Abstract
Identification of ligands that interact with nuclear receptors is both a major biological problem and an important initial step in drug discovery. Several in vitro and in vivo techniques are commonly used to screen ligand candidates against nuclear receptors; however, none of the current assays allow screening without modification of either the protein and/or the ligand in a high-throughput fashion. Differential scanning fluorimetry (DSF) allows unmodified potential ligands to be screened as 10µL reactions in 96-well format against partially purified protein, revealing specific interactors. As a proof of principle, we used a commercially-available nuclear receptor ligand candidate chemical library to identify interactors of the human estrogen receptor α ligand binding domain (ERα LBD). Compounds that interact specifically with ERα LBD stabilize the protein and result in an elevation of the thermal denaturation point, as monitored by the environmentally-sensitive dye SYPRO orange. We successfully identified all three compounds in the library that have previously been identified to interact with ERα, with no false positive results.
Introduction
Nuclear receptors (NRs) are transcription factors found in all metazoa that regulate a wide variety of cellular, physiological and developmental processes. Canonically, the activities of NRs are modulated by interactions between small-molecule ligands and a structurally-conserved ligand-binding domain (LBD) of the NR, inducing conformational changes that result in new interaction surfaces for the recruitment of transcriptional coactivators and/or corepressors (reviewed by [Glass and Rosenfeld, 2000]). The intrinsic chemical properties of known NR ligands, in particular, with most possessing the ability to diffuse readily through both aqueous and membranous environments, make them highly effective intercellular signaling molecules [Mangelsdorf et al., 1995].
Identification of ligands that bind to NRs has become an important focus of research efforts driven by two overlapping agendas. First, is the interest in identifying the “natural” ligand(s) of a given NR to understand the role of the NR within the organism, especially when the NR has no known ligand (i.e., “de-orphanization”). Second, is the perspective of the NR as a target for pharmaceutical intervention. The desire to identify interacting partners of NRs has led to a proliferation of different techniques, each with associated advantages and disadvantages (reviewed by [Mukherjee and Mani, 2010; Raucy and Lasker, 2010; Schulman and Heyman, 2004; Shi, 2006]).
Differential scanning techniques allow identification of conditions that enhance the stability of proteins. The transition from native to denatured protein is measured as a function of increasing temperature, using either light scattering (Differential Scanning Light Scattering) or the fluorescence of an environmentally-sensitive dye (Differential Scanning Fluorescence – DSF). In plate format, this allows for a large number of conditions (e.g., pH, salt, addition of small molecules) to be assayed simultaneously. Conditions that cause a protein to shift to a higher denaturation temperature are identified as “stabilizing” conditions. Vedadi et al. have pioneered this technique to determine conditions for an array of proteins used to optimize conditions for crystal growth [Vedadi et al., 2006]. Some studies have also used this technique to demonstrate that peptide or small molecule interactors have the ability to shift the denaturation point of proteins to a higher temperature [Qiagen, 2010; Reinking et al., 2005; Vedadi et al., 2006; Wan et al., 2009]. Here, we present evidence that suggests this technique successfully identifies known interactors of the estrogen receptor α (ERα) from a commercially-available compound library. Overall, we have found the technique to be relatively inexpensive and it offers some intriguing advantages over other commonly-used methodologies.
Reagents and instruments
His6-hERα LDB (302-552) expression vector (kindly provided by Dino Moras [Eiler et al., 2001]).
Terrific Broth (Fisher Scientific, Waltham, MA).
IPTG (Acros Organics, Geel, Belgium).
EDTA-Free Protease Inhibitor Tablets (Roche, Indianapolis, IN).
Branson Sonifier 450 (Branson Corporation, Danbury, CT).
Ni-NTA Superflow (Qiagen, Valencia, CA).
β-estradiol (MP Biomedicals, Solon, OH).
pET15b (EMDBiosciences, San Diego, CA).
Sypro Orange, 5000X (Sigma-Aldrich, St. Louis, MO).
CFX96 Real-Time PCR System (Bio-Rad, Hercules, CA).
Nuclear Receptor Ligand Library BML-2802 (Enzo Life Sciences, Plymouth Meeting, PA).
