Abstract
All juvenile NOD mice exhibit insulitis, but there is substantial variation in their progression to diabetes. We demonstrate that a patient-validated magnetic-resonance-imaging (MRI) strategy to non-invasively visualize local effects of pancreatic-islet inflammation can predict diabetes onset in NOD mice. MRI signals acquired during a narrow early time-window allowed pre-sorting into disease-progressors and -nonprogressors and an estimate of time-to-diabetes. We exploited this capability to identify novel elements correlated with disease protection, including CRIg (complement receptor of the immunoglobulin superfamily), which marked a subset of macrophages associated with diabetes resistance. Administration of CRIg-Fc depressed MRI signals and diabetes incidence. In addition to identifying regulators of disease progression, this study shows that diabetes is set at an early age in NOD mice.
Type-1 diabetes (T1D) is an organ-specific autoimmune disease characterized by specific destruction of the insulin-producing beta cells of the pancreatic islets of Langerhans (reviewed in 1). Autoimmune diabetes is driven primarily by T lymphocytes that respond to beta-cell-specific antigens, but other populations of immune system cells can also play an important enhancing or dampening role, notably B lymphocytes, natural killer cells, macrophages (MFs) and dendritic cells (DCs). Disease has two major checkpoints in most mouse models: initiation of insulitis at 2–5 weeks of age; and, with a highly variable delay and penetrance, conversion of insulitis to overt diabetes 2. Whether these same two checkpoints exist in human T1D has been the subject of discussion, but there have been indications that they do.
This and other issues underline our relative ignorance about the pathogenesis of autoimmune diabetes, particularly in humans. A major hindrance in both experimental and clinical contexts has been the inability to directly visualize the onset and progression of islet attack. Experimentally: investigators are constantly challenged by the scatter in disease parameters exhibited by individual animals, even those presumed to be genetically and environmentally homogeneous. For example, a particular 15-week-old NOD mouse with advanced insulitis may develop clinical diabetes in days, weeks or months, or never. Clinically: beyond the same problem of disease heterogeneity, a diagnosis of overt diabetes generally implies that most of a patient’s beta cells have already been destroyed or disabled, meaning that the early, autoimmune, phases of the process have already played out, and that strategies for disease reversal are severely limited.
In recognition of this impediment, much effort has been devoted to identifying biomarkers that indirectly signal at least the major landmarks of T1D pathogenesis. So far, serum autoantibody (autoAb) titers seem to be the best indicators of islet attack, nicely confirmed in the Diabetes Prevention Trial-Type 1 (reviewed in 3). The appearance of autoAb specificities against at least one beta cell antigen is generally considered to reflect insulitis; emergence of specificities against two or more predicts eventual conversion of insulitis to diabetes. Plasma C-peptide concentrations are still the most wildly accepted indicator of beta cell activity: secreted in equimolar concentration with insulin, C-peptide is a semi-quantitative marker of beta-cell secretion – in contrast to insulin, a marker, not subject to hepatic extraction 4. Although following autoantibody titers and C-peptide amounts certainly provides useful information, a non-invasive means of more directly following disease progression would have several important experimental and clinical applications. There are two fundamental needs: an assessment of islet infiltration, as a read-out of ongoing autoimmune attack; and quantification of beta cell mass or function, as an indicator of cumulative islet destruction. Although substantial effort has addressed these goals, no clinically reliable method is currently in place (reviewed in 5).
A non-invasive, real-time, whole-mouse imaging method was successfully used for visualizing vascular and inflammatory-cell changes that signal disease checkpoints in mouse models of T1D 6, 7. Insulitis is essentially an inflammation of the pancreatic islets and, as such, is accompanied by a range of microvascular alterations – including modifications of endothelial cells, vascular swelling and leakage, increased islet blood flow and edema 8–10 – as well as by influxes of inflammatory cell populations, in particular early infiltration of MFs 11. A novel method entailing magnetic resonance imaging (MRI) of magnetic nanoparticles (MNP) was sensitive to such changes, permitting disease progression to be monitored, genetically determined variability in disease to be quantified, imminent onset of diabetes signaled, and immunotherapy-induced disease reversal very early on 6, 7. Importantly, a small clinical trial on recent-onset T1D patients has recently validated the use of this MRI-MNP approach in the human context 12.
Here, we exploited MRI-MNP as a prediction tool in the NOD mouse model of T1D. Surprisingly, optimum predictability occurred during a narrow time-window between six and ten weeks of age, long before eventual conversion of insulitis to overt diabetes. MRI-MNP sorting of 6–10 week-old insulitic mice that should or should not go on to develop diabetes permitted us to identify molecules and cells associated with aggressive versus innocuous insulitis, which uncovered a novel pathway important in regulating diabetes progression.
RESULTS
Predicting conversion of insulitis to diabetes
Regardless of gender, all individuals in our NOD mouse colony show insulitis starting at about 3–5 weeks after birth. But overt diabetes develops in only 60–80% of females and 10–20% of males, anytime from 15–30 weeks of age. This heterogeneity in disease penetrance and kinetics compromises the accuracy of mechanistic explorations because we do not know for a given individual if and when it will eventually convert to diabetes. Thus, we set out to develop a non-invasive method to predict diabetes onset in mouse models.
Our previously described MRI-MNP technique tracks a long-circulating, dextran-coated, monocrystalline, super-paramagnetic, iron-oxide probe (MION-47) that can be detected by high-resolution MRI 6, 7. Building on these past studies, the basic protocol for the present experiments was to anesthetize groups of mice and obtain a baseline MRI scan (pre-MNP); to inject them intravenously (iv) with MNP and immediately obtain a second image (post-MNP, an indicator of vascular volume); and to perform a third scan 24 hours later (24h-post, a reflection of probe accumulation). Regions of interest (ROIs) were drawn over the pancreas and control muscle tissue (Fig. 1a, top left), and a T2 value for each organ calculated (Fig. 1a, top right), the presence of MNP within the organ diminishing its T2 value. Probe accumulation, a function of vascular leakage and macrophage uptake 7, was calculated using the T2pre-MNP and T224hr-post values (Fig. 1a, bottom).
Figure 1.
