Abstract
Objectives
Recent genome-wide association studies (GWAS) have identified more than 40 common sequence variants associated with type 2 diabetes (T2D). However, the results are not always the same in populations with differing genetic backgrounds. We evaluated a hypothesis that a North Asian population living in a geographic area with unusually harsh environmental conditions developed unique genetic risks.
Methods
We performed a population-based association study with 21 single-nucleotide polymorphisms (SNPs) in 9 genes selected according to the results of GWAS conducted in other populations. The study participants included 393 full-heritage Mongolian individuals, 177 diagnosed with T2D and 216 matched controls. Genotyping was performed by TaqMan methodology.
Results
The strongest association was detected with SNPs located within the potassium-channel coding KCNQ1 (highest OR=1.92; P=3.4×10−5) and ABCC8 (OR=1.79; P=5×10−4) genes. Genetic variants identified as strongly influencing the risk of T2D in other populations such as those in KCNJ11 or TCF7L2 genes did not show statistically significant association in Mongolia.
Conclusions
The strongest T2D risk-associated SNPs in Mongolians are located within 2 of 3 tested potassium-channel coding genes; accumulated variations in these genes may be related to environmental exposure to extreme cold.
Keywords: type 2 diabetes, genetic association, potassium channels, Mongolia, KCNQ1, ABCC8
Introduction
Genome-wide association studies have identified more than 40 genomic loci associated with the risk for type 2 diabetes mellitus.1 The results of GWAS conducted in European or American mixed populations2 were partly inconsistent with subsequent studies in Asians,3,4 suggesting that specific gene variants may independently influence risk of T2D in populations having unique demographic origins and history. The differences were most striking between the signals reported for the KCNQ1 locus that were strong in Asians (OR 1.42; p=2.5×10−40 [3]) but have not reached significance in several studies of Europeans,2 before a weaker association (OR 1.08; p=2.8×10−13) was detected with another SNP.5 Conversely, TCF7L2 considered the strongest candidate in the Europeans2 was not as prominent in at least some Asians populations.6 The new challenge in the post-GWAS era is to validate the genetic associations through replication of the candidate genes in various populations because the efficiency of individual gene-directed medications may depend on genetic background.7
Northern populations are exposed to unique ecological conditions including severe chronic cold stress, marked seasonality, sparse vegetation, and low overall energy availability.8 It has been hypothesized that survival in the harsh environment may be associated with the accumulation of risk alleles that predispose to T2D.9 The Mongolian population selected for this study has generally low but rapidly increasing prevalence of T2D (currently 8.9% of the population at age >25). 10
The purpose of this study was to replicate genetic associations previously reported in GWAS and map more densely the candidate regions in order to detect risk alleles predisposing Mongolians to T2D.
Methods
Study population
Mongolia is one of the most sparsely populated areas in the world with 1.6 inhabitants per square kilometer. The average life expectancy is 61.6 years for males and 67.8 for females. T2D prevalence in Mongolia grew among adult urban population from 3.2% in 1999 to 8.9% in 2009.10 Patients were diagnosed using the World Health Organization diagnostic criteria, officially registered, and received treatment for this condition. Each patient underwent annual follow-up that included an exam, biochemical tests, and adjustment of treatment regiments. The non-diabetic control group included age, gender and place of residence matched individuals in which diabetes has been excluded. Control individuals were not related to the T2D patients or each other. The study population included 393 full-heritage Mongolian individuals, of which 177 were patients affected with T2D at various stages of the disease and 216 were control subjects. The T2D-affected individuals in our study were on average in their mid-50s at the time of sample collection and did not differ significantly from the current age of control individuals (P=0.52). The patients had an increased body mass index (30.5±9.0 vs. 27±4.7 in controls; P=0.0008) and fasting plasma glucose level (12.45±5.6 vs. 6.5±2.1 in controls; P=0.0001) and HbA1c (9.1±2.4 vs. 5.5±0.9 in controls; P=0.0001). The study was in compliance with the Helsinki Declaration. Research protocols were approved by a local Ethics Committee and Institutional Review Boards of participating institutions. Informed consent was obtained from each participant.
