SUMMARY
Small ubiquitin-related modifier (SUMO) proteins are ~11 kDa proteins that can be covalently conjugated to lysine residues in defined target proteins. The resultant post-translational modification, SUMOylation, is vital for the viability of mammalian cells and regulates, among other things, a range of essential nuclear processes. It has become increasingly apparent in recent years that SUMOylation also serves multiple functions outside the nucleus and that it plays a critical role in the regulation of neuronal integrity and synaptic function. In particular, dysfunction of the SUMOylation pathway has been implicated in the molecular and cellular dysfunction associated with neurodegenerative and psychiatric disorders. Here, we outline current knowledge of the SUMO pathway and discuss the growing evidence for its involvement in multiple neurodegenerative disorders, with a view to highlighting the potential of the SUMO pathway as a putative drug target.
SUMO (small ubiquitin-related modifier) proteins are ~11 kDa proteins that can be covalently conjugated to lysine residues in target proteins, altering the biochemical and/or functional properties of the modified protein. SUMO conjugation occurs via an enzymatic cascade analogous to that of ubiquitin, another well-characterized protein modifier.1
SUMO was first described in 1996 as a protein involved in the partitioning of the nuclear Ras-related GTPase-activating protein (RanGAP) between the cytosol and nuclear pore complex.2,3 In the decade following its discovery, numerous studies reported the SUMO modification of other nuclear proteins, suggesting that SUMOylation is a predominantly nuclear phenomenon. However, in recent years, multiple cytosolic and plasma membrane SUMO targets have been described, many of which are essential for neuronal function. In addition, an increasing number of SUMO targets have been linked to various neuropathological conditions. This review provides a summary of recent findings related to SUMOylation and neurological disease, with a view to the potential for modulation of protein SUMOylation in the brain as a means for therapeutic intervention.
SUMO ISOFORMS
Yeast contain only one SUMO protein, ubiquitin-like protein Smt3, whereas humans possess four SUMO isoforms, designated SUMO1 to SUMO4.4,5 In their conjugatable forms, SUMO2 and SUMO3 differ only in three N-terminal residues and have yet to be functionally distinguished (hence they are usually collectively referred to as SUMO2/3). SUMO1 and SUMO2/3 are evolutionarily conserved, and SUMO1 shares 18% homology with ubiquitin (Fig. 1). However, despite the relatively low sequence homology, the proteins possess highly similar three-dimensional structures.6
Figure 1.
Alignment of mature ubiquitin and SUMO protein sequences. Residues conserved across all four proteins are shown in red. Residues conserved across all SUMO members are shown in turquoise. The conjugatable C-terminal diglycine motif is indicated. Due to the controversy over whether it is a functional protein modifier, SUMO4 is not shown.
SUMO1 and SUMO2/3 are conjugated to substrate proteins by the same enzymatic machinery7 and while some targets are exclusively modified by one family member,8 many target proteins can be modified by both SUMO1 and SUMO2/3.9,10 To date, each of the SUMO isoforms have been implicated in similar cellular functions and there is no strong consensus about the functional differences elicited by SUMO1 versus SUMO2/3 conjugation. Indeed, while ablating the SUMO system via knockout of the sole SUMO-conjugating enzyme Ubc9 is embryonically lethal,11 SUMO1 knockouts are viable and overtly normal, suggesting that SUMO2/3 can largely compensate for loss of SUMO1 in vivo.12,13 Interestingly, however, SUMO2/3 and Smt3 appear to be able to form poly-SUMO chains as they contain SUMO-conjugation motifs within their sequence7,14,15 whereas this motif is absent from SUMO1. While a general function for SUMO chains is unclear, numerous groups have reported that SUMO chains can be recognized by a class of SUMO-targeted ubiquitin ligases (STUbLs).16 These proteins recognize and ubiquitinate SUMO chains, leading to degradation of the SUMO substrate. This suggests that both SUMO and ubiquitin can act cooperatively in protein degradation. SUMO1 and SUMO2/3 also differ in their conjugation dynamics and responses to cellular stress.8 Under resting conditions, very little SUMO1 is present in an unconjugated form, yet there is a large free pool of SUMO2/3.8 However, in response to cellular stresses, such as oxidative stress, osmotic stress or heat shock, a rapid increase in SUMO2/3 conjugation is observed. This increase implies that SUMO2/3 may act as a cellular SUMO reserve that under some conditions allows rapid protein SUMOylation, which forms part of the initial response of the cell to stress.
The role of SUMO4 remains unclear. SUMO4 mRNA appears to be limited to the kidney,5 and this isoform may even be incapable of modifying substrate proteins due to a proline residue that appears to prevent its maturation.17 However, a recent report suggests that SUMO4 may be rapidly degraded in resting cells and matured only in response to severe cellular stress.18 While further work will be required to investigate its exact role, it is possible that SUMO4 also constitutes a stress-responsive cellular reserve of SUMO.18
THE SUMO CONJUGATION PATHWAY
Conjugation of SUMO to substrate proteins
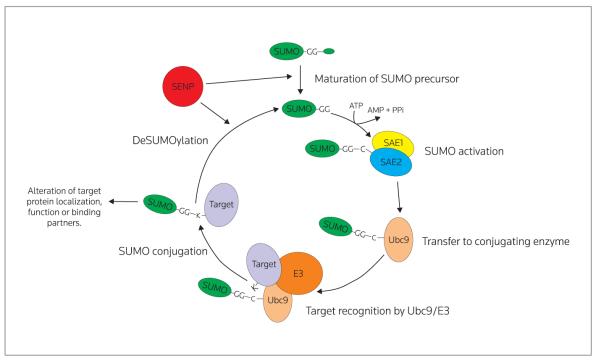
Protein SUMOylation is mediated by a reversible enzymatic cascade in a manner analogous to modification of proteins by ubiquitin. In summary, SUMO proteins are first activated by the action of an E1 “activating” enzyme, which passes the activated SUMO to an E2 “conjugating” enzyme. The E2 enzyme, often in conjunction with an E3 “ligase” enzyme, then catalyzes the conjugation of SUMO to its substrate. The detailed mechanisms of protein SUMOylation have been extensively reviewed by several groups1,19-21 and so will be only briefly described here. A schematic of the SUMOylation/de-SUMOylation pathway is shown in Figure 2.
Figure 2.
The SUMO pathway. SUMO is transcribed as an inactive precursor, which is cleaved by members of the SENP (sentrin-specific protease) family to expose a C-terminal diglycine motif. This mature form of SUMO is then activated by the ATP-dependent formation of a thioester bond with the active site of the E1 enzyme, a heterodimer of SUMO-activating enzyme (SAE)1 and SAE2. The activated SUMO is then passed to the active site cysteine of the E2 conjugating enzyme, Ubc9, which then catalyzes the transfer of SUMO to the target protein, often in conjunction with an E3 enzyme. The SUMOylated protein then displays phenotypic differences to the unmodified form, until the SUMO tag is removed, again by the SENP family of proteases, releasing the unmodified target protein (not shown), and mature SUMO, which can then undergo further rounds of conjugation to target proteins.
In mammals, SUMO activation occurs via the actions of a heterodimer of SUMO-activating enzyme (SAE)1 and SAE2.22 SAE1 catalyzes the formation of adenylated SUMO, in which the C-terminal carboxyl group of SUMO is covalently linked to adenosine monophosphate (AMP). Cleavage of this bond leads to the conjugation of SUMO to the sulfhydryl group of the C173 residue of SAE2, via a thioester bond. SUMO is then transferred by a transesterification reaction to the cysteine (C93) active site of Ubc9.23,24
In contrast to the ubiquitin pathway, for which multiple conjugating enzymes have been described,25 Ubc9 is the only conjugating enzyme in the SUMO pathway.23,24 Importantly, Ubc9 is SUMO-specific and does not conjugate ubiquitin,23 indicating that SUMO and ubiquitin are conjugated by analogous, but distinct pathways.