Prediction of T1D in NOD mice by MRI-MNP: cross-sectional cohort. (a) MNP-MRI methodology. Top left: A representative T2-weighted MRI image (coronal view). M, muscle; P, pancreas; Sp, spleen; St, stomach. Top right: Pancreas and muscle T2 values were calculated using T2 Fit-map plug-ins from OsiriX software. A screenshot of one such Fit-map is shown. Te, echo time. Bottom: Pancreatic and muscle T2 values measured at the three standard time-points (pre-MNP, post-MNP, 24hr post-MNP) for a representative mouse. Probe accumulation value was calculated as ln(T2pre-MNP/T224hr post-MNP) (b) A cohort of 22 female NOD mice 10–17 weeks of age was injected with MNP; MRI scans were performed and analyzed as illustrated in panel A and detailed in Experimental Procedures. Animals were followed until 40 weeks of age, and were placed retrospectively into “pre-diabetic” and “non-diabetic” groups that did or did not develop clinical diabetes, respectively. *P=0.02; **P=0.0023. P value was determined by the Student’s t test (Prism5, Graphpad Inc.) (c) Correlation between baseline T2 and probe accumulation values. R2=0.8397, P<0.0001. P value was determined by the F test (Prism5, Graphpad Inc.). Throughout the figure: ms, microsecond; AU, arbitrary units; NS, not significant.
Both cross-sectional and longitudinal imaging designs were explored. For the cross-sectional protocol, twenty-two NOD females ranging in age from 10 to 17 weeks were injected with MNP, pancreata and control muscles were triple-scanned as described above, and the mice were monitored for signs of diabetes until 40 weeks of age. Animals were retrospectively assigned to two groups: thirteen that eventually developed diabetes (which we have termed “pre-diabetic”) and nine that did not (termed “non-diabetic”). Pancreas and muscle T2 signals for individual mice are presented (Supplementary Fig.1), and the corresponding baseline T2pre-MNP and probe accumulation values are plotted (Fig. 1b). Two substantial differences between pre-diabetic and non-diabetic animals were observed. First, the baseline T2 signals were higher for the pancreas, but not muscle, of individuals that went on to develop diabetes (Fig. 1b, top panels), suggesting the existence of edema measurable by MRI even in the absence of MNP. Second, probe accumulation was also higher for the pancreas, but not muscle, of pre-diabetic compared with non-diabetic mice (Fig. 1b, bottom panels). There was an excellent correlation between the pancreatic T2pre-MNP and probe accumulation values across the whole cohort (Fig. 1c).
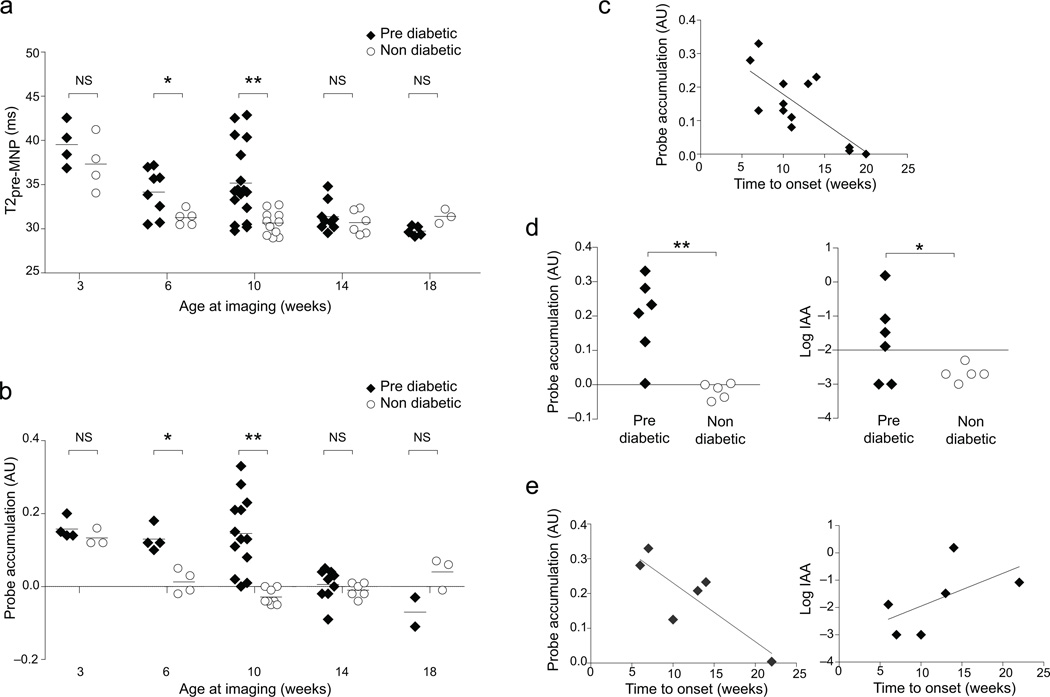
Next, we addressed how early in the course of the NOD disease our MRI-MNP approach was capable of predicting eventual onset of T1D. For this longitudinal design, synchronous cohorts of juvenile mice were sequentially scanned every 3–4 weeks until 18 weeks of age (as detailed in the legend to Fig. 2), an interval known to be adequate for probe wash-out 13. Animals were monitored for signs of diabetes until they were 40 weeks old, permitting retrospective assignment into non-diabetes and pre-diabetes groups and, for the latter, calculation of time-to-diabetes values. Surprisingly, the MRI-MNP signals of 6- and 10-week-old mice were the most predictive (Fig. 2a, b), especially the probe accumulation values at 10 weeks, which were essentially non-overlapping for non-diabetic and pre-diabetic individuals (Fig. 2b). Plots of probe-accumulation and blood-glucose values for individual animals over time solidified this observation (Supplementary Fig. 2a). There was also a significant correlation between the amount of probe accumulation and the time to diabetes onset (Fig. 2c). After 10 weeks, animals with either disease status exhibited relatively low baseline T2 and probe accumulation signals, consistent with our previous observations 6. Before 6 weeks, both non-diabetic and pre-diabetic animals showed relatively high values, likely reflecting the immature condition of the vasculature, with incomplete closure of endothelial cell junctions 14. High pancreatic MRI signals at this time-point do not reflect insulitis because like values were obtained for 3-week-old Eα16/NOD mice, which lack both insulitis and diabetes 15 (Supplementary Fig. 2b, leftmost panel). Nor are they a specific feature of the NOD strain as C57Bl/6 (B6) mice showed similar values at this young age (Supplementary Fig. 2b, left-center panel). In both Eα16/NOD and B6 animals, the elevated imaging signals at 3 weeks dropped quickly, already by 6–7 weeks, and remained low thereafter. The same pattern was seen in a group of male NOD mice, which universally manifest insulitis while very few develop diabetes (Supplementary Fig. 2b, rightmost panel).