Genotyping
For the purposes of this analysis, we selected 21 SNPs in 9 genes, ABCC8, CDKN2A/B, CDKAL1, KCNJ11, KCNQ1, HHEX, PPARg, SCL30A8, and TCF7L2, shown to be associated with T2D in GWAS or association studies with individual genetic markers performed in other populations. SNPs were genotyped using Taqman® SNP Genotyping assays with TaqMan Universal PCR Master Mix (No AmpErase® UNG) or TaqMan® Genotyping Master Mix (both from Applied Biosystems, Foster City, CA). All reactions were run on an ABI7300 system and analyzed using SDS 1.2 software (Applied Biosystems, Foster City, CA). The results were consistent with direct sequencing of selected samples and/or testing with dHPLC on automated WAVE Nucleic Acid Fragment Analysis System (Transgenomic, Omaha, NE).
Statistical analysis
We tested association between candidate gene SNPs and T2D using logistic regression. Estimated odd ratios (ORs) are equivalent to those obtainable from Cochran-Armitage trend test. Under the widely accepted additive genetic model for this disorder, trend test is more robust to deviations from Hardy-Weinberg equilibrium, hence preferred to other tests such as the ones calculated by contrasting allele frequencies or homozygosity frequencies.11 Study-wide threshold P value for association was set at 0.0024 by applying Bonferroni correction for testing 21 SNPs. Formal statistical tests and parameter estimations including 95% confidence intervals were carried out using SAS/STAT 9.2 (SAS Institute Inc, Cary, NC). Pair-wise estimates for linkage disequilibrium (r2) between neighboring markers located in the same locus were calculated using Haploveiw (Broad Institute of MIT and Harvard, USA, version 4.1).
Results
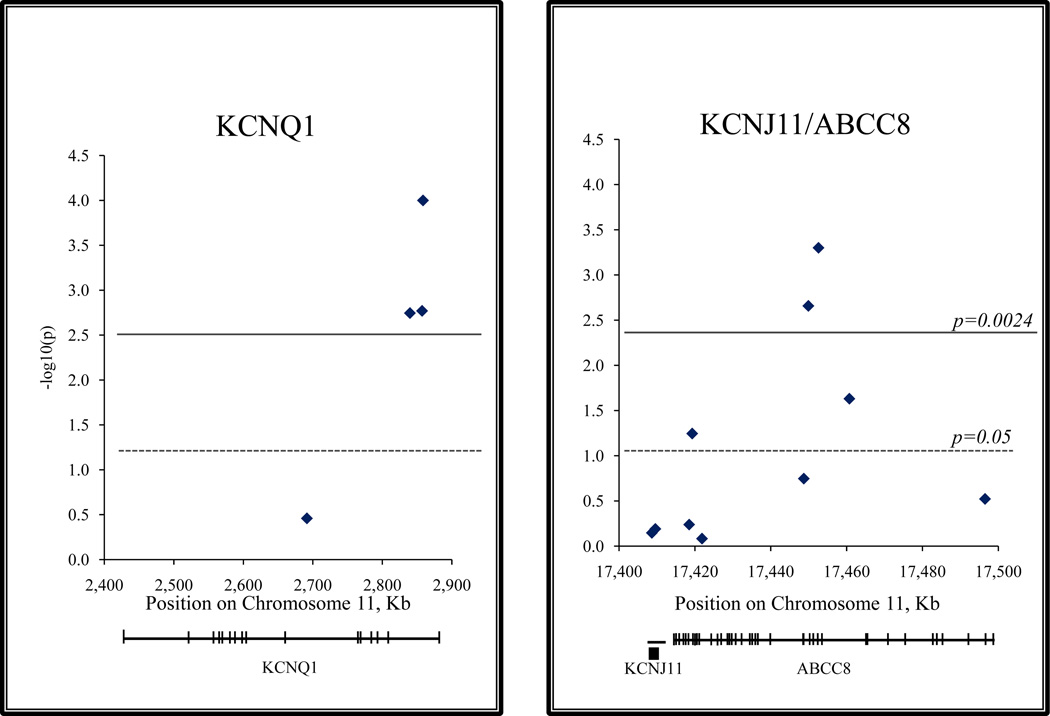
The prevalence of type 2 diabetes in Mongolia has increased three-fold in the last 10 years, with an alarming tendency of further growth. A total of 21 SNPs in 9 genes were genotyped in 177 T2D patients and genotype profiles compared to 216 controls. Initially, variations in four genes, PPARg, CDKAL1, KCNQ1, and ABCC8, have shown weak association with T2D. Denser mapping with additional follow-up SNPs was carried out within the KCNQ1 and ABCC8 candidate loci. P-values for 3 SNPs in KCNQ1 (rs2237892, rs2237895, and rs2237897) and 2 SNPs in ABCC8 (rs1799858 and rs2074308) overcame the P=0.0024 threshold adjusted for multiple testing (Table). The three positively associated KCNQ1 SNPs are located within intron 15, and the ABCC8 signals are in a relatively short fragment between intron 11 and exon 14 (Figure). The strongest signals were detected with marker rs2237897 in KCNQ1 gene (OR=1.92; p=3.4×10−5) and rs2074308 in ABCC8 (OR=1.79; p=5×10−4). Of the neighboring SNPs with positive association, the rs1799858 and rs2074308 