Interestingly, Ubc9 is capable of directly recognizing target proteins.26 As a result, SUMOylation occurs predominantly at a consensus motif in substrate proteins,27,28 which is directly bound by Ubc9.28 This consensus motif can be defined as ψKxD/E, where ψ is a large hydrophobic residue, K is the target lysine, x is any residue, and D/E is aspartate or glutamic acid (acidic residues). Although not all SUMO substrates are modified within ψKxD/E motifs, and not all ψKxD/E motifs are SUMOylated,29 the presence of a SUMOylation consensus motif is a useful starting point for the bioinformatic identification of potential novel SUMO substrates from sequence data.
In the ubiquitin pathway, E3 enzymes contribute to substrate recognition; however, because Ubc9 is capable of directly recognizing target proteins, requirement of the E3 “ligase” enzymes for SUMOylation was initially debated. Subsequently, a number of proteins have been discovered which posses E3 activity in the SUMOylation pathway (Table I).
Currently, the mechanisms by which E3 enzymes enhance the rate and efficiency of Ubc9-mediated SUMOylation are not known. Unlike the ubiquitin system, there are no known E3 enzymes that actually receive SUMO from the E2 enzyme, indicating the actual SUMO-transfer reaction is always dependent on Ubc9. SUMO E3 enzymes may therefore act by binding to either the substrate or SUMO-loaded Ubc9 and promoting a conformation more conducive to SUMO transfer. Indeed, structural studies indicate this is likely the case for the E3 SUMO-protein ligase RanBP2.30 However, despite structural similarity between the PIAS family of E3 proteins and the seeming redundancy of family members implied by knockout studies, they appear to be able to facilitate SUMO transfer to a wide array of different substrates, indicating that there is still much to learn about E3 enzyme function.
Processing of nascent SUMO and cleavage from substrates
SUMO proteins are synthesized as inactive precursors which need to be cleaved at the C-terminus to expose a diglycine motif before conjugation can occur.31 This form of SUMO can then be conjugated to substrate proteins via the actions of the E1, E2 and E3 proteins described above.
Surprisingly, the enzymes responsible for cleavage of the nascent peptide to expose the diglycine motif are the same as those responsible for removal of conjugated SUMO from substrate proteins.32 Screens for mammalian deSUMOylating enzymes have revealed 6 sentrin-specific proteases (SENPs), designated SENP1-3 and SENP5-7. The SENP enzymes differ in their subcellular location, as well as their substrate specificity32 but at present, the mechanisms that define SENP specificity and localization remain ill-defined.
CELLULAR ROLES FOR SUMO
The critical importance of the SUMO pathway is emphasized by knockdown or deletion of Ubc9, a critical component of the SUMO pathway, which results in loss of viability of mammalian cells.11,33 Whereas ubiquitin is generally involved in protein degradation pathways,25 the functions of SUMO modification appear to be more diverse. For the majority of SUMO substrates, the precise functional role and consequences of SUMOylation remain unclear. In most cases SUMOylation likely acts either by altering protein-protein interactions or by regulating other post-translational modifications such as acetylation or ubiquitination.
Modulation of protein interactions by SUMOylation
The best characterized action of SUMOylation is alteration of the binding properties of the substrate protein. There are three nonexclusive ways in which this can be mediated:
Conjugated SUMO can itself interact with SUMO-binding proteins, creating a new interface for protein interactions with the substrate. Indeed, many of the molecular consequences of SUMOylation may be dependent on cellular proteins that bind SUMO noncovalently.34 Interestingly, a number of separate research groups have identified SUMO-interacting motifs (SIMs).35-38 A prototypical example of SIM–SUMO interactions mediating the sorting of a SUMOylated protein involves the SUMOylation-induced interaction between RanGAP and RanBP2.3 Upon SUMOylation of RanGAP, RanBP2 is able to recognize the SUMO moiety via a SIM in RanBP2,38 recruiting SUMO–RanGAP from the cytosol to RanBP2 on the nuclear pore. An elegant method of regulation of SUMO–SIM interactions has recently come to light with the observation that the SIMs of PIAS1, PML and PMSCL1 (exosome component 9) are only capable of binding SUMO when phosphorylated on flanking serine residues by the kinase CK2.39 Many SIM-containing proteins contain these flanking serines, suggesting the possibility that the sorting and interactions undergone by a SUMOylated substrate could be generally regulated depending on the phosphorylation state of the SIM on the SUMO-interacting protein.
SUMO conjugation can induce a conformational change in the substrate, either exposing or masking specific protein interaction domains in the substrate protein. For instance, the DNA repair enzyme thymine-DNA glycosylase (TDG) is SUMOylated and SUMO conjugation causes a dramatic conformational change in TDG which is required to complete each catalytic cycle.40
The conjugated SUMO moiety can sterically occlude a binding site on the substrate protein. SUMOylation of the ubiquitin-conjugating enzyme E2-25K prevents its interaction with the ubiquitin E1 enzyme, inhibiting the ubiquitin-conjugating function of E2-25K.41
Regulation of other lysine-directed post-translational modifications by SUMOylation
Several post-translational modifications occur at lysine residues in target proteins. For example, there are numerous examples of proteins which can be both SUMOylated and ubiquitinated at the same lysine residue.42 While in some cases the two modifications may compete for the target lysine, SUMOylation and ubiquitination have also been reported to act cooperatively and sequentially in the activation of nuclear factor-κB (NF-κB) essential modulator (NEMO).43 In addition, SUMOylation has also been shown to compete for the same lysine residue as acetylation for the transcription factor Sp3, transcriptional coactivator p300 and myocyte-enhancer factor 2A (MEF2A), a key regulator of postsynaptic differentiation.44
REGULATION OF SUMOYLATION
Despite a growing number of SUMO substrates, relatively little is known about how SUMOylation is regulated. In many cases it appears that SUMOylation may be regulated in a substrate-specific manner. For example, SUMOylation of the kainate receptor subunit GluR6 first requires agonist stimulation of the receptor,45 presumably resulting in a conformational change more conducive to Ubc9 binding and SUMOylation. In addition, as discussed above, SUMOylation can be regulated either positively or negatively by other post-translational modification. For example, phosphorylation can either act to promote (e.g., MEF2A46) or inhibit (e.g., c-Fos47 or Ikappa-Balpha48) protein SUMOylation, depending on the target. Thus, tight regulation of other post-translational modifications can directly regulate target SUMOylation.
In addition, various stimuli have been reported to regulate the SUMOylation enzymes themselves. Some of these stimuli appear to globally regulate activity of the enzymes themselves, whereas others appear to favor particular subsets of SUMO substrates. For example, oxidative stress has been reported to globally reduce SUMOylation by leading to reversible oxidation of the SUMOylation E1 and E2 enzymes.49 Cellular stresses such as ischemic shock, heat or osmotic shock have also been reported to globally modulate SUMOylation;8 however, the direct mechanisms for this are currently unclear.
