (right panel). (C) The Rif1p protein level is not affected in mft1 and thp2 cells. The Rif1p level is analyzed in mft1 and thp2 cells using immunoblotting analysis. (D) tho2 and hpr1 reduced the RIF1 RNA levels. The RNA transcripts of GCN1, RIF1, RIF2, and TUB1 were analyzed using Northern blotting assays (left panel). The expression levels of the indicated RNA transcripts were quantified and displayed as the percentages relative to the expression level in wild-type cells (right panel). The error bars were standard deviations calculated using data from three different colonies of the indicated yeast strain.
It has been reported that the expression levels of yeast genes increase with the G+C contents in wild-type and both hpr1 and tho2 cells [44], [45]. The G+C contents of the genes encoding telomere-associated proteins were also surveyed (Table 1). Among a total of 16 genes analyzed, all of them showed a G+C content within the range of 34–44%, similar to the ∼40% average G+C content of all yeast genes. As a comparison, the E. coli lacZ gene has a G+C content of ∼56%. According to our findings, the expression of telomere-associated genes in hpr1 and tho2 cells did not appear to be affected by their G+C content (Fig. 2A).
To test if the reduction of Rif1p protein is specific to hpr1 and tho2 mutations, Rif1p protein expression was also determined in mft1 and thp2 cells. We discovered that the Rif1p protein level remained the same in mft1 or thp2 cells (Fig. 2C), indicating that the reduction in Rip1p levels is specific to hpr1 and tho2 mutations. This result is also consistent with our observation that only hpr1 and tho2 but not mft1 or thp2 cells showed long telomeres (Fig. 1A).
To determine whether the reduced Rif1p level was due to transcription defects caused by hpr1 and tho2 mutations, the RNA level of RIF1 was analyzed. Total RNAs were prepared from yeast cells and hybridized with specific probes against GCN1 (8,019 bps), RIF1 (5,751 bps), RIF2 (1,188 bps), and TUB1 (1,460 bps) genes. As shown in Figure 2D, both hpr1 and tho2 mutations showed defects in transcribing the two long genes GCN1 and RIF1. The effect is specific, as the transcript levels of short genes RIF2 and TUB1 were not affected by the mutations. Thus, our results are in agreement with the previous findings which suggest that hpr1 and tho2 mutations affect the transcription of long genes. Consequently, both the protein and RNA levels of RIF1 were significantly reduced in these cells.
THO2 and HPR1 share the same pathway of RIF1 to regulate telomere length
Given the role of RIF1 as a negative regulator of telomere length and the finding that the deletion of RIF1 in yeast causes elongation of telomere length [27], it is likely that the long telomeres observed in hpr1 and tho2 cells were due to the reduction of RIF1 expression. This simple scenario predicts that the telomere lengths of rif1 hpr1 or rif1 tho2 cells should not be longer than that in the rif1 cells. Indeed, the telomere lengths of cells carrying rif1 hpr1 and rif1 tho2 mutations were similar to those in the rif1 cells (Fig. 3A). In comparison, we found that telomere lengths were further elongated when rif2 was introduced into hpr1 or tho2 cells (Fig. 3A). Similarly, combinatorial effects were also observed when pif1-m2 mutation was introduced into tho2 cells (Fig. 3A). These results show that Hpr1p and Tho2p might affect telomere length through the same pathway as Rif1p.
Figure 3. THO2 and HPR1 function in the same pathway as RIF1 to regulate telomere length.