Figure 2.
Prediction of T1D in NOD mice: longitudinal cohorts. Cohorts of 3-week-old female NOD mice were MNP injected and imaged as per Figure. 1, and then were re-injected and re-imaged every 3–4 weeks until 18 weeks of age. Individuals were followed until 40 weeks, permitting retroactive assignment into groups that did (pre-diabetic) or did not (non-diabetic) develop clinical diabetes. (a) Pancreatic T2pre-MNP values over time. *P=0.0194; **P=0.0006. (b) Pancreatic probe accumulation values. *P=0.0037; **P=0.0001. (c) Correlation between pancreatic probe accumulation values measured at 10 weeks and the time-to-onset of diabetes (n=13). R2=0.5557, P=0.0034. (d,e) Comparison of success of MNP-MRI (left) and insulin autoantibody (IAA) (right) assays in T1D prediction. A small cohort of 10-week-old mice was assayed by both of these methods, and was then followed for diabetes development until 40 weeks of age. (d) 10 week values for individual mice. *P=0.1167; **P=0.0031. (e) Correlation between values at 10 weeks and time to diabetes onset. Left, R2=0.7073, P=0.0359. Right, R2=0.3301, P=0.0645. Each symbol represents one mouse; bars indicate the group mean. P values were determined by Student’s t test (a,b,d) or F test (c,e). ms, microsecond; AU, arbitrary units; NS, not significant.
We compared the ability of our MRI-MNP procedure and a previously reported assay of anti-insulin autoAb (IAA) titers 16 to predict conversion of insulitis to diabetes in a cohort of NOD mice. While the number of animals examined was not large, the MRI-MNP approach showed higher significance with regards to prediction of both eventual diabetes development and time-to-diabetes (Fig. 2d, e).
Thus, the MRI-MNP method is capable of distinguishing diabetes progressors from non-progressors months before clinical manifestation of disease, and also allows an estimation of time to diabetes onset for pre-diabetic individuals. This means, first of all, that eventual conversion of insulitis to diabetes is set at a young age and, secondly, that it is possible to explore what molecular and cellular processes predicate this conversion.
Over-expression of myeloid-lineage-associated genes
To exploit the new possibilities offered by being able to select a given 6–10 week-old NOD mouse and predict with quite good accuracy whether or not it will go on to develop diabetes, we first followed an unbiased, hypothesis-generating approach, performing microarray-based gene-expression profiling on pancreas-residing leukocytes from pre-diabetic and non-diabetic mice. A cohort of five females was MRI-scanned at 10 weeks of age, their pancreas was removed immediately after final imaging, and CD45+ cells were sorted from the dispersed tissue for microarray analysis of gene expression. We were interested in genes whose transcript levels were positively or negatively correlated with these signals, as they stand to be diabetes-promoting or diabetes-protective, respectively. The Pearson correlation coefficient between the normalized expression value of each gene and the T2pre-MNP signals for the five individual mice was calculated. As an illustration, the expression profiles for the ten most positively or negatively correlated genes were superimposed on the T2pre-MNP histogram (Fig. 3a). Overall, the correlation coefficients of all genes on the microarray were distributed across the spectrum (Fig. 3b, blue bars), but with a bias for those whose transcription levels negatively correlated with T2pre-MNP signals, in comparison with the null hypothesis distribution of a randomized permutation of the expression data (red dotted line). This bias was particularly marked for those transcripts with the lowest correlation coefficients (<= −0.8), and thus associated with resistance, contrasting with earlier studies of aggressive diabetes where the dominant signatures were those associated with inflammation 17. Therefore, we focused the subsequent analyses on the 100 most negatively correlated “protective” genes, which are listed in Supplementary Table 1.
Figure 3.
Gene-expression profiling of pancreas-resident hematopoietic cells as a function of MRI signal. 10-week-old NOD females were MRI-scanned, the pancreas was excised and its cells dispersed, and gene expression profiling was performed on purified CD45+ cells. Gene expression values were correlated to the T2 values across all mice. (a) Normalized expression profiles of genes whose expression was most positively or negatively correlated with T2 values (each line represents an individual gene), super-imposed on the T2pre-MNP values for the mice (blue bars). (b) Distribution of expression/T2pre-MNP correlation coefficients for all genes (blue histogram). The red dotted line shows the distribution of correlation coefficients observed in a randomized dataset. (c) Cell-type-distribution analysis. Normalized expression values of the 100 genes most negatively correlated to T2 were examined for a panel of immune cells – data retrieved from the ImmGen database 34. T.4Nve.Sp, T.8Nve.Sp, spleen naïve CD4 and CD8+ T cells; T.4FP3+25+.Sp, spleen natural Treg cells; B.T1.Sp, B.T2.Sp, B.T3.Sp, spleen transitional B cells (stages 1, 2, 3); NK, natural killer cells; DC.4+.Sp, DC.8+.Sp, DC.4-8-11b+.Sp, DC.4-8-11b-.Sp, spleen CD4+, CD8+ CD4/8-double negative CD11b+ and CD4/8/11b-triple negative DCs; Mo.6C+II-.Bl, blood Ly6C+ MHC II− monocytes; MF, macrophages; Lu, lung, RP, spleen red pulp; MF.II-480hi.PC, peritoneal MHC II−F4/80hi macrophages.
Changes in transcript levels in the CD45+ cell populations of the different mice could reflect alterations in the composition of the leukocytic infiltrate and/or modifications in gene expression programs. To bioinformatically address this issue, we generated a cell distribution profile, plotting for each of the 100 genes its normalized expression value in a panel of cell-types, exploiting datasets available from the ImmGen project (www.immgen.org) (Fig. 3c). The set of genes whose transcript levels negatively correlated with the T2pre-MNP values were characteristic of cells of the myeloid lineage, in particular tissue-resident MFs.
The over-representation of genes characteristic of cells of the myeloid lineage prompted us to explore the cellular composition of the insulitic lesion via histology and flow cytometry (Fig. 4 and Supplementary Fig. 3). These analyses were performed on 10-week-old, MRI-scanned NOD female mice. Insulitis was less extensive in the mice with lower MRI signals, ie those predicted to be refractory to diabetes (Supplementary Fig. 3a), which was also reflected in a reduced number of leukocytes infiltrating the pancreas (Supplementary Fig.3b, left), largely due to decreased numbers of CD3+ T cells (Supplementary Fig.3b, middle). There were no other obvious differences in the histological presentation of the insulitic lesions at this age (Supplementary Fig.3b, right and data not shown).
