Abstract
Retinoic acid (RA) is one of the principal active metabolites of vitamin A (retinol) which mediates a spectrum of critical physiological and developmental processes. Transcriptional regulation by RA is mediated primarily by members of the retinoic acid receptor (RAR) subfamily of the nuclear receptor (NR) superfamily of transcription factors. NRs bind specific genomic DNA sequence motifs and engage coregulators and components of the basal transcription machinery to effect transcriptional regulation at target gene promoters. Disruption of signaling by retinoic acid is thought to underlie the etiology of a number of inflammatory and neoplastic diseases including breast cancer and haematological malignancies. A meeting of international researchers in retinoid signaling was convened in Strasbourg in September 2011 under the auspices of the European Molecular Biology Organization (EMBO). Retinoids 2011 encompassed myriad mechanistic, biological and pathological aspects of these hormones and their cognate receptors, as well as setting these advances in the context of wider current questions on signaling by members of the NR superfamily.
Introduction
Vitamin A, or retinol, is an essential nutrient for vertebrates. Retinoic acid (RA), the major bioactive metabolite of retinol, is a morphogen with pleiotropic roles in cell growth and differentiation in embryonic development and adult physiology [Chambon, 1996; Mark et al., 2009]. A number of key proteins and enzymes control retinoid metabolism and regulate the bioavailability of retinoic acid to its target cells. Serum retinol-binding proteins (RBPs) and cellular retinol binding proteins (CRBPs) transport retinol in the serum and cells; retinol dehydrogenases (RDHs) and retinal dehydrogenases (RALDHs) mediate the biosynthesis of RA from dietary vitamin A, the latter group of enzymes catalyzing the rate limiting step in RA biosynthesis; and the cellular RA binding proteins (CRABPs) mediate the uptake of RA in target cells.
RA signaling in its target cells occurs primarily through binding to members of the retinoic acid receptor (RAR) subfamily of the nuclear receptor (NR) superfamily of ligand-activated transcription factors [Aagaard et al., 2011; Tsai and O'Malley, 1994], namely RARα, RARβ and RARγ. Like other NRs, RARs contain a domain that mediates interaction with all-trans RA (the ligand binding domain, or LBD), a zinc finger-containing DNA binding domain (DBD) that binds to RA response elements (RAREs) in target genes; and a dimerization domain that engages members of the retinoid X receptor (RXR) subfamily in RXR/RAR heterodimers [Rochette-Egly and Germain, 2009]. RA-bound RARs effect transcriptional regulation by recruitment of coactivators and the subsequent assembly of a transcriptional preinitiation complex at target gene promoters [McKenna and O'Malley, 2002]. In contrast, in the absence of RA, unliganded RARs exert repressive effects on target gene promoters by recruitment of corepressor complexes. Central to this model is the engagement by RARs of epigenetic modifiers of promoter chromatin architecture, creating alternately permissive and restrictive environments for transcriptional initiation [Martens et al., 2011]. In addition to direct genomic effects via RARs, RA is known to engage cellular signaling pathways that mediate pre-genomic kinase cascades that also ultimately effect regulation of gene expression.
The myriad mechanistic and biological facets of retinoid signaling pathways, and the pathologies associated with their disruption, were the focus of a recent European Molecular Biology Organization (EMBO)-sponsored conference held at the Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC) in Strasbourg, France, on September 22-24th, 2011. The conference brought together researchers from a wide variety of disciplines, including phylogenetics, cell biology, biochemistry, structural biology, developmental biology and clinical practice, who collectively conveyed a keen appreciation of the fundamental roles played by retinoids and RARs, as well as other members of the NR superfamily, in orchestrating programs of transcriptional regulation required for the integrity of cellular growth and differentiation.