(A) Yeast DNA from the indicated mutations was isolated and analyzed by Southern blotting assays using the Y′ element probe. (B) The decreased Tel1p level does not contribute to telomere lengthening in tho2 and hpr1 cells. Total mRNA from the indicated strains was isolated and analyzed for GCN1 and TEL1 RNA levels using real time RT-PCR (left panel). Results were presented as relative levels normalized to the wild-type expression level. The bars were standard deviations determined from three independent experiments. Yeast DNA from indicated strains was analyzed for telomere length (right panel). (C) Overexpressing Rif1p suppresses the telomere lengthening in tho2 and hpr1 cells. Wild-type, tho2, or hpr1 yeast cells carrying plasmids pRS426 or pRS426-RIF1 (o/e RIF1) were cultured at 30°C. Telomere lengths of these cells were then analyzed using Southern blotting assays (top panel). Immunoblotting analysis of the Rif1p level was also performed (bottom panel).
It is also interesting to note that TEL1 is a long gene (8,364 bps) and its expression level is decreased in hpr1 cells [44]. Using real time RT-PCR, we found that the amount of TEL1 RNA in hpr1 or tho2 cells was decreased to ∼10–15% of the wild-type level (Fig. 3B, left panel). In comparison, the RNA level of GCN1 was also decreased in hpr1 or tho2 cells, consistent with our previous observation (Fig. 2 and 3B). Although Tel1p is not directly associated with telomeres, it participates in telomerase recruiting and its mutation has been shown to shorten telomeres [19]. To evaluate the contribution of tel1 on telomere length maintenance in THO mutants, telomere length of tel1 tho2 cells was determined. As shown in Figure 3B (right panel), telomere length in both wild-type and tho2 mutant cells was decreased by the same extent after the introduction of a tel1 mutation. The results suggest that although TEL1 RNA level is also decreased in THO mutants, the observed telomere lengthening in hpr1 or tho2 cells is not due to the decreased Tel1p level.
Our model also predicts that increasing the Rif1p level should suppress the long telomere phenotype associated with hpr1 and tho2 mutations. Initial attempts to overexpress RIF1 using a cen plasmid did not yield a detectable elevation of Rif1p levels in hpr1 or tho2 cells. Accordingly, there was no change in telomere length in these cells (data not shown). We then increased Rif1p levels by introducing a high-copy number 2 μ plasmid carrying RIF1 into yeast cells. As shown in Figure 3C, increasing Rif1p reduced telomere length in wild-type cells. Long telomeres observed in hpr1 and tho2 cells were suppressed when Rif1p levels were brought back to the wild-type level. Significantly, there appeared to be a negative correlation between telomere lengths and the expression levels of Rif1p in the cells we examined. The results provide strong evidence supporting that the long telomeres observed in hpr1 and tho2 cells are caused by the reduction in the expression level of Rif1p.
Enhancement of telomere silencing in hpr1 and tho2 cells
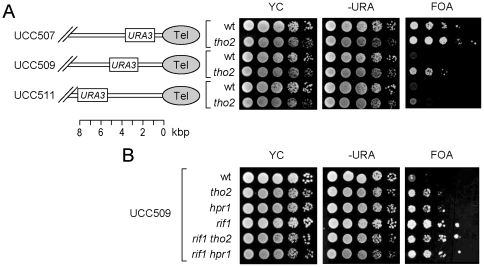
The function of RIF1 on telomeres is not limited to telomere length regulation. It has been reported that rif1 also enhances the telomere silencing effect [26], [46]. To test if mutations of THO components affect telomere silencing, expression of URA3 at varying distances from telomeres was analyzed in strains carrying THO mutations. As shown in Figure 4A, stronger repression of URA3 was observed in cells harboring a tho2 mutation. The enhancement of URA3 repression was observed in all three strains tested and was most obvious in UCC509. The effect of hpr1 and rif1 on telomere silencing was then evaluated using UCC509. We anticipated that hpr1 would affect telomere silencing in a way similar to that of tho2 mutation and that the addition of the hpr1 or tho2 mutation to the rif1 mutant should not further enhance telomere silencing. The results shown in Figure 4B, tho2 and hpr1 showed similar enhancement of telomere silencing, ant the enhancement in both THO mutants was less than that conferred by the rif1 mutation. The addition of the tho2 or hpr1 mutation had no further effect on telomere silencing in the rif1 mutant. Thus, mutations on the two THO components also affect telomere silencing through a pathway mediated by RIF1.