Figure 4.
Myeloid-lineage cell subsets infiltrating the pancreas at 10 weeks. Twenty female NOD mice were MRI-scanned at 10 weeks of age, whereupon they were euthanized, the pancreas isolated and dispersed, and cells analyzed by flow cytometry after staining with antibodies recognizing: CD45, CD11b, Ly6C, CD11c, and Gr-1. (a) Myeloid cell gating strategy: cells were pre-gated as CD45+DAPI−, and were secondarily separated according to display of CD11b and Ly6C; the Ly6C− subset was further split according to expression of Gr1 and CD11c. R1, Ly6Chi monocytes; R2, Ly6Clo monocytes; R3, MFs; R5, CD11chi DCs; R6, CD11clo pre-DCs; R7, neutrophils 35, 36. (b) Correlation between the percentage of CD11b+ in CD45+ cells and the pancreatic T2pre-MNP value. R2=0.2392, P=0.0287. (c) Correlation between the fraction of CD45+ cells comprised by each subset of myeloid cells and the pancreatic T2pre-MNP value. The R1–R3 and R5–R7 populations were those delineated in panel A. Solid rectangles highlight those plots with statistically significant P values, determined by F test.
On the other hand, a careful flow cytometric analysis of myeloid cell populations did yield some interesting associations with MRI signals. Our gating strategy (Fig 4a), distinguished between the major CD11b+ myeloid cell populations: two subsets of monocytes [Ly6Chi (R1) and Ly6Clo (R2)], mature MFs (R3), two DC subsets [CD11chi (R5) and CD11clo (R6)] and neutrophils (R7). Of note, F4/80, a marker for mature MFs, was detected only on cells within the R3 gate (Supplementary Fig. 4). It is worth mentioning that the available anti-GR1 reagent (RB6-8C5) recognizes only Ly6G on neutrophils in the NOD strain, unlike in other strains, where it also sees Ly6C on monocyte populations (data not shown). From a general perspective, there was a negative correlation between total pancreatic myeloid cells (CD11b+) and the MRI values (Fig. 4b); this increased fraction was accompanied by a decreased fraction of T cells (CD3+) in mice refractory to diabetes (Supplementary Fig. 3c). More specifically: the pancreatic infiltrates of all mice included a number of myeloid cell populations – in most cases, there was no evident correlation between their representation and MRI values; however, the fraction of Ly6Chi and of Ly6Clo CD11b+ monocytes exhibited a significant negative correlation with T2pre-MNP values, indicating an enrichment of these subsets in diabetes-resistant NOD mice (Fig. 4c).
Thus, data from gene-expression and cell-subset profiling converge to highlight increases in numbers and/or function of certain subsets of myeloid-lineage cells in 10-week-old NOD mice that exhibit insulitis but are refractory to clinical diabetes.
MRI signals versus percentage CRIg+ MFs
Pathway analysis of the subset of genes whose expression levels exhibited the strongest negative correlation with baseline T2 values, ie were best correlated with diabetes resistance, revealed enrichments of several functional classes characteristic of the monocyte-MF lineage: complement pathway components, scavenger receptors, phagocytosis-related proteins, and eicosanoids (Fig. 5a). Our attention was drawn in particular to a recently identified complement pathway component, CRIg, because it is expressed exclusively on tissue-resident MFs 18–22, it has been reported to inhibit T cell activation, proliferation and cytokine production 19, 23, 24, and Fc fusions can modulate autoimmune/inflammatory diseases 24–26. Crig transcript levels in pancreatic CD45+ cells showed a strong negative correlation with T2pre-MNP signals (Fig. 5b). Transcripts were most abundant in CD11b+ myeloid-lineage cells (Fig. 5c), and CRIg protein was displayed on a population of fully mature MFs (Fig. 5d), which could best be distinguished as CD11bhiF4/80hi (Supplementary Fig.4). As a whole, the mature MF subset within the pancreatic infiltrate showed no correlation, positive or negative, with MRI signals (Fig. 4c); however, analysis of a new cohort of mice revealed a strong negative correlation between the fraction of CD11b+F4/80+ mature MFs that were CRIg+ and baseline T2 values (Fig. 5e,f). CRIg+F4/80+ cells could also be evidenced histologically, localized mainly at the periphery of the insulitic lesion (Supplementary Fig. 5a), often in close association with CD31+ vascular endothelial cells (Supplementary Fig. 5b). Of note, CRIg+ MFs were also seen in the pancreas of Eα16/NOD mice (Supplementary Fig. 6), which are completely free of insultis and diabetes 15, pointing to the normal tissue residence of CRIg+ MFs, as has been observed in several other contexts 18, 20, 22.
Figure 5.
Enrichment of CRIg+ tissue macrophages in pancreas of NOD mice protected from diabetes. (a) Genes associated with diabetes protection, i.e., strongly anti-correlated with pancreatic T2pre-MNP values fall into distinct functional classes. Gray bars represent the correlation coefficient between transcript level and pancreatic T2pre-MNP signal. (b) Correlation between pancreatic T2pre-MNP values (dotted line) and Crig transcript levels (solid line). Determined from the same microarray dataset as in Fig. 3. (c) Crig expression in different cell types isolated from the pancreas of 10-week-old NOD mice was quantified by quantitative RT-PCR. (d) CRIg expression on pancreatic myeloid-lineage cells from 10-week-old NOD mice. R1–R4 correspond to the regions gated in the left panel. (e,f) Strong negative correlation between the fraction of CRIg+ macrophages in the pancreatic lesion and the T2pre-MNP signal from a cohort of 10-week-old NOD females independent of those used for Fig. 4. (e) The value at the upper right of each panel represents the fraction of cells that were CRIg+ in the CD45+DAPI− CD11b+F4/80+ gate, ie macrophages. CRIg-positivity was set by comparison with an isotype-matched control mAb. (f) The correlation between MRI signals and % of CRIg+ macrophages in the pancreas for this cohort. R2=0.9212, P=0.0096.
Hence, the MRI-MNP signals of young NOD mice, a biomarker of diabetes development, can be used to nominate candidate molecules, pathways or cells associated with disease protection.