Keynote Lecture: Michael G. Rosenfeld
Many models of tumor development invoke random DNA translocation events which confer selective growth advantage on the cancerous cell. Michael Rosenfeld (UCSD, La Jolla, USA) proposed an alternative model of tumor translocation in which non-random, induced DNA translocation events result in cell proliferation and selection. TMPRSS2 is an androgen-regulated gene located on human chromosome 21q22.3 that encodes an enzyme of the serine protease family. The human ERG gene, also at 21q22.3, encodes a member of the erythroblast transformation-specific (ETS) family of transcription factors. Up to 70% of all prostate tumors have a specific fusion of TMPRSS2 and ERG. Rosenfeld described work showing that dihydrotestosterone (DHT)-induced spatial proximity, involving nuclear myosin I-dependent motors, was required for interchromosomal translocation between TMPRSS2 and ERG in prostate tumors. The induced double-stranded DNA break sites were localized to intronic regions of the gene translocation partners, and were shown to involve the association of androgen receptor (AR) with an AID/Gadd45 complex, the recruitment of which was abolished by selective AR modulators (SARMs). Gadd45/GADD45A is a member of the Gadd45 family of acidic nuclear proteins, which also contains MyD118 and CR6, whose transcript levels are increased following exposure of cells to stress conditions and treatment with DNA-damaging agents. While Rosenfeld’s observations were made in the context of tumor translocation events, the invocation of long distance DNA-DNA interactions in the context of AR action has exciting implications for the model of regulation of gene expression by this receptor and conceivably other members of the NR superfamily.
PIWIL1 is a member of the PIWI subfamily of Argonaute proteins, which play important roles in stem cell self-renewal, RNA silencing and translational regulation in diverse organisms. Sketching a model casting corepressors as protectors against translocation events in cancer, Rosenfeld drew attention to decreased levels of PIWIL1 in LNCaP prostate cancer cells. Supporting such a notion, he described experiments showing that translocation events attributable to genotoxic stress were increased in the context of PIWIL1 knockdown and decreased in the context of PIWIL1 overexpression, observations that appeared to apply generally to other translocations in addition to TMPRSS2/ERG. Rosenfeld briefly alluded to a chemical library screen for inhibitors of translocation using expression of fusion transcripts as a readout and focused on one particular hit, SD70, that was shown to bind to the active site of lysine demethylase 4C (KDM4C). Rosenfeld proceeded to describe a 3-dimensional DNA annealing, selection and ligation (DASL) strategy that had been used to show that E2-induced binding of estrogen receptor α (ERα) to target gene enhancers causes activation through induction of bidirectional non-coding RNA (ncRNA) transcripts and short range enhancer-promoter looping. In support of this, the SD70 compound, which had earlier been shown to inhibit translocation events, inhibited 17βE2 induction of target genes. The principle of DNA breakage events playing a key role in NR-mediated regulation of gene expression developed into a recurring theme throughout the meeting, and would later feature in talks by Pierre Chambon and Jean-Marc Egly (IGBMC, Strasbourg, France).
While retinoids are well-known inducers of stem cell differentiation, the cellular mechanisms underlying this biology are unclear. Rosenfeld showed that RA treatment of human stem cell lines, such as Ntera-2, but not stem cell lines from other species, induced RNA Polymerase III-regulated DR2 Alu transcripts. These transcripts were stabilized through an interaction with Ago3, a member of the Argonaute AGO subfamily, and accumulated in cytoplasmic P bodies, where they underwent Dicer-dependent processing and mediated degradation of specific stem cell mRNAs and subsequent exit from the stem cell state (Figure 1). A biochemical strategy to identify Ago3-interacting proteins implicated EDC4 in the assembly of a decapping complex and the subsequent degradation of the stem cell RNAs, leading to inhibition of stem cell proliferation and progression to a neuronal stem cell fate. Placing his results in a broader context, Rosenfeld speculated that by dint of their sheer numbers – approximately 3500 identified to date – non-coding RNAs were eminently positioned to function as global modulators of the regulation of gene expression by multiple classes of transcription factors.
Figure 1. Role of non-coding RNAs in RA signaling during neuronal differentiation.
RA induces a RAR, PolIII-dependent activation of DR2 Alu repeats in embryonic stem cells, generally adjacent to active PolII transcription units, and these transcripts are processed and serve to cause Ago3-dependent degradation of stem cell mRNAs harboring complementary sequences, with biological effects on signal-dependent exit from the stem cell state.
Special Plenary Lecture: Pierre Chambon
Glucocorticoids are one of the most widely prescribed prescription drug classes and have applications in a broad range of therapeutic indices, including cancer, osteoporosis and inflammatory disorders such as asthma and eczema. Their anti-inflammatory properties derive in part from the ability of agonist-bound GR to effect transrepression of transcription factors such as AP-1, which are key components of the inflammatory signaling response (Figure 2). Despite their clinical success, the myriad side-effects that accompany their use have restricted their more widespread use, and the pursuit of dissociated glucocorticoids, which retain the beneficial anti-inflammatory properties at the expense of undesirable side effects of GR transactivation, is the subject of intensive investigation in the pharmaceutical industry. Pierre Chambon (IGBMC, Strasbourg, France) described studies into an inflammatory skin condition that shed light on fundamental mechanisms of transcriptional regulation by GR and other NRs.