Figure 4. Deletion of THO complex components Hpr1p or Tho2p enhances the telomere position effect.
(A) tho2 enhances the telomere position effect. Wild-type or tho2 cells with URA3 at 1.1 kbp (UCC507), 2.5 kbp (UCC509), or 5.5 kbp (UCC511) from the telomere of chromosome VII–L were analyzed. Cells in 10-fold serial dilutions were spotted on YC, YC lacking uracil, or plates containing 5-FOA, and incubated until colonies formed. (B) THO2 and HPR1 function in the same pathway as RIF1 to regulate the telomere position effect. Samples of serial dilutions of UCC509 cells with the indicated mutations were spotted on plates as indicated.
Overexpressing SUB2 cannot restore the Rif1p level or telomere length in tho2 and hpr1 cells
A high-copy suppressor screen using lacZ fused to a strong promoter has found that SUB2 suppresses the transcriptional defect of hpr1 [47]–[49]. SUB2 encodes a DEAD-box RNA helicase that is involved in splicesosome assembly and mRNA exporting [50]–[52]. Sub2p binds to Yra1p and the THO complex to form a TREX (TRanscription/EXport) complex that couples elongation and exporting of mRNA [35], [52], [53]. To test whether overexpressing SUB2 also recovers RIF1 expression and restores telomere length in THO mutants, tho2 and hpr1 cells with a plasmid carrying SUB2 or sub2-5 were analyzed for Rif1p levels and telomere length. The sub2-5 allele expresses a mutant Sub2p with a Q308R mutation near the helicase motif IV, which does not cause any detectable defects in splicing activity or spliceosome assembly [54]. However, the expression level of sub2-5 is higher than that of wild-type SUB2 in cells for unknown reasons [55]. As shown, overexpressing SUB2 or sub2-5 could rescue the growth defect of tho2 and hpr1 at 37°C (Fig. 5A), but did not restore the Rif1p level (Fig. 5B). Our results of real time RT-PCR experiments also showed that the RIF1 RNA in both tho2 and hpr1 cells was not restored to the wild-type level by overexpressing SUB2 (Fig. 5C). Consequently, telomere length in the tho2 and hpr1 cells remained long (Fig. 5D). Thus, while it is unclear why the Rif1p level cannot be restored through overexpressing SUB2 or sub2-5, the results support our conclusion that the lowered Rif1p level is the cause for long telomeres in tho2 and hpr1 cells.
Figure 5. Overexpressing SUB2 cannot restore the Rif1p level and telomere length in tho2 and hpr1 cells.
(A) Overexpressing SUB2 suppressed the growth defect of tho2 or hpr1 cells. Strains of the indicated genotypes derived from THO2/tho2, or HPR1/hpr1 diploids carrying SUB2 or sub2-5 overexpressing plasmids were grown on YC plates at 30° or 37°C. (B) SUB2 or sub2-5 overexpression did not restore the Rif1p level in tho2 or hpr1 cells. Immunoblotting assays were carried out as described. (C) SUB2 overexpression did not restore the RIF1 level in tho2 or hpr1 cells. Total mRNA from the indicated strains was isolated and analyzed for the RIF1 RNA level using real time RT-PCR. Results were presented as relative levels normalized to the wild-type expression level. The bars were standard deviations calculated using data from three independent experiments. (D) SUB2 or sub2-5 overexpression did not restore the telomere length in tho2 or hpr1 cells. Telomere length analyses were performed as previously described.