Methods
Production and purification of estrogen receptor
BL21 (DE3) E. coli cells were transformed with an expression vector containing coding information for His6-hERα LBD (302-552) [Eiler et al., 2001]. Freshly transformed cells were grown at 37°C in Terrific Broth until reaching mid-log phase of growth. The culture was cooled to 15°C and induced with 1mM IPTG overnight (18 hours). Cells were pelleted at 1800 x g and resuspended in Binding Buffer (500mM NaCl, 10mM Imidazole, 5% glycerol @ pH 8.0). Two tablets of EDTA-free protease inhibitor cocktail were added, and the cells were lysed by sonication. The lysate was clarified by centrifugation at 40,000 x g for 30 minutes. The supernatant was loaded onto 2mL bed volume of nickel affinity resin in a gravity-flow column, pre-equilibrated with binding buffer. After allowing the clarified lysate to flow-through, the column was washed with 400mL of Binding Buffer. Purified ERα LBD was eluted with elution buffer (500mM NaCl, 30mM Imidazole, 5% glycerol @ pH 8.0) and dialyzed into Analysis Buffer (500mM NaCl, 50mM HEPES, 5% glycerol, 10mM DTT @ pH 7.5). “Empty Vector” protein product was produced as above, except pET15b with no insert was used.
Protein “melting” curves
The RT-PCR machine was programmed to equilibrate samples at 25°C for 5 minutes and then increase temperature to 95°C at a rate of 1°C/minute, taking a fluorescence reading every 0.2°C using a LED/photodiode set matched to the excitation and emission wavelengths of SYPRO orange. The melting point of the protein was obtained as the lowest point of first derivative plot, as calculated by the software included with the RT-PCR machine.
Optimal concentrations of ERα LBD and SYPRO orange dye were determined by performing DSF on a grid of varying concentrations of protein and SYPRO orange in Analysis Buffer. For ERα, the optimal conditions were ~1 mg/mL protein (as determined by Bradford Assay) in Analysis Buffer and 2X SYPRO orange (diluted from 5000X concentrate) within the reaction well, as determined by maximum range of relative Fluorescence Units observed in the “melt curve”.
In a single well of a 96-well pCR plate, a 10µL reaction was conducted by combining 6µL of protein solution (in Analysis Buffer), 2µL of 10X Sypro orange (diluted from 5000X stock in DMSO with Analysis Buffer) and 2µL of solution containing either 1mM ligand (diluted from 10mM DMSO stock with Analysis Buffer) or 10% DMSO (v/v) in Analysis Buffer (as vehicle controls). The final conditions in the experimental well were 2X Sypro orange, 200µM compound (if any), protein concentration ranging from 08-1.2 mg/mL (depending on preparation) and 2% (v/v) DMSO in Analysis Buffer.
Ligand titrations
A series of Estradiol (E2) dilutions was created in 10% (v/v) DMS on Analysis Buffer ranging from 1 to 500µM. Purified ERα LBD was assayed against the E2 dilution series at two different protein concentrations by combining either [6µL protein + 2µL ligand solution + 2µL 10X SYPRO orange] or [3µL protein + 3µL Analysis Buffer + 2µL ligand solution + 2µL 10X SYPRO orange]. DSF was performed to determine melting points. This process was repeated for estrone (E1) and tamoxifen citrate.
Results
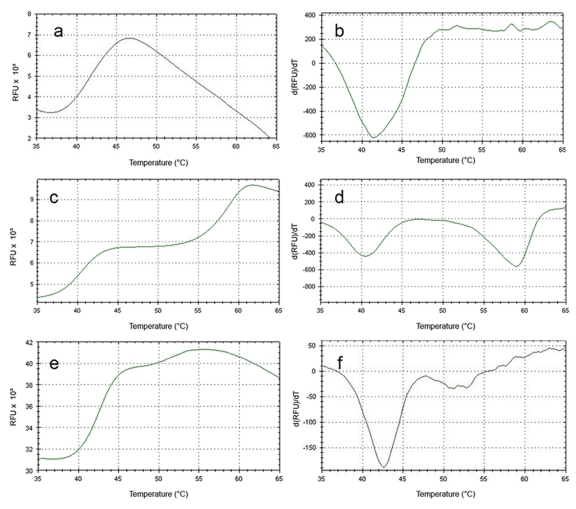
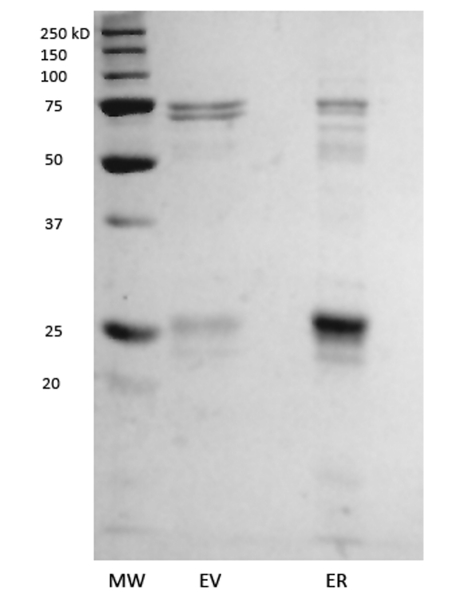