CRIg modulates MRI signals and diabetes
Next, we explored whether the anti-correlation of pancreatic CRIg expression and MRI signals in juvenile NOD mice had functional relevance. First, we verified that a CRIg-Fc fusion protein could suppress the proliferation of NOD T cells when cultured in vitro, in a dose-dependent manner (Supplementary Fig. 7), as has been reported in other contexts for other mouse strains 19, 23, 24. To determine whether CRIg expression by pancreas-resident MFs might have a causal relationship with MRI signal intensity and eventual diabetes development, we injected cohorts of female NOD mice with either CRIg-Fc or a control Fc fusion protein from 3–10 weeks of age, an interval that spans the initiation and establishment of insulitis. MRIs were taken at 7–8 weeks of age, the predictive window, and diabetes development was monitored from 10–32 weeks (protocol schematized in Fig. 6a). As expected, the range of probe-accumulation values for mice administered control-Fc proteins spanned values typical of non-diabetic (−0.05 to 0.05) and pre-diabetic (0 to 0.33) mice at that age (Fig. 6b, left). In clear contrast, the values for mice given CRIg-Fc protein were almost all within the range expected of non-diabetic mice (Fig. 6b, left); a significant difference between mice treated with CRIg-Fc versus control-Fc was also seen when T2pre-MNP values were plotted (Fig. 6b, right). As would be predicted from such a reduction in MRI signals, diabetes development was significantly delayed and less frequent (20% versus 67%) in mice that had been injected with CRIg-Fc vs control-Fc protein (Fig. 6c).
Figure 6.
Inhibition of pancreatic MRI-MNP signal and protection from diabetes after CRIg-Fc treatment of NOD female mice. (a) Experimental design: 3-week-old female NOD mice were ip-injected with CRIg-Fc or control-Fc twice a week until 10 weeks of age. They were MRI-MNP screened at 7–8 weeks and followed for diabetes development until 32 weeks. (b) Probe accumulation (left) and pancreatic T2 pre-MNP values (right) and in CRIg-Fc treated mice (n=10) compared with control-Fc treated mice (n=9). *P=0.0012; **P=0.0001. (c) Diabetes development in the same cohort. *P=0.0196. P value was determined by the Gehan-Breslow-Wilcoxon test (Prism5, Graphpad Inc.).
Therefore, CRIg and CRIg+ MFs can be considered newly identified players in T1D pathogenesis, protective elements whose enhancement has potential therapeutic benefit.
DISCUSSION
Our previous studies demonstrated that MRI-MNP can be used to monitor the unfolding of diabetes in NOD mice, quantify genetically determined variability in disease severity in derivative models, foretell imminent onset of clinical diabetes, and monitor disease reversal subsequent to immunotherapy 6, 7. More recently, the approach was translated to humans, successfully discriminating normal controls from patients recently diagnosed with T1D, and opening the door to non-invasive monitoring of response to immunotherapy in clinical trials 12. Here, we focused on optimizing MRI-MNP as a prediction tool in the NOD model. The most striking finding was that MRI signals (T2pre-MNP and, even better, probe accumulation values) measured during a discrete early time-window distinguished between mice that would or would not go on to develop clinical diabetes and permitted an estimation of time to onset. This observation has important theoretical and practical ramifications.
First, the fact that it is already set at 6–10 weeks of age whether or not a given NOD mouse will eventually develop overt diabetes (usually 2–5 months later) has implications for reigning theories of T1D pathogenesis, at least as concerns the NOD mouse model. Scenarios that require a “second-hit” for progression from insulitis to diabetes – for example, some microbial, dietary or stress-related insult – appear to be inconsistent with such an early window of determinism. Also seemingly disfavored are scenarios calling for delayed stochastic generation of the ultimately pathogenic T or B cell specificities. Rather, our findings argue that some early event predicates clinical diabetes development in a fraction of the NOD mouse colony – some event that occurs before 6 weeks of age – but that regulatory processes keep the insulitic lesion in check for variable lengths of time in different individuals. This conclusion is consistent with a report that early development of IAAs could predict eventual diabetes development in NOD mice 16, though later studies seemed not to support the generality of this finding 27.
The practical implication of the MRI-MNP data is that we are now able to sort individual young NOD mice into those that should or should not progress to clinical T1D. Besides permitting more meaningful interpretations of results from diabetes prevention trials, this ability allows us to address pathogenetic mechanisms in a much more accurate manner. Mechanistic dissections in the NOD model have always been compromised by the heterogeneity in disease penetrance and kinetics; MNP-MRI signals will allow us to correlate chosen experimental parameters with the likelihood and proximity of diabetes onset. While the current fairly high cost of this technique precludes high-throughput use, we anticipate price reductions in time, as is often seen with sophisticated technologies of this nature. Exploiting this approach, we could identify specific immune cells and molecules in the islet infiltrate of 10 week-old NOD mice whose representation was correlated with protection from clinical diabetes.
Both the molecular and cellular analyses pointed to a role for cells of the myeloid lineage in disease attenuation. The set of loci whose expression levels in islet-infiltrating leukocytes had the strongest negative correlation with MRI signals (ie high levels reflected a low probability of progression to diabetes) was highly enriched for genes expressed by monocytes and MFs, in particular tissue-resident MFs. Similarly, the only cell-types with a significant negative correlation with MRI signals were Ly6Chi monocytes, Ly6Clo monocytes and CRIg+ tissue-resident MFs. A regulatory role for cells of the myeloid lineage would be consistent with the growing number of this lineage’s members recognized to have immunosuppressive activity 28, 29. As concerns the diabetes context, it was reported long ago that MFs are the earliest detectable hematopoietic cells in the islets of NOD mice and certain other T1D models 11, and it was generally assumed that they promote insulitis and its progession to diabetes. However, from today’s vantage point, it is not known what particular subsets of monocyte-MFs participate in the insulitic lesion, at what specific disease stages, and whether they have a positive or negative role in diabetes progression.