Discussion
Telomere length homeostasis is mainly achieved through regulation of telomerase and telomere-associated proteins. Here we discovered that THO complex components Hpr1p and Tho2p also affect telomere length and telomere silencing. Long telomeres and enhanced telomere silencing were observed in hpr1 and tho2 cells. We also discovered that RIF1 expression was greatly reduced in hpr1 and tho2 cells, which is in agreement with a previous observation that showed selective reduction of long transcripts by THO mutations [36]. Moreover, our genetic analyses using double mutations and overexpression demonstrated that telomere phenotypes observed in hpr1 and tho2 cells were caused by reduction of Rif1p levels. Thus, we have provided evidence that THO mutations exert their effects on telomeres through affecting the expression of Rif1p, the largest telomere-associated protein.
It has been shown that the hpr1 and tho2 mutations have the highest impact on gene expression and recombination among THO mutations [56]. Similarly, here we found that mutations on HPR1 and THO2 affected both telomere length and telomere silencing, whereas mutations on the other two THO components, Mft1p and Thp2p, did not affect Rif1p level and telomere lengths. Thus, it appears that the roles of Hpr1p and Tho2p are functionally distinct from those of Mft1p and Thp2p. Although the function of each component within the THO complex and the detailed mechanism underlying the differential impacts of individual THO mutations are not clearly defined, there appears to be a hierarchical difference within these components in terms of their in vivo biological relevance. Among these four components, the Tho2p and Hpr1p subunits are more important for telomere homeostasis.
Mutations of components of the THO complex cause a reduction in the transcription of yeast genes [34], [36]–[38], [45], [57]. Using the E. coli lacZ gene as a reporter, it has been shown that the level of gene transcription decreases with increasing gene length [39]. Further analysis has revealed that high G+C content might contribute to the reduced expression of lacZ [39]. Here we demonstrate a preferential reduction of RIF1 expression in hpr1 or tho2 cells. The result is consistent with the prediction of preferential reduction of long genes in THO mutants. We did not find noticeable reduction in the expression of high G+C content genes in hpr1 or tho2 cells. It is possible that because the overall G+C contents of the genes encoding telomere-associated proteins are low, the effect of G+C content becomes less apparent. In any case, the G+C content did not appear to have a role in modulating the expression of genes encoding telomere-associated proteins in cells depleted of the THO complex.
A genome-wide expression analysis using microarray has showed that the TEL1 level was significantly decreased in hpr1 cells [44]. Our analysis also shows that the TEL1 RNA levels are decreased in both tho2 and hpr1 cells. Because cells harboring a tel1 mutation have short telomeres [19], a combinatory effect of telomere shortening and lengthening was observed in the tho2 tel1 double mutant cells (Fig. 3B). This suggests that the telomere lengthening observed in tho2 or hpr1 cells is not due to the reduced TEL1 RNA levels. It is possible that although TEL1 expression is reduced in THO mutants, the amount of Tel1p is still sufficient to maintain its function on telomeres. In contrast, a ∼40–50% reduction of RIF1 RNA level causes a ∼75–80% reduction of Rif1p level, which is sufficient to contribute to an increase of telomere length. The results implicate that the Rif1p level has to be tightly regulated to stably maintain the telomere length. Of note, although we found significant reduction of RIF1 expression in both tho2 and hpr1 cells, RIF1 was not identified among the repressed genes in THO mutants in the genome-wide expression analyses. The microarray analyses considered the genes with expression levels at least 1.5-fold above or below the wild-type values as significantly different [44]. It is possible that the ∼40–50% reduction of the RIF1 RNA level in tho2 and hpr1 cells evaded the detection by microarray analyses.