SUMOylation can also be regulated by post-translational modification of the SUMOylation enzymes. For example, the viral protein Gam1 has been reported to globally reduce SUMOylation by targeting the SUMOylation E1 complex for ubiquitin-mediated degradation.50,51 Additionally, a recent study reported that Ubc9 is itself SUMOylated at Lys14, thus regulating target discrimination.52 While the activity of Ubc9 for some targets is not altered, SUMOylation of Ubc9 impairs its ability to SUMOylate RanGAP and strongly activates SUMOylation of the transcriptional regulator Sp100. Given the rapidly growing number of known SUMO targets, these data provide new insights into the mechanisms of target-specific SUMOylation with a limited number of enzymes, as particular modifications of the enzymes involved appear to specifically increase or decrease the modification of a subset of SUMO targets. Thus, it appears that both cellular conditions and modification of the SUMO-pathway enzymes can globally regulate SUMOylation, in addition to regulation of individual substrates. It is therefore possible that any of these pathways could show potential for therapeutic intervention.
SUMO IN NEUROPATHOLOGICAL CONDITIONS
Neuropathological conditions resulting from abnormalities in neuronal pathways can give rise to a host of deleterious effects which manifest in a diverse range of characterized disease states. Studies on the effects of SUMOylation in the central and peripheral nervous systems have begun to elucidate a number of molecular mechanisms by which SUMO might contribute to a host of neuropathologies. Much of the focus of neuronal SUMOylation has been on aberrant protein aggregation and cellular inclusion bodies or on polyglutamine-related pathologies, both common to many neurological disease states.53 In addition, reports of novel patterns of SUMO conjugation in diseases such as ischemia have begun to emerge (Table II). The known and putative roles of SUMO in neuropathologies have been reviewed recently by other groups20,54 and are presented here in brief.
Cellular stress and ischemia
As outlined above, SUMO1 and SUMO2/3 display different conjugation dynamics in response to cellular stress; levels of free SUMO2/3 are higher compared to SUMO1,8 suggesting that free SUMO2/3 might act as a pool for immediate response to cellular stresses. Indeed, there is substantial evidence for rapid changes in SUMO conjugation in response to various cellular stresses such as oxidative stress, osmotic stress, or heat shock.55-58 One characteristic of brain ischemia, an extreme metabolic stress due to oxygen and glucose deprivation (OGD), is excitotoxic neuronal death mediated, at least in part, by massive glutamate release. During hibernation animals undergo ischemic insult but do not appear to suffer damaging long-term consequences. In hibernating ground squirrels, the severe reduction in blood flow is correlated with a massive increase in global protein SUMOylation, suggesting that SUMO conjugation may be protective against oxygen and glucose deprivation;59,60 however, the cellular mechanisms and target proteins underlying this effect remain to be determined.
Multiple studies have shed light on the dynamics of SUMOylation in various middle cerebral artery occlusion (MCAO) models of ischemia.61 In rat and mouse models of both focal and global ischemia and transient versus permanent ischemia, increases in both SUMO1 and SUMO2/3 have been reported.62,63 Another recent study examined changes in SUMO1 and SUMO2/3 conjugation in a transient rat and permanent mouse MCAO model of focal ischemia.64 Increases were reported in SUMO1 conjugation in both the infarct and nonischemic hippocampus in the permanent mouse MCAO model, but SUMO2/3 conjugation was only increased outside the infarct area. Furthermore, MCAO caused a decrease in levels of excitatory AMPA and kainate receptors. Coupled with the recent finding that SUMOylation regulates agonist-induced endocytosis of the kainate receptor subunit GluR6,45 these data are consistent with the possibility that increased SUMO conjugation during ischemia might be neuroprotective by reducing glutamate receptor-mediated excitoxicity and resulting neuronal death. A recent study provided additional support to this hypothesis. In cultured cell lines and primary cortical neurons, it was reported that overexpression of SUMO1 or SUMO2 significantly protects cells against OGD-induced cell death.65 In addition, knockdown of SUMO1 led to a decreased survival in response to severe OGD.65 These results suggest that the elevated levels of SUMOylation seen in various ischemia models may well be a neuroprotective mechanism, rather than a nonspecific stress response.
SUMO and Parkinson’s disease
Parkinson’s disease (PD) is a degenerative disorder of the CNS that often impairs motor function and speech. PD is the most common movement disorder and is characterized by the progressive loss of dopaminergic neurons in the substantia nigra that project to the striatum. It is further characterized by the accumulation of Lewy bodies, cytosolic protein aggregates composed of α-synuclein, which form in the neurons of PD patients.66-68 The function of α-synuclein is as yet undefined, but recent evidence suggests that α-synuclein acts as a molecular chaperone, assisting in the folding and refolding of synaptic SNARE proteins, which mediate the fusion of synaptic vesicles with the cell membrane at the synapse.69 A report that SUMO1 marks subdomains within glial cytoplasmic inclusions of multiple system atrophy (MSA)70 and the recent finding that α-synuclein is a substrate for SUMO171 raise intriguing questions as to the identity of SUMO targets within the aggregates.
Several genes involved in the pathogenesis of PD, including α-synuclein and ubiquitin carboxy-terminal-hydrolase-L1 (UCH-L1), are linked to the ubiquitin-proteasome system.72 Another such protein is E3 ubiquitin ligase parkin, which noncovalently interacts with SUMO1 in vitro and in vivo.73 Importantly, parkin mutations account for many of the familial cases of PD.74 In an interesting example of the interplay between the ubiquitin and SUMO systems, it has been reported that parkin ubiquitinates the SUMO E3 ligase RanBP2, leading to its degradation.75 SUMO1 interacts with parkin to enhance its ubiquitin-ligase activity in a negative feedback loop. This underscores the importance of conjugation-independent SUMO functions and might implicate noncovalent SUMO interactions in PD pathogenesis.
Another role of SUMOylation in PD relates to the multifunctional protein DJ-1, which is involved in the transcriptional regulation of a variety of genes, many of which are involved in the cellular response to oxidative stress.76,77 Loss of function of DJ-1 results in the onset of PD. DJ-1 is a SUMO substrate78 and has also been shown to interact with SUMO E3 ligases.77 Two DJ-1 mutants have been identified which demonstrate the importance of the SUMO pathway in DJ-1 function and PD. Mutation of K130, the SUMO acceptor lysine, abolishes all known functions of DJ-1 in cultured cells.78 A second mutation, the disease-associated missense mutant L166P, shows increased levels of SUMOylation compared to wild-type DJ-1.79 The authors suggest that this phenomenon might be a result of either polySUMOylation on selected lysines or incorrect SUMOylation on multiple lysines. It is possible that improper SUMOylation of the L166P mutant may promote aggregation, causing the observed increase in protein insolubility and leading to an increase in protein degradation by the proteasome.78
SUMO and Alzheimer’s disease
Alzheimer’s disease (AD) is an age-dependent neurodegenerative disorder that is the cause of the most common type of dementia associated with progressive cognitive dysfunction. AD is characterized histopathologically by the presence of neuritic plaques and neurofibrillary tangles80 in the cerebral cortex. Plaques are the result of extracellular accumulation of amyloid-β (Aβ) peptides, which are cleaved from the amyloid precursor protein (APP) by β- and γ-secretases.81 APP was initially speculated to be a SUMO substrate in an in vitro expression cloning screen,82 and recent evidence suggests it can be modified on two lysine residues both in cultured cells and in brain by both SUMO1 and SUMO2/3.83 Interestingly, these lysine residues flank the γ-secretase cleavage site in APP, implicating SUMOylation in the regulation of APP cleavage to Aβ. The authors provided further evidence that SUMOylation of APP, or overexpression of the SUMO conjugating enzyme Ubc9, negatively regulates Aβ production., Another molecular screening approach has also identified SUMO3 as a regulator of Aβ expression,84 though the mechanisms by which SUMO3 modulates Aβ remain unclear. Different groups have reported either an increase85 or decrease84 in the production of neurotoxic Aβ in the presence of exogenously expressed SUMO3. The authors reporting an increase in Aβ production also found that the increase was maintained when nonconjugatable SUMO3 was expressed,85 suggesting that the SUMO3-dependent effect on Aβ production might not be due to covalent modification by SUMO.