were in low linkage disequilibrium (r2=0.134), and the three SNPs located within the KCNQ1 exon 15, rs2237892, rs2237895, and rs2237897, showed moderate correlation, r2=0.324 and r2=0.444, respectively. Marker rs7903146 in TCF7L2 frequently replicated in large-scale T2D studies showed no evidence of association in the Mongolians (OR=1.17; P=0.488). The second most replicated marker rs5219 of KCNJ11 also failed to show signs of association (OR=1.07; P=0.645). Signals in PPARg and CDKAL1 genes did not overcome the P-value threshold adjusted for multiple testing. We were likewise unable to confirm the association with two SNPs within the retinol binding protein 4 gene reported in the only previous study of T2D in Mongolia.12
Table.
Results of genotyping with 21 SNPs in Mongolian patients with type 2 diabetes
| Chromo some number |
rs_number | Gene | SNP position within the gene |
Risk allele |
Risk allele frequency |
OR | 95% CI | P-value | |
|---|---|---|---|---|---|---|---|---|---|
| Cases/Controls | |||||||||
| 3 | rs1801282 | PPARg | P12A | C | 0.89 | 0.94 | 1.93 | 1.16–3.34 | 0.015 |
| 3 | rs3856806 | PPARg | H477H | T | 0.16 | 0.14 | 1.13 | 0.77–1.67 | 0.962 |
| 6 | rs7754840 | CDKAL1 | Intron 5 | C | 0.32 | 0.39 | 1.30 | 1.02–1.88 | 0.038 |
| 8 | rs13266634 | SCL30A8 | Exon R276W |
C | 0.57 | 0.64 | 1.31 | 0.96–1.72 | 0.091 |
| 9 | rs10811661 | CDKN2 A/B |
3’-flanking region |
T | 0.67 | 0.68 | 1.04 | 0.74–1.38 | 0.940 |
| 10 | rs1111875 | HHEX | 3’-flanking region |
C | 0.32 | 0.32 | 1.00 | 0.77–1.39 | 0.820 |
| 10 | rs7903146 | TCF7L2 | Intron 2 | C | 0.94 | 0.93 | 1.17 | 0.68–2.08 | 0.488 |
| 11 | rs231361 | KCNQ1 | Intron 11 | G | 0.31 | 0.34 | 1.15 | 0.86–1.54 | 0.348 |
| 11 | rs2237892 | KCNQ1 | Intron 15 | C | 0.63 | 0.73 | 1.69 | 1.21–2.32 | 0.0020 |
| 11 | rs2237895 | KCNQ1 | Intron 15 | C | 0.29 | 0.34 | 1.70 | 1.22–2.38 | 0.0020 |
| 11 | rs2237897 | KCNQ1 | Intron 15 | C | 0.52 | 0.67 | 1.92 | 1.41–2.59 | 3.4×10−5 |
| 11 | rs5215 | KCNJ11 | Exon 1 V337I |
T | 0.66 | 0.68 | 1.06 | 0.79–1.42 | 0.714 |
| 11 | rs5219 | KCNJ11 | Exon 1 K23E |
C | 0.67 | 0.68 | 1.07 | 0.80–1.44 | 0.645 |
| 11 | rs757110 | ABCC8 | Exon 33 A1369S |
A | 0.66 | 0.68 | 1.09 | 0.81–1.46 | 0.577 |
| 11 | rs1799859 | ABCC8 | Exon 31 R1273R |
G | 0.67 | 0.72 | 1.35 | 0.99–1.84 | 0.057 |
| 11 | rs2074311 | ABCC8 | Intron 29 | G | 0.68 | 0.69 | 1.03 | 0.77–1.39 | 0.827 |
| 11 | rs1799854 | ABCC8 | Exon 16 | G | 0.56 | 0.51 | 1.22 | 0.92–1.62 | 0.179 |
| 11 | rs1799858 | ABCC8 | Exon 14 K649K |
T | 0.16 | 0.23 | 1.78 | 1.23–2.58 | 0.0022 |
| 11 | rs2074308 | ABCC8 | Intron 11 | T | 0.22 | 0.33 | 1.79 | 1.29–2.50 | 5×10−4 |
| 11 | rs2237982 | ABCC8 | Intron 10 | C | 0.79 | 0.85 | 1.54 | 1.07–2.24 | 0.023 |
| 11 | rs1048099 | ABCC8 | Exon 2 P69P |