Of the set of transcripts enriched is the islet infiltrates of diabetes-nonprogressors, we were particularly intrigued by those encoding CRIg because this molecule is expressed exclusively by tissue-resident MFs 18–22, can inhibit T cell activation, proliferation and cytokine production 19, 23, 24, and derivative reagents modulate autoimmune/inflammatory diseases, notably experimental arthritis and uveitis 24–26. Indeed, CRIg proved relevant in the current context: it marked a specific subset of MFs whose representation in the insulitic infiltrate showed a significant negative correlation with 10-week MRI values and most importantly, administration of a CRIg-Fc fusion protein both lowered the MRI values and inhibited development of T1D. How might CRIg+ MFs, CRIg and CRIg-Fc be operating to control progression to overt diabetes? To date, CRIg has been ascribed three functions (reviewed in 30): in the phagocytosis of particles, microbes and cells, via binding to C3b and iC3b; as an inhibitor of the alternative pathway of complement, through blockade of the C3 and C5 convertases; and as a T cell suppressor, reflecting its membership in the B7 family of co-stimulatory/co-inhibitory receptors. That CRIg’s activity as a negative regulator of T cell activation is important in the diabetes context is supported by the reduced fraction of CD3+ T cells observed in the insulitic lesions of non-progressor NOD mice. According to such a scenario, then, CRIg-Fc would deliver a negative signal directly to T cells. CRIg, as a B7 homologue, is expressed by APCs, not T cells. So, CRIg-Fc would not be expected to block APC-T cell interactions as, for example, T-cell-displayed CTLA4-Ig does, but rather to engage an as-yet-unidentified ligand on T cells, just like PD-L1.Ig delivers a negative signal directly to T cells by engaging PD-1 31. Indeed, we and others 19 found CRIg-Fc to directly inhibit T cell proliferation in an in vitro assay.
It is also possible that CRIg’s activity in promoting phagocytosis plays a role in keeping diabetes in check in unmanipulated NOD mice. Effective clearing of apoptotic cells, whether engendered by a wave of physiological beta cell death known to take place at 3 weeks of age in rodents (reviewed in 32) or by some microbial, dietary or stress-inducing insult, could promote tolerance over immunity. Of note, the set of transcripts negatively correlated with diabetes development included a number of others encoding proteins involved in opsonization or phagocytosis. For example, Msr1 and CD14 outfit tissue-resident MFs to clear apoptotic cells and dampen inflammation. In addition, transcripts encoding Timd4 (T cell immunoglobulin and mucin domain containing 4), a recently identified phosphatidylserine receptor 33, were up-regulated in diabetes-resistant mice. Timd4 is also highly expressed on CRIg+ macrophages, suggesting a potential molecular basis for recognition of apoptotic cells by CRIg+ MFs. The message would be reinforced by local dampening of pathogenic anti-beta cell T lymphocyte reactivity, as discussed above.
Thus, the studies described herein provide proof-of-principle that the ability to distinguish closely matched NOD diabetes progressors and nonprogressors can yield novel mechanistic insights. A next step will be to apply the MRI-MNP approach to at-risk humans to see whether it can be used as a T1D prediction tool.
METHODS
Mice
NOD/Lt, Eα16/NOD, and C57BL/6 mice were bred under specific-pathogen-free conditions in the animal facility of the Joslin Diabetes Center (JDC). They were cared for in accordance with the ethical guidelines of the JDC and Harvard Medical School (HMS), all studies having been approved by the JDC’s Institutional Animal Care and Use Committee (#99-20). Mice were monitored for diabetes development as described previously 37, with a serum-glucose higher than 350 mg/dl for two consecutive measurements being considered diabetic.
MRI-MNP
The MNP used in these experiments was MION-47, which has a size of about 22 nm and an R2 relaxivity of 69 (mM.sec)-1 at 37°C (0.47 T). Mice were anesthetized by inhalation of isoflurane, and were placed in a birdcage radio-frequency coil with an inner diameter of 38 mm. A 4.7 Tesla microimaging system (PharmaScan; Bruker BioSpin) was used, the imaging protocol including multiple-slice and multiple-echo spin-echo sequences (MSME). 16 different echo times for the MSME scan were used as: 8.68, 17.36, 26.04, 34.72, 43.40, 52.08, 60.76, 69.44, 78.12, 86.80, 95.48,104.16, 112.84, 121.52, 130.20 and 138.88 microsecond (ms). Other parameters include: Tr, 2325 ms; FOV, 4×4; matrix size, 128×128; slice thickness, 0.6 mm interleaved; number of excitations, 4. A total of 16 sequential coronal images was obtained to cover the entire pancreatic region. Before MNP injection, mice were scanned to get a baseline T2 value. Without removing the mouse from the coil, we iv-injected MNP at 20 mg/kg Fe to allow quantification of microvascular changes associated with pancreatic inflammation 7. A third scan was performed 24 hours after MNP administration to permit quantification of probe accumulation, a function of microvascular leakage and macrophage uptake. Image analysis was performed using Osirix software (http://www.osirix-viewer.com) with in-house-developed plug-ins. ROIs for analyses were defined manually on the pancreas or muscle tissue on 3 consecutive slices. To ensure that there were no volume-averaging effects with adjacent organs on calculated T2 values, the ROIs were propagated to adjacent slices, and were modified as needed such that the windows on original and adjacent slices contained only the tissue of interest. T2 values for individual organs were calculated by fitting a standard exponential relaxation model to the data averaged over the entire ROI on each slice. The quantitative probe accumulation was calculated as [ln(T2pre-MNP/T224hr post-MNP)] 7. Analysis was performed independently by two blinded scorers.
Microarrays
After MRI of experimental mice, pancreata were collected and digested at 37°C for 30 min with collagenase IV (1 mg/ml) and DNase I (10 U/ml). Single-cell suspensions were prepared, stained, and CD45+ cells were sorted into 500 µl TRIzol (Invitrogen) for RNA isolation. RNA was amplified in two rounds using MessageAmp aRNA (Ambion), labeled with biotin (BioArray High Yield RNA Transcription Labeling, Enzo), and the resulting cRNAs hybridized to MoGene 1.0 ST arrays (Affymetrix). Raw data were normalized with the RMA algorithm implemented in the “ Expression File Creator” module of the GenePattern suite 38. Pearson correlation to the T2 values was computed in S-Plus, on the real or row-wise randomized datasets. Pathway enrichment analysis was performed using Ingenuity (Ingenuity Systems, Inc., Redwood city, CA).
Histology
Pancreata were removed and fixed in 10% neutral-buffered formalin (Sigma-Aldrich). Paraffin-embedded step-sections were stained with hematoxylin and eosin (H&E) and insulitis was scored as described previously 37.