The neurofibrillary tangles (NFTs) associated with AD are composed of intracellular aggregates of the hyperphosphorylated form of microtubule-associated tau protein.86,87 Tau is involved in the stabilization of microtubules and regulates axonal development, functions that are negatively regulated by its phosphorylation state.88 Interestingly, tau is a target for both SUMO171 and ubiquitin.89 In cultured cells, inhibition of the proteasome pathway has been shown to increase tau ubiquitination and decrease tau SUMOylation,71 suggesting that a competitive mechanism between ubiquitin and SUMO may regulate tau stability. A recent study examined the colocalization of tau and SUMO1 in transgenic mice expressing AD-causing APP mutations or an AD-causing, hyperphosphorylated mutant tau.90 SUMO1 was shown to colocalize with phospho-tau in APP transgenic mice, but not with hyperphosphorylated mutant tau; however, the pathophysiological relevance of these observations is still unclear.
Polyglutamine diseases
Polyglutamine (polyQ) disorders are a family of neurodegenerative disorders that include Huntington’s disease (HD), dentatorubral pallidoluysian atrophy (DRPLA), spinocerebellar ataxias (SCAs) and spinobulbar muscular atrophy (SBMA).91 In each of these disorders, expression of an aberrant protein with a toxic stretch of polyglutamine repeats, ranging from 35 to over 300, has been shown to be the causative factor. Although the proteins involved have unique neurological functions and localizations, they share the common phenotype of abnormal aggregation of polyQ and formation of cytoplasmic and/or intranuclear inclusions in neurons.
HD is characterized by the gradual atrophy of striatal neurons,92 and is the result of an extended polyQ stretch in the N-terminal domain of the Huntingtin (HTT) protein, which causes accumulation of the protein in affected neurons. The interplay between SUMOylation, ubiquitination and HTT is not yet clear; however, a recent study demonstrated that HTT can be SUMOylated and that in a Drosophila melanogaster model, SUMOylation increases HTT solubility and reduces aggregation, but also increases its abundance in neurons.93 As SUMO and ubiquitin share the target lysine in HTT, it is possible that a balance exists between HTT SUMOylation and ubiquitination. Interestingly, disruption of the target lysine (disrupting both SUMOylation and ubiquitination) reduces HD pathology, suggesting that the function of SUMOylation is not merely to block ubiquitination.
DRPLA is a neurodegenerative disorder caused by polyQ expansion in the atrophin-1 protein. Atrophin-1 can be found in both the nuclear and cytoplasmic compartments of neurons. Although its function is not yet known,94,95 atrophin-1 has been identified as a SUMO1 substrate.96 Coexpression of polyQ atrophin-1 with wild-type SUMO1 in PC12 cells resulted in an acceleration in the formation of nuclear aggregates and increased apoptosis, while coexpression of nonconjugatable SUMO with polyQ atrophin-1 resulted in a reduction of aggregates of polyQ atrophin-1 and promoted cell survival.96 The authors suggest that either SUMO disrupts or competes with proteasome-mediated degradation, or that SUMOylation of polyQ atrophin-1 results in aberrant trafficking or sequestration of SUMO-modified proteins and promotes aggregation in the nucleus.
SBMA presents another example of the interplay between SUMOylation and ubiquitination. SBMA is a neuromuscular disease, the result of a pathogenic polyQ expansion in the androgen receptor (AR). Wild-type AR is a known SUMO1 substrate and its transcriptional activity is inhibited by SUMOylation.97 The mutant AR forms nuclear and cytoplasmic aggregates, and results in progressive neurodegeneration. In a D. melanogaster model,98 disruption of the SUMO pathway by overexpression of a mutant E1 enzyme incapable of activating SUMO led to significantly enhanced polyQ-induced degeneration.98 Similarly, compromising the proteasome pathway with the overexpression of an inactive proteasome β-subunit mirrored the degenerative phenotype, suggesting that both pathways are functional in diminishing the pathogenic nature of the polyQ AR in SBMA.
The SCAs are a family of progressive neurodegenerative disorders characterized by the atrophy of the cerebellar Purkinje layer. SCA type 1 (SCA1) is the result of an extended polyQ in ataxin-1.99 Wild-type ataxin-1 has been shown to be SUMOylated on at least five lysine residues,100 and SUMOylation is reduced in the presence of the mutant polyQ stretch. Ataxin-1 is a phosphoprotein, and SUMOylation levels of a phosphorylation-deficient mutant (S776A) polyQ ataxin-1 were similar to that of wild-type ataxin-1. The implications of SUMOylation of ataxin-1 in SCA are not yet fully understood, though this provides an interesting example of inhibitory interplay between phosphorylation and the SUMO pathway.
Neuronal intranuclear inclusion disease
Neuronal intranuclear inclusion disease (NIID) is a rare neurodegenerative disorder characterized by ataxia in younger patients and progressive ataxia and dementia in adults. NIID is characterized by the occurrence of insoluble, intranuclear inclusions in almost all types of central, peripheral and autonomic neurons. Extensive SUMO1 immunostaining has been observed in fibrillar inclusions in unrelated cases of familial101 and juvenile NIID,102 as well as in sporadic cases of NIID.103 No specific SUMO substrates have yet been identified in the pathology of NIID, though potential targets have been suggested based on reports of major protein components of NIID aggregates.103,104 For example, anti-SUMO immunocapture followed by mass spectroscopy identified the proteins dynamin 1, NSF, Unc-18-1 and heat shock protein 90 (Hsp90) as proteins associated with inclusions in a familial case of NIID.104 However, further work will be required to determine whether these proteins are bona fide SUMO substrates associated with NIID inclusions. In addition, the transcriptional corepressor histone deacetylase HDAC4, a known SUMO substrate,105,106 is also associated with the intranuclear inclusions present in NIID.103 Interestingly, HDAC4 may also act as a SUMO E3 ligase,106 suggesting the possibility that SUMOylation in the aggregates could be due to excessive E3 ligase activity of HDAC4 in the inclusions.
SUMOYLATION AS A POTENTIAL DRUG TARGET
Given the role of SUMOylation in various disease pathways, it is tempting to speculate that SUMOylation may prove a useful target for therapeutic intervention. For example, the massive increase in protein SUMOylation seen in neurons after ischemia appears to be a neuroprotective response,65 and it may therefore be therapeutically beneficial to temporarily enhance SUMOylation in the area surrounding the infarct region.61 In addition, in diseases characterized by hyper-SUMOylation of target proteins, specifically decreasing SUMOylation of those proteins may also prove useful.
Given the importance of the SUMO pathway in multiple cellular processes, care must be taken in designing modulators of SUMOylation. However, it is worth noting that various modulators of the analogous ubiquitin system have shown some promise in drug trials.107 There are numerous ways in which the SUMOylation pathway could be modulated by pharmacological tools: by manipulation of any of the four enzymatic activities associated with SUMOylation, or by preventing binding of specific SUMO substrates to members of the SUMOylation/deSUMOylation machinery.