Figure 2. Beneficial and undesirable clinical effects of glucocorticoids are mediated by distinct transcriptional mechanisms.
See text for details.
Human atopic dermatitis (AD) is a familial skin condition affecting up to 20% of children and 3% of adults in Western society and is characterized by erythema, scaling and epidermal hyperplasia, among other symptoms. While glucocorticoid therapy is effective for the disease, chronic treatments using these drugs carry with them debilitating side-effects. Reviewing previous findings in his laboratory [Li et al., 2009; Li et al., 2006], Chambon described a mouse model, in which RXRα and RXRβ were selectively ablated in epidermal keratinocytes, that developed a skin and systemic syndrome mimicking AD that was attributable to ectopic expression of the inflammatory cytokine thymic stromal lymphopoietin (TSLP). Under normal physiological conditions in epidermal keratinocytes, multiple signaling pathways exert repressive effects on the TSLP promoter. Unliganded RXRα/VDR and RXRβ/RARγ heterodimers, for example, were shown to bind to VDREs (DR3-d, DR3-f and DR3-g) and a RARE (DR2-b) respectively, in the upstream region of the TSLP promoter. However, topical treatment with vitamin D3 or its hypocalcemic analog MC903, along with RA, results in the clearance of corepressors and recruitment assembly of coactivators to the TSLP promoter, induction of the gene and generation of an AD-like condition. The pivotal role of TSLP in the etiology of AD led Chambon to speculate whether the beneficial effects of glucocorticoid therapy in AD might be mediated in part by repression of TSLP. He demonstrated that the GR agonist fluocinolone acetonide repressed TSLP in epidermal keratinocytes in a GR-dependent manner. A tandem manual and bioinformatically-assisted search of the TSLP promoter identified an IR1 (consensus inverted repeat separated by a single nucleotide) that was shown to bind liganded GR in vivo, along with a cadre of corepressors and associated repressive histone writers, including HDAC2 and HDAC3. Referring to this sequence as an inverted repeat negative GRE (IR nGRE), he described a chromosome conformation capture assay, similar to that discussed earlier by Rosenfeld, which demonstrated that binding of GR to the nGRE precluded looping of the region containing the activating VDRE with the proximal promoter and subsequent initiation of transcription. Placing the mechanistic studies in a wider clinical context, Chambon noted not only that side effects of glucocorticoid-based therapy were attributable in certain cases to nGRE-dependent repression, but that available dissociated glucocorticoids retained the ability to directly transrepress through IR nGREs. These provocative findings have profound consequences for the direction of efforts to fine-tune glucocorticoid therapy, and point to the possible development of a new class of “improved” dissociated GCs which lack all GR functions, including transactivation and direct transrepression through nGREs, but not tethered transrepression [Surjit et al., 2011].
Phylogeny of retinoid signaling
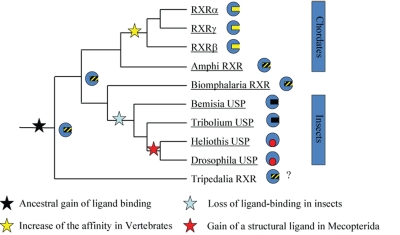
While the evolutionary functions of RARs have been well studied in chordates, outside this phylum, evidence for roles of RA signaling is scarce and the evolutionary origin of the RA pathway itself thus remains elusive. Vincent Laudet (IGFL, Lyon, France) highlighted the great variability in the NR complement of different organisms in the same subphyla. While vertebrates, he observed, have 50 NR genes, fish have up to 70; similarly, invertebrates share 25 known receptor genes, but nematodes have 250, while sponges have only a pair. Laudet suggested that NR phylogeny, placed in the context of metazoan evolution, was needed to detect NR orthologs. Such an approach was used to show that the genome of the Urbilaterian, the hypothetical last common ancestor of the bilaterian clade, contained about 25 NR genes, considerably more than anticipated. This number included orthologs of liganded receptors, which were previously thought to be specific to vertebrates. Moreover, he suggested that the NR phylogenetic tree, previously comprising 6 known subfamilies, could be updated to include 3 new subfamilies, namely NR7, NR8 and NR9, which are present in molluscs and annelids, but which have been lost in Drosophila, nematodes and vertebrates. Laudet suggested that the ancestral NR was most likely to be a low affinity fatty acid sensor (Figure 3). Turning to a discussion of the evolution of retinoid signaling, he pointed out that the DNA binding domains of metazoan RARs and RXRs are well conserved, and the RAR/RXR heterodimer was present in ancestral bilaterians. Laudet highlighted the good conservation of residues contacting RA in the LBD of RARs from various metazoans, although LBDs of early metazoans such as Amphioxus exhibited more variability than those of vertebrates. Laudet stressed the fact that there is a marked difference between the species tree and the NR tree: ultraspiracle in locusts, for example, is more similar to its human ortholog than that in Drosophila.