Upon purification by a single affinity column, the purification product containing human ERα LBD produced a denaturation curve with a melting point (Tm) of ~41°C (representative curve in Figure 1a-b). Addition of β-estradiol (E2) produced a denaturation curve with two transitions, at ~41°C and ~59°C (Figure 1c-d). The presence of two peaks is unlikely to be caused by the presence of bound and unbound populations of ERα, since E2 was present at a concentration well above the dissociation constant of ERα/E2 and in molecular excess to ERα. Additionally, titration of higher concentrations of E2 did not alter the magnitude or presence of 41°C transition (data not shown). The 41°C Tm that remains in the ERα + E2 in vitro samples is therefore likely to be caused by non-ERα protein contaminants, as can be observed in the SDS-PAGE analysis of apo ERα (Figure 2). A mock purification of the bacteria transformed with an expression vector with no coding DNA inserted in the multiple cloning site results in purification of several endogenous bacterial proteins that closely match the contaminants seen in the ERα purification (Figure 2). When the denaturation experiment is run on the products of the “empty vector” purification, the resultant curve displays a melting curve that closely matches the melting point seen for apo ERα LBD and the first peak of ERα LBD plus E2 (Figure 1e-f), but does not change upon addition of E2. These results indicate that the co-purified proteins associated with the partial purification of ERα LBD have a collective melting point of ~41°C. Apo ERα LBD either also denatures at this range, or does not display a signal that is otherwise discernable from the contaminants. However, upon addition of E2, an easily discernable secondary melting point arises at a significantly higher temperature, indicating the presence of a specific ERα/E2 complex.
Figure 1. Using DSF to determine “melting” points of protein preparations.
Representative DSF output from the RT-PCR machine as matched sets of graphs displaying Relative Fluorescence Units (RFU) vs. temperature and the first derivative of RFU vs. temperature for ERα (a-b), ERα plus 200µM E2 (c-d) , and “empty vector” protein (e-f). Tm is calculated from the first derivative plots on the right.
Figure 2. “Dirty” protein preparations.
SDS-PAGE analysis of “empty” pET 15 vector (EV) and ERα LBD (ER).
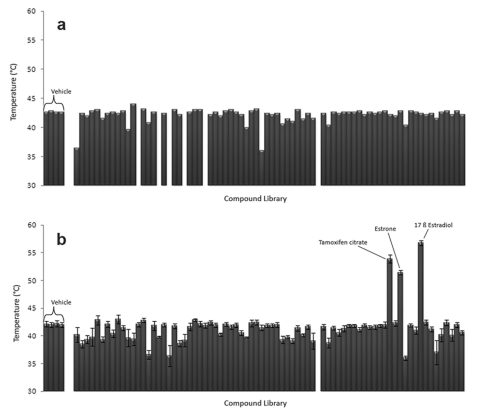
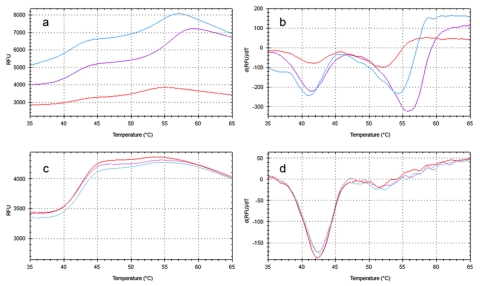
Differential scanning fluorimetry was used in a high-throughput fashion to screen a commercially-available compound library containing 76 known or suspected nuclear receptor ligands (Supplementary File 1). The 96-well format of the RT-PCR machine allowed all compounds to be tested simultaneously, as well as the addition of four wells containing vehicle solution as a baseline. When screened against the “empty vector” protein purification, most wells displayed melting points similar to the vehicle wells (Figure 3a). Nonspecific interactions can lead to a decrease in melting point seen for some compounds. None of the compounds resulted in higher melting points for the “empty vector” proteins. When ERα LBD was screened against the library, three compounds displayed significant secondary melting points well above that observed for the vehicle (Figure 3b, Figure 4a-b). All three compounds are known ERα agonists (β-estradiol, estrone) or antagonists (tamoxifen citrate). These compounds induced no secondary melting points in the “empty vector” protein sample (Figure 4c-d).
Figure 3. Using DSF to screen a compound library.