SAE1/SAE2 and Ubc9 are the only E1/E2 enzymes for the SUMOylation pathway, and pharmacological intervention at this stage would cause a global modulation of cellular SUMOylation. Given the integral roles of SUMOylation in cell viability, this would likely be undesirable, though a brief therapeutic window might exist in certain disease states. Currently, there are no reported chemical modulators of either of these enzymatic activities, however, structural data on both SAE1/SAE2108 and Ubc930,109,110 has shed light on their active site regions and on the interaction interface between Ubc9 and substrate proteins. These findings are leading the way for the possible structure-based design of small molecule inhibitors. Analogous studies in the ubiquitin pathway have led to compounds that have E1 inhibitory actions.111 E1 inhibition could potentially take a number of forms—by blocking ATP binding to SAE1, SUMO binding to SAE1/SAE2 or SAE1/SAE2 binding to Ubc9. Occlusion of ATP binding sites has proven to be particularly efficacious, with the development of imatinib, for example, in the treatment of chronic myelogenous leukemia.112
A preferential mode of interference with SUMOylation lies in targeting steps in the pathway for which there is more than one enzyme, to favor SUMOylation target-driven modulation over global changes in SUMOylation. Indeed, the most successful ubiquitin pathway drugs have targeted the E3 ligase complexes and deubiquitinating (DUB) enzymes.107 In addition, structural data for the SUMO E3 enzyme RanBP2 in complex with the other components of the SUMO pathway is now available,30,109 making possible the structure-based design of specific inhibitors.
Another potential target for intervention involves the SENP enzymes. This approach is problematic, however, as the SENP enzymes are responsible for both the maturation of SUMO as well as the cleavage of SUMO from substrate proteins,32 and inhibition of a specific SENP would not necessarily lead to the desired increase or decrease in SUMOylation of a given target protein. However, the existence of 6 SENP family members specific to SUMO, each with specific subcellular locations and preferences between SUMO paralogues, makes them a more attractive drug target than the E1/E2 complexes. Furthermore, although the catalytic subunits of the SENPs are highly conserved, the different isoforms display a high level of specificity to SUMO paralogues,32 suggesting that it may be possible to select for inhibition of deconjugation of SUMO1 versus SUMO2/3. Recent structural data has shed light on the basis for the difference in substrate preference and catalytic activity of the SENPs,113,114 which will certainly be useful in the design of small molecule SENP inhibitors.
Targeting of individual substrates presents possibly the most attractive point of intervention in the SUMO pathway. Despite the success of E3-targeted and DUB enzyme inhibitors in the ubiquitin system, there are considerably more ubiquitin E3/DUB enzymes in the ubiquitin pathway than the SUMO pathway. Therefore, the potential to inhibit a single pathway or a small number of pathways with minimal off-target effects by virtue of specific E3 or DUB inhibitors is greatly increased in the case of ubiquitin. Attempts to inhibit interactions between the ubiquitination machinery and substrate proteins have recently shown promise. Drugs targeting the interaction between the E3 ubiquitin ligase Mdm2 and its substrate p53 have been shown to trigger apoptosis in vitro and inhibit xenograft tumor growth in vivo.115,116 However, the situation is once again more complex for the SUMO pathway. Given that a single E2 enzyme, Ubc9, is responsible for the direct transfer of SUMO to the substrate, most substrates are modified within the SUMO consensus sequence with which Ubc9 directly interacts.28 Therefore, blocking the interaction of Ubc9 with its substrate will likely interfere with SUMOylation of a large number of targets. It is theoretically possible to design substrate-specific blockers of Ubc9 interaction by targeting regions on the substrate close to the SUMOylation site, though the potential to develop such inhibitors is currently limited as detailed structural information will be required for each proposed target. Attempts to block E3-substrate interactions may therefore prove more fruitful, although the E3 enzyme responsible for SUMO transfer to many substrates is not known, and in many cases interaction with Ubc9 alone is sufficient for SUMO conjugation to occur.
A final potential route for therapeutic intervention may actually lie in leaving the SUMOylation of a particular protein intact and attempting instead to interfere with the downstream consequences of SUMOylation. For many SUMO substrates, the downstream effects of SUMOylation are likely to involve the recruitment of protein interactors to the SUMOylated substrate, potentially via SUMO–SIM interactions. While compounds that disrupt SUMO–SIM interactions would likely have seriously deleterious effects, it may be possible to intervene at this step if the SUMO–SIM interaction is regulated in some way. For example, the SIM of PIAS1 only becomes capable of binding SUMO when phosphorylated.39 Potentially, blocking the phosphorylation of PIAS1, either by blocking the kinase responsible or by specifically preventing the PIAS1-kinase interaction, may counteract deleterious effects of substrate SUMOylation by preventing the downstream interaction with PIAS1. However, this is likely to be particularly difficult given that the downstream proteins responsible for the majority of effects of SUMOylation are unknown. Furthermore, as the similarity between the defined SIMs and the observation that phosphoregulation of SUMO–SIM interactions may be general,39 achieving a sufficient level of specificity would pose a significant challenge.
There are therefore many potential points in the SUMOylation cascade which might be amenable to therapeutic manipulation. However, further advances in this field will require greater characterization of the enzymes involved, as well as detailed analysis of the E3 enzymes responsible for the efficient SUMOylation and the SENPs responsible for deSUMOylation of substrates of interest.
CONCLUSIONS AND PERSPECTIVES
In recent years there has been rapid and substantial progress towards greater understanding of the mechanisms and consequences of SUMOylation. The vast majority of SUMO research has concentrated on the nuclear functions of SUMO. However, an extranuclear role for SUMO in neuronal function and dysfunction is beginning to emerge. SUMO therefore represents a new and exciting field for understanding of both normal neuronal function, and as a potential target for intervention in neurodegenerative disease. We anticipate that, like phosphorylation and ubiquitination, SUMOylation will prove to be an intricate and fundamentally important regulator of neuronal biology. Furthermore, the growing body of evidence for a role of SUMOylation in the pathogenesis of neurological disease suggests the potential for modulation of SUMOylation with therapeutic effects. Indeed, the generation of pharmacological agents targeting the ubiquitin–proteasome system is particularly encouraging. Future research into the mechanisms, regulation and targets for protein SUMOylation, coupled with targeted screens for chemical modulators of the SUMOylation pathway will no doubt enhance our understanding of this modification and potentially lead to promising compounds for the treatment of the various neurological disorders in which SUMOylation has been implicated.
The growing body of evidence for a role of SUMOylation in the pathogenesis of neurological disease suggests the potential for modulation of SUMOylation with therapeutic effects.
Table I. SUMOylation enzymes.
| SUMO protein | E1 activating enzyme | E2 conjugating enzyme | E3 enzymes | SENP enzymes |
|---|---|---|---|---|
| SUMO1 | PIAS1, PIAS3 | |||
| SUMO2 | SAE1/SAE2 | Ubc9 | PIASxα, PIASxβ, PIASy, RanBP2, Pc2, Mms21, TOPORS, TRAF7 HDAC4 |
SENP1-3 |
| SUMO3 | Complex | SENP5-7 | ||
| SUMO4 |
SUMO, small ubiquitin-related modifier; SAE, SUMO-activating enzyme; Ubc9, ubiquitin conjugating protein 9; PIAS, protein inhibitor of activated STAT; RanBP2, Ran GTPase binding protein 2; TOPORS, topoisomerase I-binding arginine-serine rich protein; Pc2, polycomb protein 2; Mms21, methyl methanesulfonate-sensitive; TRAF7, tumor necrosis factor receptor associated factor 7; HDAC4, histone deacetylase 4; SENP, sentrin protease.