Figure 3. Evolution of retinoid signaling.
See text for details.
Classical models of retinoid signaling
Classical retinoid signaling involves the RAR-mediated transcriptional activation or repression of target genes in response to the presence or absence of RA. The binding of retinoic acid to RARs results in their formation of heterodimers with RXRs and their interaction with RAREs in the enhancer regions of target genes, primarily direct repeats separated by one (DR1), two (DR2) or five (DR5) nucleotides. These heterodimers associate with coactivator complexes and effect long-distance communications with the promoters of target genes, culminating in the recruitment of basal transcription factors such as TFIIH, the stabilization of a RNA Polymerase II-nucleated preinitiation complex and the transcription of these genes. In contrast, the establishment of repressive complexes in the absence of agonist is associated with silencing of target genes. Interactions of coregulators with NRs are known to involve specific α-helical sequences referred to as NR boxes, or LXXLL motifs in the case of coactivators [Heery et al., 1997] and CoRNR boxes in the case of corepressors [Hu and Lazar, 1999]. A solid body of evidence supports the notion that acetylation and methylation reactions catalyzed by coregulator-associated enzymes target a variety of substrates, including histones, and contribute to specific epigenomic signatures across target promoters that are critical determinants of their transcriptional productivity.
Ligand, receptor and coregulator interactions
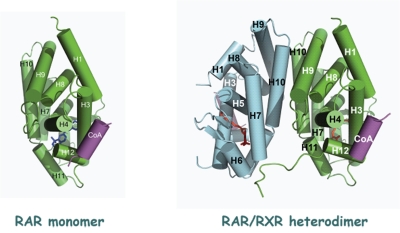
Small angle X-ray scattering (SAXS) and small angle neutron scattering (SANS) are complementary techniques used for structural characterization of crystallized molecules that yield data in the nanometer range. Dino Moras (IGBMC, Strasbourg, France) discussed a series of SAXS and SANS-based crystal structures of RARβ in solution that painted an intimate portrait of this receptor’s association with several coregulators. Moras observed that while the RARβ LBD was stable as a monomer in solution (Figure 4A), the functional form in vitro was a heterodimer with a member of the RXR subfamily. He demonstrated that the stoichiometry of the interaction of MED1/TRAP220 with RARβ was 1 coregulator:1 RARβ/RXR heterodimer (Figure 4B) – a ratio that held true for other coregulators such as SRC-1/NCOA1 and TIF2/NCOA2, and that mutation of the RXR moiety had no effect on the binding of TRAP220/MED1 to the heterodimer. By inducing asymmetry in the complex, DNA sequence and spacing proved to be critical modulators of the association of coactivator with receptor, suggesting a structural basis for the well-known effect of promoter context on receptor and coactivator function [Korzus et al., 1998]. Moras went on to show that in solution, RAR homodimers were asymmetrical, and that ligand alone did not induce dimerization and that the presence of a coactivator peptide established an equilibrium between the RARβ monomer and heterodimer. Moreover, in the context of a longer domain of SRC-1, the dimeric form was even more stable, such that a complex comprising RARβ and a 20kD SRC-1 receptor interaction domain (RID) was principally in the dimeric form. Moras sketched a model in which binding of coactivator induces a conformational change in the liganded LBD, resulting in increased stability of the dimeric form, and in which negative co-operativity reduces the affinity of the complex for a second coactivator moiety (Figure 3).
Figure 4. Stable forms of RARα in solution and in vivo.