Maximum melt temperature (Tmax) recorded for chemical compound library (200µM ligand concentration) with either empty vector protein preparation (a) or ERα LBD (b). When multiple Tm are present in a sample, only the higher value is plotted. Error bars in (b) represent the average of seven replicate data sets ± Standard Error of the Mean (SEM).
Figure 4. “Hits” from the compound library.
Representative DSF output of wells containing E2 (purple), estrone (red) and tamoxifen citrate (blue) for screens containing ERα LBD (a-b) or empty vector protein preparation (c-d).
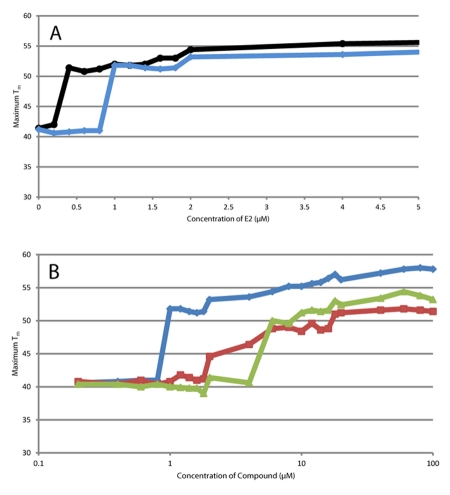
Titrations of the E2 (Figure 5a) yielded an apparent EC50 that is well above the sub-nanomolar reported values for ER/E2 associations. Detection limit of this assay is correlated to the protein concentration (higher protein concentration yields a higher apparent EC50), which is not surprising since the status of the protein is the readout of the DSF experiment. The apparent EC50 of the other lower affinity “hits” from the screen were also determined (Figure 5b). Both ligands demonstrated similar protein concentration - EC50 correlations (data not shown). When viewed over a broader ligand concentration range (Figure 5b), all three ligands are detected at 10µM, but also demonstrate subtle increases of the maximum Tm at higher concentrations of ligand.
Figure 5. Ligand titrations.
a) Titrations of E2 against ER at “high” (6µL protein in 10µL reaction well, blue trace) and “low” (3µL of protein in 10µL reaction well, black trace) concentrations, demonstrating that the apparent EC50 is dependent on protein concentration. b) Titrations of three “hits” of the screen, E2 (blue trace), E1 (red trace) and tamoxifen citrate (green trace) against “high” concentration of ER (6µL protein in 10µL reaction well).
Discussion
NR LBDs are generally difficult to purify and concentrate in the absence of a ligand, but are more tractable when the ligand is combined with the protein. Many crystal structures of liganded NR LBDs are produced by introducing the ligand into the heterologous expression system prior to overexpression [Hassell et al., 2007]. The relative poor behavior (from the perspective of the purifier) of apo NR LBDs can be accounted for by a high degree of conformational mobility which is alleviated upon introduction of the ligand [Hamuro et al., 2006; Johnson et al., 2000; Yan et al., 2004]. Accordingly, the ERα LBD has been reported to be easier to purify to homogeneity and concentrate when purified in the presence of a ligand [Eiler et al., 2001; Pike, 2009]. In this assay, we also demonstrate that upon ligation, the stabilization of ERα LBD can be easily measured using differential scanning fluorimetry.
Herein, we demonstrate DSF has the ability to identify all known interactors of ERα contained in a commercially-available compound library with a high degree of reproducibility and minimal-to-no background in a 96-well plate format using 10µL reaction volumes. We expect the assay should also be able to function with 384-well format versions of RT-PCR machines with the appropriate channel that are currently available on the market. Like some other commonly used in vitro ligand identification assays, DSF requires relatively small amounts of both ligand and protein and has the ability to detect (but not differentiate) interactors that can potentially activate or repress transcription of the genes under the regulation of the NR. Unlike other in vitro assays, DSF does not require that either the protein or the ligand need be tagged (radioactively, fluorescently or with other adducts). Also, unlike in vivo transactivation or yeast two-hybrid assays, DSF does require a separate fusion construct. The same expression construct used to produce protein for DSF can also be used for other in vitro investigations such as crystallization trials or other biophysical characterizations. The detection limit of this technique appears to be determined by combination of both the affinity of the protein-ligand interaction, as well as the concentration of the protein used in the assay. Lower protein concentration actually improves the detection limit, but quality of the signal sets a practical lower limit on how dilute a protein can be used.
For many nuclear receptors, especially among non-vertebrates, no ligands are known, making it impossible to incorporate ligands to improve the stability and behavior of a NR LBD. As we have shown, DSF tolerates relatively low protein concentration and preparations that are not completely pure, making it especially amenable to qualitative identification of ligands of orphan NRs. We believe this technique has the potential to be applicable to other NR LBDs than can be at least partially purified.