A | 0.65 | 0.69 | 1.16 | 0.87–1.56 | 0.301 |
Figure.
Plots of T2D association signals in KCNQ1 (Panel A) and KCNJ11/ABCC8 (Panel B) genes detected in the Mongolian population. P-values for the association (in −log10 scale) are shown for each SNP as diamonds and plotted according to their location on chromosome 11. The strongest P-values for 3 SNPs in KCNQ1 (rs2237892, rs2237895, and rs2237897) and 2 SNPs in ABCC8 (rs1799858 and rs2074308) overcame the P-value threshold adjusted for multiple testing (P=0.0024).
Discussion
We found variants rs2237892, rs2237895, and rs2237897 located in intron 15 of the KCNQ1 gene to be associated with T2D in Mongolians. Similar results have previously been obtained in Japanese cohorts and replicated in Chinese, Korean, Swedish and Danish populations.3,4,5 Our studies in the Mongolian population confirm these earlier reports. KCNQ1 located on chromosome 11p15.5 encodes a pore-forming subunit of voltage-gated potassium channel that is expressed in β-cells. The channel has a role in insulin secretion. Follow-up genetic studies have shown that the intron 15 KCNQ1 risk alleles are associated with impaired insulin secretion presumably through altering the KCNQ1 gene expression in β-cells.3,13 Rare mutations in KCNQ1 are known to cause potassium channel dysfunction leading to a form of cardiac long QT syndrome (LQT1) and serious arrhythmias, ventricular fibrillation and cardiac arrest.14
The second strong signal associated with risk of T2D in Mongolians was detected in the KCNJ11/ABCC8 region located on chromosome 11p15.1. KCNJ11 encodes the inwardly rectifying potassium channel Kir6.2, and ABCC8 codes for sulfonylurea receptor 1 (SUR1); they together form a β-cell adenosine triphosphate–sensitive potassium channel (KATP). KATP channels regulate insulin release from β-cells. Rare heterozygous activating mutations in these genes are the cause of permanent neonatal diabetes.15 The KCNJ11 E23K variant was consistently found to be associated with T2D in multiple studies, but this SNP is in complete linkage disequilibrium with a non-synonymous S1369A variant in the neighboring ABCC8 gene.16 In comparative experiments it was the ABCC8 A1369 and not the KCNJ11 K23 allele that was responsible for the effect of decreased ATP inhibition.17 These and other studies led to a hypothesis that the role of ABCC8 in T2D development is underestimated and that the 11p15.1 chromosomal region needs to be analyzed for further contributing variants.18 Indeed, rare mutations in exons 4,5, and 7 of ABCC8 exerting a relatively minor or no effect on KATP channel activity still caused type 2 diabetes in adult patients.19 Our data contribute a new observation indicating that two SNPs in ABCC8 are in fact associated with the disease in Mongolians.