Immunostaining and microscopy
6 µm cryo-sections of pancreas were cut and fixed with cold acetone. Immunostaining was performed under standard procedures. Before adding primary mAbs, sections were blocked with 5% normal donkey serum (Jackson ImmunoResearch). The following mAbs were used in different combinations as indicated in the figure legends: anti-CRIg mAb (17C9, a gift from Genentech, South San Francisco, CA); anti-F4/80 (BM8), and -CD11b (M1/70), all from BioLegend; anti-CD31 (MEC13.3) from BD Pharmingen. Nuclei were stained with DAPI (4',6-Diamidino-2-phenylindole dihydrochloride). Images were acquired on an Axiovert 200M confocal microscope (Zeiss) using a xenon arc lamp in a Lambda DG-4 wavelength switcher (Sutter Instrument), and were processed with Slidebook imaging software (Intelligent Imaging).
Flow cytometry
Single-cell suspensions of pancreata were prepared as above. All staining began with an incubation with the anti-FcγR mAb, 2.4G2. Abs used for subsequent staining were: anti-CD45 (30-F11) and -Ly6C (Al-21), both from BD Pharmingen; anti-CD11b (M1/70), -CD11c (N418), -F4/80 (BM8), and -Gr1 (RB6-8C5) (all from BioLegend); and anti-CRIg (17C9, a gift from Genentech, South San Francisco, CA). Samples were acquired with an LSR II (BD Bioscience) and data were analyzed with FlowJo software (Tree Star, Inc)
T cell proliferation assay
Spleen naïve T cells were sorted as CD4+CD45R−CD8−CD11b−CD11c−CD25−CD62Lhi, and were labeled by incubation at 106/ml in RPMI1640 with 10 µmol/l CFSE (Molecular Probes, Eugene, Oregon) at 37° for 20 min, washed and resuspended in complete medium (RPMI 1640, 10% fetal calf serum, 2 mmol/L L-glutamine, penicillin/streptomycin). They were cultured at 5 × 104 cells/well in round-bottom, 96-well plates (Corning, Corning, NY).
For T cell activation, anti-CD3 and -CD28 beads (Dynabeads, Oslo, Norway) were added at a 1:1 ratio of bead: cell, with addition of IL-2 (20 U/ml, Proleukin, Chiron). CRIg-Fc or control Fc (anti-gp120 IgG1) (both gifts from Genentech) was added into the culture medium at the final concentration indicated in the relevant figure legend. Proliferation was assessed by flow-cytometric analysis of CFSE dilution. MD (mean division number) was calculated as a proliferation index 39.
In vivo effect of CRIg-Fc
Three-week-old female NOD mice were randomly divided into two groups, and were ip-treated twice a week until 10 weeks of age with 4 mg/kg CRIg-Fc or control-Fc (anti-gp120 IgG1). MRI-MNP was performed at 7–8 weeks of age. All mice were monitored to 32 weeks of age or longer, as indicated in the figure legends, for diabetes development.
Statistical analysis
Statistical analyses were carried out using the methods specified in individual figure legends. P ≤0.05 was considered to be statistically significant.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. M. van Lookeren Campagne (Genentech, South San Francisco, CA) for helpful discussions and for generously providing the anti-CRIg mAb and CRIg-Fc fusion protein; Drs. G. Eisenbarth and L. Yu (Barbara Davis Center for Childhood Diabetes, CO) for measuring the IAA titers; Drs. U. Mahmood and A. Guimaraes for MRI protocol setup and valuable discussions; C. Kaufman and J. Chan for assistance with MRI; J. Pagan, K. Hattori, P Waterman and A Sumski for assistance with mice; Dr. J. Hill, K. Leatherbee, M. Painter and S. Davis for microarray experimentation and analysis; Dr. J. Nishio for histology and diabetes monitoring; and J. LaVecchio and G. Buruzala for help with flow cytometry. This work was supported by grants from the NIH (P01 AI054904) to D.M., C.B. and R.W; by the Core facilities of the Joslin Diabetes Center (P30 DK036836); by the NOD Mouse Manipulated Core of the Juvenile Diabetes Research Foundation Center on Immunological Tolerance at Harvard Medical School (4-2007-1057); and by the Mouse Imaging Resource at the Massachusetts General Hospital (U24CA092782). W.F. was funded by a Takeda Pharmaceuticals Beta Cell Award from the American Diabetes Association (7-07-BETA-14).
Footnotes
AUTHOR CONTRIBUTIONS
W.F., R.W., C.B. and D.M. designed experiments; W.F. and G.W. performed experiments; all authors analyzed the data; W.F., R.W., C.B. and D.M. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Accession code
GEO: microarray data GSExxxxx.