Through a high-copy suppressor screen, it has been discovered that overexpressing SUB2 suppressed both the growth and transcription defects of tho2 and hpr1 mutants [47]–[49]. It is surprising to us that the Rif1p levels were not restored by overexpressing SUB2 or sub2-5 in tho2 and hpr1 cells, although the growth defects were abrogated. The cause of the temperature-sensitive growth in tho2 and hpr1 cells is unclear. Given roles of the THO complex in transcription and mRNA export, it is possible that the growth defect is due to the impairment of bulk transcription-, mRNA export, or the generation of transcriptional byproducts upon the collapse of these gene expression processes. Elucidation of the involving molecular mechanism awaits the identification of target yeast genes affected by the THO complex mutations. Alternatively, it is also possible that due to a separation of function conferred by the sub2 alleles, SUB2 overexpression leads to suppression of temperature sensitivity but not telomere phenotypes. Indeed, it has been reported that SUB2 overexpression in tho2 cells only recovers ∼20–30% of the lacZ activity driven by a GAL1 promoter [49], further supporting that SUB2 might function differently in suppressing growth defect and telomeres. No matter what mechanism is used by Tho2p and Hpr1p in controlling gene expression, our results still clearly indicate that the lowered Rif1p level is the cause of long telomeres in tho2 and hpr1 cells.
Materials and Methods
Strains and plasmids
All of the strains in this study were constructed in the YPH499 (MATa ura3-52 lys3-5 ade2-10 trp1-Δ63 his3-Δ200 leu2-Δ1) background [58]. YPH499-derived strains carrying SIR4-TAP-TRP1, tlc1Δ::LEU2, tho2Δ::HIS3, hpr1Δ::HIS3, mft1Δ::HIS3, thp2Δ::HIS3, tel1Δ::HIS3, rif1Δ::TRP1, rif2Δ::TRP1 and pif1–m2 were constructed in this study or obtained from S.-C. Teng (National Taiwan University College of Medicine, Taipei, Taiwan). MS179 (Rif1-G8-myc9) and MS206 (Rif2-G8-myc9) yeast strains were kindly provided by V. A. Zakian (Princeton University). In these two strains, the chromosomal copies of RIF1 and RIF2 were tagged with 9-myc, respectively. A spacer with 8-glycine was also introduced in between RIF1 or RIF2 and the tag [59]. Both strains were back crossed four times to YPH499 to minimize differences in genetic backgrounds. UCC507, UCC509, and UCC511 strains were kindly provided by D. E. Gottschling [60]. In these strains, URA3 was inserted at 1.2, 2.5, 5.5 kbp away from the left telomere of chromosome VII, respectively. Plasmid pRS426-RIF1-myc9 was generated by PCR amplification of the 3′ DNA fragment of RIF1-G8-myc9 using the genomic DNA of MS179 as the template and subsequent subcloning of the fragment into plasmid pRS426-RIF1 (2µ, URA3, D. Shore, University of Geneva, Sciences III, Geneva, Switzerland [26]). Plasmids pRS424-SUB2 and pRS424-sub2-5 were provided by E. Lahue [55].
Telomere length determination
To determine telomere lengths, yeast DNA was prepared, digested with XhoI, and separated on 1% agarose gels. Next, DNA fragments were transferred to a Hybond N+ paper (Amersham) for hybridization using a random primed Y′ DNA probe [61].
Immunoblotting analysis
Immunoblot analysis was carried out according to standard procedures using ECL detection (Perkin Elmer). Polyclonal antibodies against Rap1 were kindly provided by S.-C. Teng. Polyclonal antibodies against Cdc13(1–252)p were raised in rabbits [62]. Anti-α-tubulin (T9026) and anti-actin (MAB1501) antibodies were purchased from Sigma-Aldrich and Millipore, respectively. The anti-myc antibody (for Rif1-G8-myc9 and Rif2-G8-myc9) was purchased from LTK-Biolab. The anti-TAP antibody (CAB1001, for Sir4-TAP) was purchased from Open Biosystems. Horseradish peroxidase-conjugated donkey anti-rabbit or sheep anti-mouse antibodies (Amersham) were used as the secondary antibodies.