Populations living in cold environments require higher energy use and can significantly enhance fat utilization by increasing insulin sensitivity20 through minute changes in genes that could have been critical for survival. The KCNQ1 encoded potassium channel is known to be reacting to cold environment: swimming in cold water causes prolongation of the QT interval and results in syncope and frequent drawning in patients with long QT syndrome associated with mutations in KCNQ1.21 Dramatic improvements within the past 10–20 years of living conditions and work requirements promoting sedentary lifestyle in combination with the traditional habits of meat and animal fat consumption resulted in an abrupt and extensive exposure to the risks of type 2 diabetes.10
The estimated effects of the associated variants seem larger than in other reports, which may reflect the fact that the Mongolian population is smaller and more homogeneous than the previously studied populations. At the same time, as the sample size in this study is small and statistical power limited, negative results on several loci such as KCNJ11 or TCF7L2 associated with T2D in other populations (including Asians22,23) are not strong enough for exclusion and need further evaluation.
Genetic variants predisposing to T2D are expected to influence the choice of therapeutic strategies. A recent study24 evaluated the effects of widely used T2D treatment modalities in individuals possessing intron 15 KCNQ1 risk alleles. After Repaglinide treatment, the TT homozygotes at rs2237892 showed lower 2-h glucose levels than the C allele carriers. Repaglinide is a fast-acting insulin secretagogue that initiates insulin secretion by closing the ATP-dependent potassium channels. Rosiglitazone showed a less pronounced effect. In infants with neonatal diabetes due to mutations in the ATP-sensitive potassium channel encoded by ABCC8 and KCNJ11 genes the disease can be successfully treated with oral sulfonylurea-based medications rather than lifelong insulin injections.25
In conclusion, this study replicates data regarding the role of KCNQ1 variants in the development of type 2 diabetes in an independent ancient Asian population. The association with ABCC8 genetic variants indicates their participation in the causation of type 2 diabetes in Mongolians, most likely as a result of functional interaction with the effects of other potassium channel gene.
The significant finding of the study is establishing of an association between genetic variants in potassium-channel genes KCNQ1 and ABCC8 and the development of type 2 diabetes in the Mongolian population.
The study adds that potassium channels known to be affecting the adjustment to harsh environmental conditions may accumulate genetic defects that predispose to type 2 diabetes.
Acknowledgements
The authors would like to thank the patients, their family members, and healthy volunteers for their participation in the study. This work was supported in part by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, NIH.
Footnotes
Disclosure
None declared.
References
- 1.Bonnefond A, Froguel P, Vaxillaire M. The emerging genetics of type 2 diabetes. Trends in Molecular Medicine. 2010;16:407–416. doi: 10.1016/j.molmed.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Zeggini E, Scott LJ, Saxena R, Voight BF. Meta-analysis of genome-wide association data andlarge-scale replication identifies additional susceptibility loci for type 2 diabetes. Nature Genetics. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yasuda K, Miyake K, Horikawa Y, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nature Genetics. 2008;40:1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 4.Unoki H, Takahashi A, Kawaguchi T, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nature Genetics. 2008;40:1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 5.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature Genetics. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng MC, Park KS, Oh B, et al. Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2 and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes. 2008;57:2226–2233. doi: 10.2337/db07-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox CB, Moore PD. Biogeography: an ecological and evolutionary approach. 5th ed. Oxford: Blackwell Scientific Publications; 1993. [Google Scholar]