A. The RARα LBD exists as a stable monomeric form in solution. B. The functional form of RARα is as a heterodimer with RXR. The stoichiometry of its interaction with its heterodimeric partner and coactivator is 1:1:1 RAR:RXR:coactivator peptide.
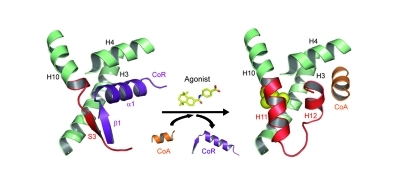
While the centrality of coregulator exchange as a general mechanism in the dual activation and repressive transcriptional properties of NRs has been accepted for over a decade [Glass and Rosenfeld, 2000], structure-function studies probing the molecular basis of RAR-coregulator interactions have been few in number. William Bourguet (Montpelier, France) described a number of crystallographic studies of RARs which illuminated the role of specific interaction interfaces on both receptor and corepressor in mediating gene silencing. Bourguet pointed out that RARs possess a strong basal repression function which RARα antagonists such as BMS614 can induce by disrupting the coactivator recognition surface of RARα by displacing helix H12. Bourguet presented two structures representing the opposing functionalities of RARα, a repressive form in complex with the antagonist BMS493 and CoRNR1 of SMRT/NCOR2 (to which RARα exhibits preferential binding), and an active form in complex with the agonist Am580 and an LXXLL motif-containing peptide. While broad similarities are evident between the two structures, they are distinguishable by secondary structures adopted by residues in helix H12 of the receptor, namely α-helical in the active form and adopting a predominantly β-strand character in the repressive form (Figure 5). Bourguet proceeded to discuss mammalian two-hybrid data indicating that residues adopting a β-strand conformation in CORNR1 were essential for corepressor/RAR interactions. Similarly, residues in β-strand 3 (S3) of the RARα LBD were critical for corepressor recruitment: a single mutation (I396E) prevents corepressor binding while preserving coactivator recruitment in the presence of agonist, and confers a much higher basal activation function than that of the wild type receptor. Bourguet suggested that by uncoupling the repressive and activational functions of RARα, the I396E mutant might be a valuable molecular tool with which to probe the molecular mechanism of RAR-mediated gene silencing more deeply.
Figure 5. A secondary-structure switch controls constitutive gene repression by RAR.
The constitutive recruitment of corepressor (CoR) by RAR is conferred by an extended β-strand (S3) that forms an antiparallel β-sheet with CoR residues (strand β1). This specific interaction allows docking of helix α1 of the CoR into the coregulator binding groove of RAR otherwise masked by helix H12. Upon agonist binding, a β-strand-to-α-helix transition allows for helix H11 formation which in turn provokes CoR release, repositioning of the activation helix H12 and coactivator (CoA) recruitment. For clarity, only the regions involved in the interaction between RAR, CoR and CoA are depicted.
TFIIH is a conserved multi-subunit complex involved primarily in transcriptional initiation and elongation and is comprised of a 7-subunit core, associated with a 3-subunit CDK-activating kinase (CAK) module. Mutations in genes encoding subunits of TFIIH involved in nucleotide excision repair (NER) have been characterized in a variety of genetic diseases, including xeroderma pigmentosum (involving XPB/TCRR3), trichothiodystrophy (involving XPD/TTD/ERCC2) and Cockayne syndrome (involving XPG/ERCC5). These diseases share a spectrum of common symptoms involving the skin (photosensitivity, skin cancer), development (growth and skeletal retardation) and neurological deficits. In addition to phosphorylating RNA Polymerase II, TFIIH has been shown to phosphorylate the N-terminal domains of several nuclear receptors, including RARα. Jean-Marc Egly (Illkirch, France) showed data demonstrating that residues in XPD played an essential role in anchoring the binding of the CAK moiety to the core TFIIH. Egly went on to show that transfection of wild type XPD into the HD2 cell line, which contains a XPD R683G mutation, restored RARα phosphorylation concomitant with restoring the responsiveness of these cells to all-trans retinoic acid, indicating a critical functional link between TFIIH phosphorylation of the receptor and its transcriptional function. Moreover, a RARα containing a phosphomimetic mutation at one of the serine residues discussed in Bruno Kieffer’s talk (S77E) was transcriptionally-competent in HD2 cells. Egly next used a ChIP-based approach to clarify the mechanistic basis of the involvement of NER factors in transcription. He described the recruitment of NER proteins at target gene promoters and showed that transient siRNA-mediated knockdown of NER factors disrupted patterns of histone modifications (H3K9/14) and DNA methylation (H3K4/H3K9) at RAR-regulated promoters. Recalling Rosenfeld’s earlier talk, Egly showed that XPG mutations prevented the establishment of a chromatin loop between the promoter and the terminator during RAR activation, suggesting that these factors were required for relaxing DNA and establishing conditions favorable for acetylation and demethylation. Concluding, he speculated that the pathological consequences of mutations in NER proteins in genetic disorders could be ascribed in large part to their role in establishing transcriptionally-permissive environments at promoters regulated by a broad range of transcription factors.