Table II. SUMO substrates and neuropathological disorders.
| Neurological condition | Modified SUMO substrate | Refs. |
|---|---|---|
| Parkinson’s disease | α-Synuclein | 71 |
| Parkin (noncovalent) | 73, 75 | |
| DJ-1 | 78 | |
|
| ||
| Alzheimer’s disease | Amyloid beta | 83, 117 |
| Tau | 71 | |
|
| ||
| Ischemia | Multiple targets | 62, 64, 65 |
|
| ||
| Huntington’s disease | Huntingtin | 93 |
|
| ||
| DRPLA | Atrophin-1 | 96 |
|
| ||
| SBMA | Androgen receptor | 98 |
|
| ||
| SCA-1 | Ataxin-1 | 118 |
|
| ||
| NIID | Unknown | 119 |
SUMO, small ubiquitin-related modifier; DRPLA, dentatorubral pallidoluysian atrophy; SBMA, spinobulbar muscular atrophy; SCA, spinocerebellar ataxia; NIID, neuronal intranuclear inclusion disease.
ACKNOWLEDGEMENTS
Dina Anderson is a Ph.D. student funded by UCB Pharma S.A. We thank Stephane Martin and Helena Cimarosti for critical reading of the manuscript. We are grateful to the Wellcome Trust, BBSRC and the MRC for financial support.
REFERENCES
- 1.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73(1):355–82. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 2.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135(6 Pt 1):1457–70. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88(1):97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 4.Lapenta V, Chiurazzi P, van der Spek P, Pizzuti A, Hanaoka F, Brahe C. SMT3A, a human homologue of the S. cerevisiae SMT3 gene, maps to chromosome 21qter and defines a novel gene family. Genomics. 1997;40(2):362–6. doi: 10.1006/geno.1996.4556. [DOI] [PubMed] [Google Scholar]
- 5.Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D. A M55V poly-morphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J Biol Chem. 2004;279(26):27233–8. doi: 10.1074/jbc.M402273200. [DOI] [PubMed] [Google Scholar]
- 6.Bayer P, Arndt A, Metzger S, et al. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol. 1998;280(2):275–86. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 7.Tatham MH, Jaffray E, Vaughan OA, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276(38):35368–74. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 8.Saitoh H, Hinchey J. Functional hetero-geneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275(9):6252–8. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 9.Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. Embo J. 2002;21(6):1456–64. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann H, Floss S, Stamminger T. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J Virol. 2000;74(6):2510–24. doi: 10.1128/jvi.74.6.2510-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nacerddine K, Lehembre F, Bhaumik M, et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9(6):769–79. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121(Pt 24):4106–13. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- 13.Zhang FP, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Janne OA. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 2008;28(17):5381–90. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278(45):44113–20. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 15.Bencsath KP, Podgorski MS, Pagala VR, Slaughter CA, Schulman BA. Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J Biol Chem. 2002;277(49):47938–45. doi: 10.1074/jbc.M207442200. [DOI] [PubMed] [Google Scholar]
- 16.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33(5):201–8. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337(2):517–20. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- 18.Wei W, Yang P, Pang J, et al. A stress-dependent SUMO4 sumoylation of its substrate proteins. Biochem Biophys Res Commun. 2008;375(3):454–9. doi: 10.1016/j.bbrc.2008.08.028. [DOI] [PubMed] [Google Scholar]
- 19.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8(12):947–56. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 20.Martin S, Wilkinson KA, Nishimune A, Henley JM. Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat Rev Neurosci. 2007;8(12):948–59. doi: 10.1038/nrn2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anckar J, Sistonen L. SUMO: getting it on. Biochem Soc Trans. 2007;35(Pt 6):1409–13. doi: 10.1042/BST0351409. [DOI] [PubMed] [Google Scholar]
- 22.Gong L, Li B, Millas S, Yeh ET. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 1999;448(1):185–9. doi: 10.1016/s0014-5793(99)00367-1. [DOI] [PubMed] [Google Scholar]
- 23.Desterro JM, Thomson J, Hay RT. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417(3):297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 24.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272(43):26799–802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 25.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–72. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 26.Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H. In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem Biophys Res Commun. 1999;254(3):693–8. doi: 10.1006/bbrc.1998.9995. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276(16):12654–9. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 28.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276(24):21664–9. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson KA, Nishimune A, Henley JM. Analysis of SUMO-1 modification of neuronal proteins containing consensus SUMOylation motifs. Neurosci Lett. 2008;436(2):239–44. doi: 10.1016/j.neulet.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435(7042):687–92. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–82. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 32.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32(6):286–95. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi T, Seki M, Maeda D, et al. Ubc9 is essential for viability of higher eukaryotic cells. Exp Cell Res. 2002;280(2):212–21. doi: 10.1006/excr.2002.5634. [DOI] [PubMed] [Google Scholar]
- 34.Kerscher O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8(6):550–5. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hannich JT, Lewis A, Kroetz MB, et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280(6):4102–10. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 36.Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281(23):16117–27. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 37.Minty A, Dumont X, Kaghad M, Caput D. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem. 2000;275(46):36316–23. doi: 10.1074/jbc.M004293200. [DOI] [PubMed] [Google Scholar]
- 38.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101(40):14373–8. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stehmeier P, Muller S. Phospho-Regulated SUMO Interaction Modules Connect the SUMO System to CK2 Signaling. Mol Cell. 2009;33(3):400–9. doi: 10.1016/j.molcel.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi H, Hatakeyama S, Saitoh H, Nakayama KI. Noncovalent SUMO-1 binding activity of thymine DNA glycosylase (TDG) is required for its SUMO-1 modification and colo-calization with the promyelocytic leukemia protein. J Biol Chem. 2005;280(7):5611–21. doi: 10.1074/jbc.M408130200. [DOI] [PubMed] [Google Scholar]
- 41.Pichler A, Knipscheer P, Oberhofer E, et al. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat Struct Mol Biol. 2005;12(3):264–9. doi: 10.1038/nsmb903. [DOI] [PubMed] [Google Scholar]