- 9.Chakravarthy MV, Booth FW. Eating, exercise, and "thrifty" genotypes: connecting the dots toward an evolutionary understanding of modern chronic diseases. J Applied Physiology. 2004;96:3–10. doi: 10.1152/japplphysiol.00757.2003. [DOI] [PubMed] [Google Scholar]
- 10.WHO Mongolian STEPS survey on the prevalence of noncommunicable disease risk factors in 2009. WHO, MOH, Public Health Institute; 2010. [Google Scholar]
- 11.Elston RC, Olson JM, Palmer L. Biostatistical genetics and genetic epidemiology. John Wiley and Sons; 2002. p. 208. [Google Scholar]
- 12.Munkhtulga L, Nakayama K, Utsumi N, et al. Identification of a regulatory SNP in the retinol binding protein 4 gene associated with type 2 diabetes in Mongolia. Human Genetics. 2007;120:879–888. doi: 10.1007/s00439-006-0264-4. [DOI] [PubMed] [Google Scholar]
- 13.Jonsson A, Isomaa B, Tuomi T, et al. A variant in the KCNQ1 gene predicts future type 2 diabetes and mediates impaired insulin secretion. Diabetes. 2009;58:2409–2413. doi: 10.2337/db09-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neyroud N, Tesson F, Denjoy I, et al. A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange-Nielsen cardioauditory syndrome. Nature Genetics. 1997;15:186–189. doi: 10.1038/ng0297-186. [DOI] [PubMed] [Google Scholar]
- 15.Flanagan SE, Clauin S, Bellanne-Chantelot C, et al. Update of mutations in the genes encoding the pancreatic beta-cell K(ATP) channel subunits Kir6.2 (KCNJ11) and sulfonylurea receptor 1 (ABCC8) in diabetes mellitus and hyperinsulinism. Human Mutation. 2009;30:170–180. doi: 10.1002/humu.20838. [DOI] [PubMed] [Google Scholar]
- 16.Florez JC, Burtt N, de Bakker PI, et al. Haplotype structure and genotype-phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes. 2004;53:1360–1368. doi: 10.2337/diabetes.53.5.1360. [DOI] [PubMed] [Google Scholar]
- 17.Hamming KS, Soliman D, Matemisz LC, Niazi O, Gloyn AL, Light PE. Coexpression of the type 2 diabetes susceptibility gene variants KCNJ11 E23K and ABCC8 S1369A alter the ATP and sulfonylurea sensitivities of the ATP-sensitive K(+) channel. Diabetes. 2009;58:2419–2424. doi: 10.2337/db09-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Bunt M, Gloyn AL. Genetic association to molecular mechanism. Curr. Diabetes Reports. 2010;10:452–466. doi: 10.1007/s11892-010-0150-2. [DOI] [PubMed] [Google Scholar]
- 19.Tarasov AI, Nicolson TJ, Riveline JP, et al. A rare mutation in ABCC8/SUR1 leading to altered ATP-sensitive K+ channel activity and beta-cell glucose sensing is associated with type 2 diabetes in adults. Diabetes. 2008;57:1595–1604. doi: 10.2337/db07-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vlasova NV, Gitel'zon II, Okladnikov IuN. Lipid metabolism in Far North aborigines. Vopr. Pitan. 1975;5:53–56. [PubMed] [Google Scholar]
- 21.Ackerman MJ, Tester DJ, Porter CJ. Swimming, a gene-specific arrhythmogenic trigger for inherited long QT syndrome. Mayo Clin Proc. 1999;74:1088–1094. doi: 10.4065/74.11.1088. [DOI] [PubMed] [Google Scholar]
- 22.Hu C, Zhang R, Wang C, et al. PPARG, KCNJ11, CDKAL1, CDKN2A-CDKN2B, IDE-KIF11-HHEX, IGF2BP2 and SLC30A8 are associated with type 2 diabetes in a Chinese population. PLoS One. 2009;4:7643. doi: 10.1371/journal.pone.0007643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu M, Bi Y, Xu Y, et al. Combined effects of 19 common variations on type 2 diabetes in Chinese: results from two community-based studies. PLoS One. 2010;5:e14022. doi: 10.1371/journal.pone.0014022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu W, Hu C, Zhang R, et al. Effects of KCNQ1 polymorphisms on the therapeutic efficacy of oral antidiabetic drugs in Chinese patients with type 2 diabetes. Clin Pharmacol Ther. 2011;89:437–442. doi: 10.1038/clpt.2010.351. [DOI] [PubMed] [Google Scholar]
- 25.Gloyn AL, Siddiqui J, Ellard S. Mutations in the genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) in diabetes mellitus and hyperinsulinism. Human Mutations. 2006;27:220–231. doi: 10.1002/humu.20292. [DOI] [PubMed] [Google Scholar]