Northern Blotting analysis
Yeast cells were grown to an OD600 of 0.6 and then RNA was extracted using the hot phenol extraction protocol. Briefly, yeast cells were harvested by centrifugation and quickly resuspended in cold TES (10 mM Tris pH 7.5, 10 mM EDTA, 0.5% SDS). Equal volumes of acid phenol (Phenol ∶ Chloroform = 5 ∶ 1, pH 4.5, Sigma) were added and the samples were incubated at 65°C for 60 min. Acid phenol extraction was repeated several times until the interface was clear. The extracted RNA was extracted with equal volumes of CHCl3 and precipitated by ethanol. Ten µg of the extracted RNAs were separated by 1.2% formaldehyde agarose gels, transferred onto Hybond N+ membranes (Amersham), and hybridized with DNA probes prepared by random priming. The primers used to prepare PCR amplified DNA probes were: Rif1 forward (5′-ATCGAATTGATCAGCATATCCTCCGTAGGTCTCAACACGA-3′), Rif1 reverse (5′-GCTACACTTTTTGTAACCGG-3′), Rif2 forward (5′-ATGGAGCATGTAGATTCCGATTTTGCACCTATAAGGAGAT-3′), Rif2 reverse (5′-GAAACTTACTTAAGCTCGGA-3′), Tub1 forward (5′-AGTATTAATGGTATGTTCGATTTGCCCGTCCAGGCTAGAT-3′), Tub1 reverse (5′-GTAAATAGCCCTTGGAACGA-3′), Gcn1 forward (5′-GCGAATCACACGTGTCTAAAAGAGTTCCCTTTTTGCAAGA-3′), and Gcn1 reverse (5′-GCGACACAAAAATCAAGAAA GA-3′).
Real time RT-PCR analysis
Five micrograms of RNA were first treated with RNase-free DNase (Qiagen #79254) at 37°C for 30 min. Reverse transcription of the RNA was conducted using the RevertAid M-MuLV reverse transcriptase (Fermentas #EP0442) with oligo-dT as the primer. Real time PCR analyses were conducted using the ABI Step one real-time PCR system. Primer pairs used in this analysis were: GCN1 (5′-TGCGTGATCCTGTTATCCCA-3′, 5′-CGACAGCCCATATTTCTCTTTCATCTTCAAATCTGTCACT-3′), RIF1 (5′-ATTCACCTCCTCGAATGACA-3′, 5′-AGTTTCTTCTTTCTTCCACAGATATTCCGTTCAAGTCGCC-3′), TEL1 (5′-AGGCGGAGTTGTGAAAGAGTTTACGCAGTA-3′, 5′-AAATTTGATGGATCCGTGGCTTGCTGAATC-3′), and TUB1 (5′-AAGAAGACCGTCCAATTGGT-3′, 5′-GGCACCCACTTCGATGTAATCTCTTTCTAAAGCAGCCAAA-3′). The TUB1 RNA level was used as an internal control in each set of the experiments.
Assay for telomere position effect
Strains UCC507, UCC509, and UCC511 were used to evaluate the effect of telomere silencing. These strains were crossed with YPH500 carrying tho2, hpr1, or rif1 mutations. The resulting diploid strains were induced into meiosis and the resulting tetrads were dissected. Haploid strains with URA3 placed near the telomere and tho2, hpr1, or rif1 mutations were then selected. These cells were streaked on YC plates and grown for 3 days at 30°C. Colonies were resuspended in water, and aliquots of different dilutions were spotted on YC plates, YC plates lacking uracil or plates containing 5-FOA. Plates were incubated at 30°C until colonies formed. Usually, it takes 3 days for colonies to form on YC plates, and it takes 6–7 days on plates containing 5-FOA.