- 42.Ulrich HD. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. 2005;15(10):525–32. doi: 10.1016/j.tcb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115(5):565–76. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 44.Shalizi A, Gaudilliere B, Yuan Z, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311(5763):1012–7. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 45.Martin S, Nishimune A, Mellor JR, Henley JM. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447(7142):321–5. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25(21):5083–93. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bossis G, Malnou CE, Farras R, et al. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Mol Cell Biol. 2005;25(16):6964–79. doi: 10.1128/MCB.25.16.6964-6979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2(2):233–9. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 49.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21(3):349–57. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 50.Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. A mechanism for inhibiting the SUMO pathway. Mol Cell. 2004;16(4):549–61. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Boggio R, Passafaro A, Chiocca S. Targeting SUMO E1 to ubiquitin ligases: a viral strategy to counteract sumoylation. J Biol Chem. 2007;282(21):15376–82. doi: 10.1074/jbc.M700889200. [DOI] [PubMed] [Google Scholar]
- 52.Knipscheer P, Flotho A, Klug H, et al. Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell. 2008;31(3):371–82. doi: 10.1016/j.molcel.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 53.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–7. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 54.Dorval V, Fraser PE. SUMO on the road to neurodegeneration. Biochim Biophys Acta. 2007;1773(6):694–706. doi: 10.1016/j.bbamcr.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 55.Navascues J, Bengoechea R, Tapia O, Casafont I, Berciano MT, Lafarga M. SUMO-1 transiently localizes to Cajal bodies in mammalian neurons. J Struct Biol. 2008;163(2):137–46. doi: 10.1016/j.jsb.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 56.Sramko M, Markus J, Kabat J, Wolff L, Bies J. Stress-induced inactivation of the c-Myb transcription factor through conjugation of SUMO-2/3 proteins. J Biol Chem. 2006;281(52):40065–75. doi: 10.1074/jbc.M609404200. [DOI] [PubMed] [Google Scholar]
- 57.Wuerzberger-Davis SM, Nakamura Y, Seufzer BJ, Miyamoto S. NF-kappaB activation by combinations of NEMO SUMOylation and ATM activation stresses in the absence of DNA damage. Oncogene. 2007;26(5):641–51. doi: 10.1038/sj.onc.1209815. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J, Goodson ML, Hong Y, Sarge KD. MEL-18 interacts with HSF2 and the SUMO E2 UBC9 to inhibit HSF2 sumoylation. J Biol Chem. 2008;283(12):7464–9. doi: 10.1074/jbc.M707122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee YJ, Hallenbeck JM. Insights into cytoprotection from ground squirrel hibernation, a natural model of tolerance to profound brain oligaemia. Biochem Soc Trans. 2006;34(Pt 6):1295–8. doi: 10.1042/BST0341295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee YJ, Miyake S, Wakita H, et al. Protein SUMOylation is massively increased in hibernation torpor and is critical for the cytoprotection provided by ischemic preconditioning and hypothermia in SHSY5Y cells. J Cereb Blood Flow Metab. 2007;27(5):950–62. doi: 10.1038/sj.jcbfm.9600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang W, Sheng H, Homi HM, Warner DS, Paschen W. Cerebral ischemia/stroke and small ubiquitin-like modifier (SUMO) conjugation - a new target for therapeutic intervention? J Neurochem. 2008;106(3):989–99. doi: 10.1111/j.1471-4159.2008.05404.x. [DOI] [PubMed] [Google Scholar]
- 62.Yang W, Sheng H, Warner DS, Paschen W. Transient global cerebral ischemia induces a massive increase in protein sumoylation. J Cereb Blood Flow Metab. 2008;28(2):269–79. doi: 10.1038/sj.jcbfm.9600523. [DOI] [PubMed] [Google Scholar]
- 63.Yang W, Sheng H, Warner DS, Paschen W. Transient focal cerebral ischemia induces a dramatic activation of small ubiquitin-like modifier conjugation. J Cereb Blood Flow Metab. 2008;28(5):892–6. doi: 10.1038/sj.jcbfm.9600601. [DOI] [PubMed] [Google Scholar]
- 64.Cimarosti H, Lindberg C, Bomholt SF, Ronn LC, Henley JM. Increased protein SUMOylation following focal cerebral ischemia. Neuropharmacology. 2008;54(2):280–9. doi: 10.1016/j.neuropharm.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 65.Lee YJ, Castri P, Bembry J, Maric D, Auh S, Hallenbeck JM. SUMOylation participates in induction of ischemic tolerance. J Neurochem. 2009;109(1):257–67. doi: 10.1111/j.1471-4159.2009.05957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baba M, Nakajo S, Tu PH, et al. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am J Pathol. 1998;152(4):879–84. [PMC free article] [PubMed] [Google Scholar]
- 67.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388(6645):839–40. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 68.Wakabayashi K, Tanji K, Mori F, Takahashi H. The Lewy body in Parkinson’s disease: molecules implicated in the formation and degradation of alpha-synuclein aggregates. Neuropathology. 2007;27(5):494–506. doi: 10.1111/j.1440-1789.2007.00803.x. [DOI] [PubMed] [Google Scholar]
- 69.Chandra S, Gallardo G, Fernandez-Chacon R, Schluter OM, Sudhof TC. Alpha-synuclein cooperates with CSPalpha in preventing neurodegeneration. Cell. 2005;123(3):383–96. doi: 10.1016/j.cell.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 70.Pountney DL, Chegini F, Shen X, Blumbergs PC, Gai WP. SUMO-1 marks subdomains within glial cytoplasmic inclusions of multiple system atrophy. Neurosci Lett. 2005;381(1-2):74–9. doi: 10.1016/j.neulet.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 71.Dorval V, Fraser PE. Small ubiquitin-like modifier (SUMO) modification of natively unfolded proteins tau and alpha-synuclein. J Biol Chem. 2006;281(15):9919–24. doi: 10.1074/jbc.M510127200. [DOI] [PubMed] [Google Scholar]
- 72.Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40(2):427–46. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 73.Um JW, Chung KC. Functional modulation of parkin through physical interaction with SUMO-1. J Neurosci Res. 2006;84(7):1543–54. doi: 10.1002/jnr.21041. [DOI] [PubMed] [Google Scholar]
- 74.Tan EK, Skipper LM. Pathogenic mutations in Parkinson disease. Hum Mutat. 2007;28(7):641–53. doi: 10.1002/humu.20507. [DOI] [PubMed] [Google Scholar]
- 75.Um JW, Min DS, Rhim H, Kim J, Paik SR, Chung KC. Parkin ubiquitinates and promotes the degradation of RanBP2. J Biol Chem. 2006;281(6):3595–603. doi: 10.1074/jbc.M504994200. [DOI] [PubMed] [Google Scholar]
- 76.Taira T, Saito Y, Niki T, Iguchi-Ariga SM, Takahashi K, Ariga H. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004;5(2):213–8. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takahashi K, Taira T, Niki T, Seino C, Iguchi-Ariga SM, Ariga H. DJ-1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor. J Biol Chem. 2001;276(40):37556–63. doi: 10.1074/jbc.M101730200. [DOI] [PubMed] [Google Scholar]
- 78.Shinbo Y, Niki T, Taira T, et al. Proper SUMO-1 conjugation is essential to DJ-1 to exert its full activities. Cell Death Differ. 2006;13(1):96–108. doi: 10.1038/sj.cdd.4401704. [DOI] [PubMed] [Google Scholar]
- 79.Moore DJ, Zhang L, Dawson TM, Dawson VL. A missense mutation (L166P) in DJ-1, linked to familial Parkinson’s disease, confers reduced protein stability and impairs homo-oligomerization. J Neurochem. 2003;87(6):1558–67. doi: 10.1111/j.1471-4159.2003.02265.x. [DOI] [PubMed] [Google Scholar]
- 80.Tiraboschi P, Hansen LA, Thal LJ, Corey-Bloom J. The importance of neuritic plaques and tangles to the development and evolution of AD. Neurology. 2004;62(11):1984–9. doi: 10.1212/01.wnl.0000129697.01779.0a. [DOI] [PubMed] [Google Scholar]
- 81.Wolfe MS. The gamma-secretase complex: membrane-embedded proteolytic ensemble. Biochemistry. 2006;45(26):7931–9. doi: 10.1021/bi060799c. [DOI] [PubMed] [Google Scholar]