Acknowledgments
The RT-PCR machine was provided by NSF MRI grant #1039966. The compound library was purchased in part with a SUNY New Paltz AYURE award. AR was supported by NSF CCLI grant #0817337. RR was supported by a Merck/AAAS Undergraduate Science Research Program grant.
Abbreviations
- DSF
differential scanning light fluorimetry
- E2
β-estradiol
- LBD
ligand binding domain
- NR
nuclear receptor
- RT-PCR
real-time polymerase chain reaction
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
Supplementary Material
References
- Eiler S., Gangloff M., Duclaud S., Moras D., Ruff M. Overexpression, purification, and crystal structure of native ER alpha LBD. Protein Expr Purif. 2001;22:165–73. doi: 10.1006/prep.2001.1409. [DOI] [PubMed] [Google Scholar]
- Glass C. K., Rosenfeld M. G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- Hamuro Y., Coales S. J., Morrow J. A., Molnar K. S., Tuske S. J., Southern M. R., Griffin P. R. Hydrogen/deuterium-exchange (H/D-Ex) of PPARgamma LBD in the presence of various modulators. Protein Sci. 2006;15:1883–92. doi: 10.1110/ps.062103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassell A. M., An G., Bledsoe R. K., Bynum J. M., Carter H. L., 3rd, Deng S. J., Gampe R. T., Grisard T. E., Madauss K. P., Nolte R. T., Rocque W. J., Wang L., Weaver K. L., Williams S. P., Wisely G. B., Xu R., Shewchuk L. M. Crystallization of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2007;63:72–9. doi: 10.1107/S0907444906047020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. A., Wilson E. M., Li Y., Moller D. E., Smith R. G., Zhou G. Ligand-induced stabilization of PPARgamma monitored by NMR spectroscopy: implications for nuclear receptor activation. J Mol Biol. 2000;298:187–94. doi: 10.1006/jmbi.2000.3636. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Mani S. Orphan nuclear receptors as targets for drug development. Pharm Res. 2010;27:1439–68. doi: 10.1007/s11095-010-0117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike A. C. X-ray crystallography of agonist/antagonist-bound receptors. Methods Mol Biol. 2009;505:51–66. doi: 10.1007/978-1-60327-575-0_4. [DOI] [PubMed] [Google Scholar]
- Qiagen Rapid, high-throughput assessment of protein stability on the Rotor-GeneQ cycler. In QIAGEN News. 2010 [Google Scholar]
- Raucy J. L., Lasker J. M. Current in vitro high throughput screening approaches to assess nuclear receptor activation. Curr Drug Metab. 2010;11:806–14. doi: 10.2174/138920010794328896. [DOI] [PubMed] [Google Scholar]
- Reinking J., Lam M. M., Pardee K., Sampson H. M., Liu S., Yang P., Williams S., White W., Lajoie G., Edwards A., Krause H. M. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell. 2005;122:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Schulman I. G., Heyman R. A. The flip side: Identifying small molecule regulators of nuclear receptors. Chem Biol. 2004;11:639–46. doi: 10.1016/j.chembiol.2003.12.021. [DOI] [PubMed] [Google Scholar]
- Shi Y. Orphan nuclear receptors, excellent targets of drug discovery. Comb Chem High Throughput Screen. 2006;9:683–9. doi: 10.2174/138620706778700125. [DOI] [PubMed] [Google Scholar]
- Vedadi M., Niesen F. H., Allali-Hassani A., Fedorov O. Y., Finerty P. J., Jr., Wasney G. A., Yeung R., Arrowsmith C., Ball L. J., Berglund H., Hui R., Marsden B. D., Nordlund P., Sundstrom M., Weigelt J., Edwards A. M. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci U S A. 2006;103:15835–40. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan K. F., Wang S., Brown C. J., Yu V. C., Entzeroth M., Lane D. P., Lee M. A. Differential scanning fluorimetry as secondary screening platform for small molecule inhibitors of Bcl-XL. Cell Cycle. 2009;8:3943–52. doi: 10.4161/cc.8.23.10114. [DOI] [PubMed] [Google Scholar]
- Yan X., Broderick D., Leid M. E., Schimerlik M. I., Deinzer M. L. Dynamics and ligand-induced solvent accessibility changes in human retinoid X receptor homodimer determined by hydrogen deuterium exchange and mass spectrometry. Biochemistry. 2004;43:909–17. doi: 10.1021/bi030183c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.