Acknowledgments
We thank Drs. S.-C. Teng, V. A. Zakian, D. Shore, E. Lahue, and D. E. Gottschling for providing yeast strains and reagents. We also thank Drs. C. Wang and M.-Y. Chen for critical reading of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the National Health Research Institute under grant NHRI-EX100-10050SI and the National Science Council under grants 99-3112-B-010-001 and 100-2311-B-010-001. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 3.Collins K, Mitchell JR. Telomerase in the human organism. Oncogene. 2002;21:564–579. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- 4.Counter CM, Meyerson M, Eaton EN, Weinberg RA. The catalytic subunit of yeast telomerase. Proc Natl Acad Sci USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, et al. Reverse transcriptase motifs on the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 6.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 7.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 8.Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;17:1–12. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, et al. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 10.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gravel S, Larrivee M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 12.Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porter SE, Greenwell PW, Ritchie KB, Petes TD. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bianchi A, de Lange T. Ku binds telomeric DNA in vitro. J Biol Chem. 1999;274:21223–21227. doi: 10.1074/jbc.274.30.21223. [DOI] [PubMed] [Google Scholar]
- 15.Boule J-B, Vega LR, Zakian VA. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 16.Zhou J-Q, Monson EM, Teng S-C, Schulz VP, Zakian VA. The Pif1p helicase, a catalytic inhibitor of telomerase lengthening of yeast telomeres. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 17.Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 18.Tseng S-F, Lin J-J, Teng S-C. The telomeraserecruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 2006;34:6327–6336. doi: 10.1093/nar/gkl786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lustig AJ, Petes TD. Identification of yeast mutants with altered telomere structure. Proc Natl Acad Sci U S A. 1986;83:1398–1402. doi: 10.1073/pnas.83.5.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie KB, Mallory JC, Petes TD. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craven RJ, Petes TD. Involvement of the checkpoint protein Mec1p in silencing of gene expression at telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:2378–2384. doi: 10.1128/mcb.20.7.2378-2384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shore D, Nasmyth K. Purification and cloning of a DNA-binding protein that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 23.Conrad MN, Wright JH, Wolf AJ, Zakian VA. RAP1 protein interacts with yeast telomeres in vivo: Overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- 24.Lustig AJ, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 25.Kyrion G, Boakye KA, Lustig AJ. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 27.Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 28.McEachern MJ, Blackburn EH. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 29.Krauskopf A, Blackburn EH. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature. 1996;383:354–357. doi: 10.1038/383354a0. [DOI] [PubMed] [Google Scholar]
- 30.Krauskopf A, Blackburn EH. Rap1 protein regulates telomere turnover in yeast. Proc Natl Acad Sci U S A. 1998;95:12486–12491. doi: 10.1073/pnas.95.21.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, et al. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, et al. Telomere length as a quantitative trait: Genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2006;2:e35. doi: 10.1371/journal.pgen.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ungar L, Yosef N, Sela Y, Sharan R, Ruppin E, et al. A genome-wide screen for essential yeast genes that affect telomere length maintenance. Nucleic Acids Res. 2009;37:3840–3849. doi: 10.1093/nar/gkp259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, et al. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 2000;19:5824–5834. doi: 10.1093/emboj/19.21.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 36.Chavez S, Aguilera A. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 1997;11:3459–3470. doi: 10.1101/gad.11.24.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prado F, Piruat JI, Aguilera A. Recombination between DNA repeats in yeast hpr1Δ cells is linked to transcription elongation. EMBO J. 1997;16:2826–2835. doi: 10.1093/emboj/16.10.