- 82.Gocke CB, Yu H, Kang J. Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates. J Biol Chem. 2005;280(6):5004–12. doi: 10.1074/jbc.M411718200. [DOI] [PubMed] [Google Scholar]
- 83.Zhang YQ, Sarge KD. Sumoylation of amyloid precursor protein negatively regulates Abeta aggregate levels. Biochem Biophys Res Commun. 2008;374(4):673–8. doi: 10.1016/j.bbrc.2008.07.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Y, Wang H, Wang S, Quon D, Liu YW, Cordell B. Positive and negative regulation of APP amyloidogenesis by sumoylation. Proc Natl Acad Sci U S A. 2003;100(1):259–64. doi: 10.1073/pnas.0235361100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dorval V, Mazzella MJ, Mathews PM, Hay RT, Fraser PE. Modulation of Abeta generation by small ubiquitin-like modifiers does not require conjugation to target proteins. Biochem J. 2007;404(2):309–16. doi: 10.1042/BJ20061451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goedert M, Wischik CM, Crowther RA, Walker JE, Klug A. Cloning and sequencing of the cDNA encoding a core protein of the paired helical filament of Alzheimer disease: identification as the microtubule-associated protein tau. Proc Natl Acad Sci U S A. 1988;85(11):4051–5. doi: 10.1073/pnas.85.11.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Suh YH, Checler F. Amyloid precursor protein, presenilins, and alpha-synuclein: molecular pathogenesis and pharmacological applications in Alzheimer’s disease. Pharmacol Rev. 2002;54(3):469–525. doi: 10.1124/pr.54.3.469. [DOI] [PubMed] [Google Scholar]
- 88.Taniguchi T, Kawamata T, Mukai H, et al. Phosphorylation of tau is regulated by PKN. J Biol Chem. 2001;276(13):10025–31. doi: 10.1074/jbc.M007427200. [DOI] [PubMed] [Google Scholar]
- 89.Perry G, Friedman R, Shaw G, Chau V. Ubiquitin is detected in neurofibrillary tangles and senile plaque neurites of Alzheimer disease brains. Proc Natl Acad Sci U S A. 1987;84(9):3033–6. doi: 10.1073/pnas.84.9.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takahashi K, Ishida M, Komano H, Takahashi H. SUMO-1 immunoreactivity colocalizes with phospho-Tau in APP transgenic mice but not in mutant Tau transgenic mice. Neurosci Lett. 2008;441(1):90–3. doi: 10.1016/j.neulet.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 91.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6(10):743–55. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 92.Gil JM, Rego AC. Mechanisms of neurodegeneration in Huntington’s disease. Eur J Neurosci. 2008;27(11):2803–20. doi: 10.1111/j.1460-9568.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- 93.Steffan JS, Agrawal N, Pallos J, et al. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304(5667):100–4. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 94.Schilling G, Wood JD, Duan K, et al. Nuclear accumulation of truncated atrophin-1 fragments in a transgenic mouse model of DRPLA. Neuron. 1999;24(1):275–86. doi: 10.1016/s0896-6273(00)80839-9. [DOI] [PubMed] [Google Scholar]
- 95.Yazawa I, Nukina N, Hashida H, Goto J, Yamada M, Kanazawa I. Abnormal gene product identified in hereditary dentatorubral-pallidoluysian atrophy (DRPLA) brain. Nat Genet. 1995;10(1):99–103. doi: 10.1038/ng0595-99. [DOI] [PubMed] [Google Scholar]
- 96.Terashima T, Kawai H, Fujitani M, Maeda K, Yasuda H. SUMO-1 co-localized with mutant atrophin-1 with expanded polyglutamines accelerates intranuclear aggregation and cell death. Neuroreport. 2002;13(17):2359–64. doi: 10.1097/00001756-200212030-00038. [DOI] [PubMed] [Google Scholar]
- 97.Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc Natl Acad Sci U S A. 2000;97(26):14145–50. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chan HY, Warrick JM, Andriola I, Merry D, Bonini NM. Genetic modulation of polyglutamine toxicity by protein conjugation pathways in Drosophila. Hum Mol Genet. 2002;11(23):2895–904. doi: 10.1093/hmg/11.23.2895. [DOI] [PubMed] [Google Scholar]
- 99.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–47. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 100.Riley BE, Zoghbi HY, Orr HT. SUMOylation of the polyglutamine repeat protein, ataxin-1, is dependent on a functional nuclear localization signal. J Biol Chem. 2005;280(23):21942–8. doi: 10.1074/jbc.M501677200. [DOI] [PubMed] [Google Scholar]
- 101.Lieberman AP, Robitaille Y, Trojanowski JQ, Dickson DW, Fischbeck KH. Polyglutamine-containing aggregates in neuronal intranuclear inclusion disease. Lancet. 1998;351(9106):884. doi: 10.1016/S0140-6736(05)70296-8. [DOI] [PubMed] [Google Scholar]
- 102.McFadden K, Hamilton RL, Insalaco SJ, et al. Neuronal intranuclear inclusion disease without polyglutamine inclusions in a child. J Neuropathol Exp Neurol. 2005;64(6):545–52. doi: 10.1093/jnen/64.6.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Takahashi-Fujigasaki J, Arai K, Funata N, Fujigasaki H. SUMOylation substrates in neuronal intranuclear inclusion disease. Neuropathol Appl Neurobiol. 2006;32(1):92–100. doi: 10.1111/j.1365-2990.2005.00705.x. [DOI] [PubMed] [Google Scholar]
- 104.Pountney DL, Raftery MJ, Chegini F, Blumbergs PC, Gai WP. NSF, Unc-18-1, dynamin-1 and HSP90 are inclusion body components in neuronal intranuclear inclusion disease identified by anti-SUMO-1-immunocapture. Acta Neuropathol. 2008;116(6):603–14. doi: 10.1007/s00401-008-0437-4. [DOI] [PubMed] [Google Scholar]
- 105.David G, Neptune MA, DePinho RA. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J Biol Chem. 2002;277(26):23658–63. doi: 10.1074/jbc.M203690200. [DOI] [PubMed] [Google Scholar]
- 106.Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005;25(19):8456–64. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5(7):596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 108.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. Embo J. 2005;24(3):439–51. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13(6):491–9. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 110.Capili AD, Lima CD. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J Mol Biol. 2007;369(3):608–18. doi: 10.1016/j.jmb.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guedat P, Colland F. Patented small molecule inhibitors in the ubiquitin proteasome system. BMC Biochem. 2007;8(Suppl 1):S14. doi: 10.1186/1471-2091-8-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5(3):172–83. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 113.Reverter D, Lima CD. Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat Struct Mol Biol. 2006;13(12):1060–8. doi: 10.1038/nsmb1168. [DOI] [PubMed] [Google Scholar]
- 114.Shen L, Tatham MH, Dong C, Zagorska A, Naismith JH, Hay RT. SUMO protease SENP1 induces isomerization of the scissile peptide bond. Nat Struct Mol Biol. 2006;13(12):1069–77. doi: 10.1038/nsmb1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Issaeva N, Bozko P, Enge M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10(12):1321–8. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 116.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 117.Li J, Fici GJ, Mao CA, et al. Positive and negative regulation of the gamma-secretase activity by nicastrin in a murine model. J Biol Chem. 2003;278(35):33445–9. doi: 10.1074/jbc.M301288200. [DOI] [PubMed] [Google Scholar]
- 118.Ueda H, Goto J, Hashida H, et al. Enhanced SUMOylation in polyglutamine diseases. Biochem Biophys Res Commun. 2002;293(1):307–13. doi: 10.1016/S0006-291X(02)00211-5. [DOI] [PubMed] [Google Scholar]
- 119.Pountney DL, Huang Y, Burns RJ, et al. SUMO-1 marks the nuclear inclusions in familial neuronal intranuclear inclusion disease. Exp Neurol. 2003;184(1):436–46. doi: 10.1016/j.expneurol.2003.07.004. [DOI] [PubMed] [Google Scholar]