REFERENCES
- 1.Bluestone JA, Herold K, Eisenbarth G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. 2010;464:1293–1300. doi: 10.1038/nature08933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andre-Schmutz I, Hindelang C, Benoist C, Mathis D. Cellular and molecular changes accompanying the progression from insulitis to diabetes. Eur. J Immunol. 1999;29:245–255. doi: 10.1002/(SICI)1521-4141(199901)29:01<245::AID-IMMU245>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 3.Orban T, et al. Pancreatic islet autoantibodies as predictors of type 1 diabetes in the Diabetes Prevention Trial-Type 1. Diabetes Care. 2009;32:2269–2274. doi: 10.2337/dc09-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polonsky KS, Rubenstein AH. C-peptide as a measure of the secretion and hepatic extraction of insulin. Pitfalls and limitations. Diabetes. 1984;33:486–494. doi: 10.2337/diab.33.5.486. [DOI] [PubMed] [Google Scholar]
- 5.Mathis D, Gaglia J. Autoimmune Diabetes. In: Weissleder R, Ross BD, Rehemtulla A, Gambhir SS, editors. Molecular Imaging. Shelton, CT: People's Medical Publishing House-USA; 2010. pp. 1130–1146. [Google Scholar]
- 6.Denis MC, Mahmood U, Benoist C, Mathis D, Weissleder R. Imaging inflammation of the pancreatic islets in type 1 diabetes. Proc Natl. Acad Sci. U.S.A. 2004;101:12634–12639. doi: 10.1073/pnas.0404307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turvey SE, et al. Noninvasive imaging of pancreatic inflammation and its reversal in type 1 diabetes. J Clin Invest. 2005;115:2454–2461. doi: 10.1172/JCI25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papaccio G, Latronico MV, Pisanti FA, Federlin K, Linn T. Adhesion molecules and microvascular changes in the nonobese diabetic (NOD) mouse pancreas. An NO-inhibitor (L-NAME) is unable to block adhesion inflammation-induced activation. Autoimmunity. 1998;27:65–77. doi: 10.3109/08916939809008037. [DOI] [PubMed] [Google Scholar]
- 9.Papaccio G, Pisanti FA, Di Montefiano R, Graziano A, Latronico MVG. Th1 and Th2 cytokines exert regulatory effects upon islet microvascular areas in the NOD mouse. J. Cell. Biochem. 2002;86:651–664. doi: 10.1002/jcb.10250. [DOI] [PubMed] [Google Scholar]
- 10.Carlsson PO, Sandler S, Jansson L. Pancreatic islet blood perfusion in the nonobese diabetic mouse: diabetes-prone female mice exhibit a higher blood flow compared with male mice in the prediabetic phase. Endocrinology. 1998;139:3534–3541. doi: 10.1210/endo.139.8.6153. [DOI] [PubMed] [Google Scholar]
- 11.Jansen A, et al. Immunohistochemical characterization of monocytes-macrophages and dendritic cells involved in the initiation of the insulitis and beta-cell destruction in NOD mice. Diabetes. 1994;43:667–675. doi: 10.2337/diab.43.5.667. [DOI] [PubMed] [Google Scholar]
- 12.Gaglia JL, et al. Noninvasive imaging of pancreatic islet inflammation in type 1A diabetes patients. J Clin Invest. 2011;121:442–445. doi: 10.1172/JCI44339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weissleder R, et al. MR lymphography: study of a high-efficiency lymphotrophic agent. Radiology. 1994;191:225–230. doi: 10.1148/radiology.191.1.8134576. [DOI] [PubMed] [Google Scholar]
- 14.Mills AN, Haworth SG. Greater permeability of the neonatal lung. Postnatal changes in surface charge and biochemistry of porcine pulmonary capillary endothelium. J Thorac. Cardiovasc. Surg. 1991;101:909–916. [PubMed] [Google Scholar]
- 15.Bohme J, Schuhbaur B, Kanagawa O, Benoist C, Mathis D. MHC-linked protection from diabetes dissociated from clonal deletion of T cells. Science. 1990;249:293–295. doi: 10.1126/science.2115690. [DOI] [PubMed] [Google Scholar]
- 16.Yu L, et al. Early expression of antiinsulin autoantibodies of humans and the NOD mouse: evidence for early determination of subsequent diabetes. Proc Natl. Acad. Sci U.S.A. 2000;97:1701–1706. doi: 10.1073/pnas.040556697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirot L, Benoist C, Mathis D. Natural killer cells distinguish innocuous and destructive forms of pancreatic islet autoimmunity. Proc Natl. Acad Sci U.S.A. 2004;101:8102–8107. doi: 10.1073/pnas.0402065101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmy KY, et al. CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell. 2006;124:915–927. doi: 10.1016/j.cell.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 19.Vogt L, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J. Clin. Invest. 2006;116:2817–2826. doi: 10.1172/JCI25673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka M, et al. Expansion of a unique macrophage subset in rheumatoid arthritis synovial lining layer. Clin Exp. Immunol. 2008;154:38–47. doi: 10.1111/j.1365-2249.2008.03733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo S, et al. Down-regulation of Z39Ig on macrophages by IFN-gamma in patients with chronic HBV infection. Clin Immunol. 2010;136:282–291. doi: 10.1016/j.clim.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Gorgani NN, et al. Complement receptor of the Ig superfamily enhances complement-mediated phagocytosis in a subpopulation of tissue resident macrophages. J Immunol. 2008;181:7902–7908. doi: 10.4049/jimmunol.181.11.7902. [DOI] [PubMed] [Google Scholar]
- 23.Xu S, et al. Induction of T cells suppression by dendritic cells transfected with VSIG4 recombinant adenovirus. Immunol. Lett. 2010;128:46–50. doi: 10.1016/j.imlet.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Chen M, Muckersie E, Luo C, Forrester JV, Xu H. Inhibition of the alternative pathway of complement activation reduces inflammation in experimental autoimmune uveoretinitis. Eur. J Immunol. 2010;40:2870–2881. doi: 10.1002/eji.201040323. [DOI] [PubMed] [Google Scholar]
- 25.Katschke KJ, Jr, et al. A novel inhibitor of the alternative pathway of complement reverses inflammation and bone destruction in experimental arthritis. J Exp. Med. 2007;204:1319–1325. doi: 10.1084/jem.20070432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Crispin JC, Dalle LJ, Tsokos GC. A novel inhibitor of the alternative pathway of complement attenuates intestinal ischemia/reperfusion-induced injury. J Surg. Res. 2011;167:e131–e136. doi: 10.1016/j.jss.2009.05.041. [DOI] [PubMed] [Google Scholar]
- 27.Robles DT, Eisenbarth GS, Dailey NJ, Peterson LB, Wicker LS. Insulin autoantibodies are associated with islet inflammation but not always related to diabetes progression in NOD congenic mice. Diabetes. 2003;52:882–886. doi: 10.2337/diabetes.52.3.882. [DOI] [PubMed] [Google Scholar]
- 28.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev. Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geissmann F, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He JQ, Wiesmann C, van Lookeren CM. A role of macrophage complement receptor CRIg in immune clearance and inflammation. Mol. Immunol. 2008;45:4041–4047. doi: 10.1016/j.molimm.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 31.Freeman GJ, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathis D, Vence L, Benoist C. beta-Cell death during progression to diabetes. Nature. 2001;414:792–798. doi: 10.1038/414792a. [DOI] [PubMed] [Google Scholar]
- 33.Miyanishi M, et al. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450:435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 34.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 35.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 36.Swirski FK, et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 2009;325:612–616. doi: 10.1126/science.1175202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katz JD, Wang B, Haskins K, Benoist C, Mathis D. Following a diabetogenic T cell from genesis through pathogenesis. Cell. 1993;74:1089–1100. doi: 10.1016/0092-8674(93)90730-e. [DOI] [PubMed] [Google Scholar]
- 38.Reich M, et al. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 39.D'Alise AM, et al. The defect in T-cell regulation in NOD mice is an effect on the T-cell effectors. Proc Natl. Acad Sci U S. A. 2008;105:19857–19862. doi: 10.1073/pnas.0810713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.