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piruat JI, Aguilera A. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 1998;17:4859–4872. doi: 10.1093/emboj/17.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chavez S, Garcia-Rubio M, Prado F, Aguilera A. Hpr1 is preferentially required for transcription of either long or GC-rich DNA sequences in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:7054–7064. doi: 10.1128/MCB.21.20.7054-7064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 41.Chan CSM, Tye B-K. A family of Saccharomyces cerevisiae repetitive autonomously replicating sequences that have very similar genomic environments. J Mol Biol. 1983;168:505–523. doi: 10.1016/s0022-2836(83)80299-x. [DOI] [PubMed] [Google Scholar]
- 42.Betz JL, Chang M, Washburn TM, Porter SE, Mueller CL, et al. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol Genet Genomics. 2002;268:272–285. doi: 10.1007/s00438-002-0752-8. [DOI] [PubMed] [Google Scholar]
- 43.Sinha H, David L, Pascon RC, Clauder-Munster S, Krishnakumar S, et al. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics. 2008;180:1661–1670. doi: 10.1534/genetics.108.092932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomez-Gonzalez B, Garcia-Rubio M, Bermejo R, Gaillard H, Shirahige K, et al. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011;30:3106–3119. doi: 10.1038/emboj.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marin A, Gallardo M, Kato Y, Shirahige K, Gutierrez G, et al. Relationship between G+C content, ORF-length and mRNA concentration in Saccharomyces cerevisiae. Yeast. 2003;20:703–711. doi: 10.1002/yea.992. [DOI] [PubMed] [Google Scholar]
- 46.Kyrion G, Liu K, Liu C, Lustig AJ. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993;7:1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- 47.Fan H-Y, Merker RJ, Klein HL. High-copy-number expression of Sub2p, a member of the RNA helicase superfamily, suppresses hpr1-mediated genomic instability. Mol Cell Biol. 2001;21:5459–5470. doi: 10.1128/MCB.21.16.5459-5470.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jimeno S, Rondon AG, Luna R, Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West RW, Jr, Milgrom E. DEAD-box RNA helicase Sub2 is required for expression of lacZ fusions in Saccharomyces cerevisiae and is a dosage-dependent suppressor of RLR1 (THO2). Gene. 2002;288:19–27. doi: 10.1016/s0378-1119(02)00482-1. [DOI] [PubMed] [Google Scholar]
- 50.Libri D, Graziani N, Saguez C, Boulay J. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev. 2001;15:36–41. doi: 10.1101/gad.852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jensen TH, Boulay J, Rosbash M, Libri D. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr Biol. 2001;11:1711–1715. doi: 10.1016/s0960-9822(01)00529-2. [DOI] [PubMed] [Google Scholar]
- 52.Strasser K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- 53.Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kistler A, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lahue E, Heckathorn J, Meyer Z, Smith J, Wolfe C. The Saccharomyces cerevisiae Sub2 protein suppresses heterochromatic silencing at telomeres and subtelomeric genes. Yeast. 2005;22:537–551. doi: 10.1002/yea.1231. [DOI] [PubMed] [Google Scholar]
- 56.Garcia-Rubio M, Chavez S, Huertas P, Tous C, Jimeno S, et al. Different physiological relevance of yeast THO/TREX subunits in gene expression and genome integrity. Mol Genet Genomics. 2008;279:123–132. doi: 10.1007/s00438-007-0301-6. [DOI] [PubMed] [Google Scholar]
- 57.Gallardo M, Aguilera A. A new hyperrecombination mutation identifies a novel yeast gene, THP1, connecting transcription elongation with mitotic recombination. Genetics. 2001;157:79–89. doi: 10.1093/genetics/157.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sabourin M, Tuzon CT, Fisher TS, Zakian VA. A flexible protein linker improves the function of epitope-tagged proteins in Saccharomyces cerevisiae. Yeast. 2007;24:39–45. doi: 10.1002/yea.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 61.Yamada M, Hayatsu N, Matsuura A, Ishikawa F. Y′-Help1, a DNA helicase encoded by the yeast subtelomeric Y′ element, is induced in survivors defective for telomerase. J Biol Chem. 1998;273:33360–33366. doi: 10.1074/jbc.273.50.33360. [DOI] [PubMed] [Google Scholar]
- 62.Wang M-J, Lin Y-C, Pang T-L, Lee J-M, Chou C-C, et al. Telomere-binding and Stn1p-interactinf activities are required for the essential function of Saccharomyces cerevisiae Cdc13p. Nucleic Acids Res. 2000;28:4733–4741. doi: 10.1093/nar/28.23.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]