In fibroblasts undergoing in vitro differentiation into adipocytes, the CRABP family member CRABPI is subject to alternate up- and downregulation by thyroid hormone (T3). Li-Na Wei (University of Minnesota, Minneapolis, USA) discussed recent work in her laboratory investigating the role of coregulators in mediating this bidirectional regulation. During the initial phase of adipogenesis, CRABPI is upregulated in a TRAP220/MED1-dependent manner, in a mechanism involving chromatin looping and nucleosome sliding. In a subsequent second phase of differentiation, CRABPI is downregulated by T3 in a RIP140/NRIP1-dependent manner. Wei demonstrated that knockdown of RIP140 during this phase results in T3 activation of the CrabpI promoter and that its re-expression restored RIP140 repression. Contrasting the loose nucleosomal organization of the CrabpI promoter during Phase I with its condensed conformation in Phase II, Wei suggested that the bidirectional regulation of the CrabpI promoter was a function of its higher order chromatin organization. Wei went on to attribute the induction of RIP140 in the latter stages of adipocyte differentiation to the downregulation of a microRNA, miR-33, which targets the 3’-UTR of RIP140. Finally, post-translational modification of RIP140 was shown to play a role in mediating its interaction with TRα. Wei showed that RIP140 was subject to acetylation during adipogenesis, and that acetylation of K158 and K287 were required for repression of CrabpI and efficient interaction with TRα.
The identification and characterization of RAR family DNA binding events in response to retinoid signaling in model systems is an area of active investigation. Irwin Davidson (Illkirch, France) described recent genome-wide location analysis of RARα in mouse embryonic fibroblasts (MEFs) and embryonic stem (ES) cells [Delacroix et al., 2010]. Using a stably-transfected FLAG-tagged RARα-based strategy, he demonstrated that there were unique genomic occupancy sites in MEFs versus ES cells and, surprisingly, that there were relatively few RAR-occupied sites – those that were identified were DR1, DR2 or DR5 RAREs.
Davidson progressed to describe ChIP-Seq analysis of RAR genomic DNA binding events during the RA-induced development of neural progenitors from embryonic stem cells via embryoid body cultures, a well-characterized model system for retinoid signaling. Comparing his laboratory’s dataset with a similar study in F9 cells [Lalevee et al., 2011], he found that there were both overlapping and distinct cistromic signatures for RARα in both model systems. The most commonly occurring RAREs were DR0, DR2, DR5 and IR0 in addition to a novel DR8 motif which was a linear composite of DR2 and DR0 elements. In vitro analysis demonstrated that DR0 and IR0 elements bound RAR/RXR heterodimers and that IR0s and DR8s, but not DR0s, acted as independent RAREs on minimal promoters. Parallel mRNA-seq analysis showed that only a small subset of RAR-occupied genes were actually regulated by retinoic acid in embryoid bodies, and that a high frequency of the induced genes were in fact DR0s – which begs the question: what is RAR co-operating with if DR0 is not an independent RAR binding site? Most likely, elements in neighboring DNA sequences play a role, and future work will investigate this possibility.
Alternative models of retinoid signaling
Alternative pre-genomic (or “non-genomic”) models of NR signaling invoke the interface between receptors and the cellular kinase cascade-mediated signaling pathways that are initiated by the binding of peptide hormones and growth factors to cell membrane-bound G-protein coupled receptors. Many proteins involved in these pathways contain Src-homology (SH) domains which have well-characterized roles in signal transduction and tumorigenesis. Work in the laboratory of Cécile Rochette-Egly (Illkirch, France) previously demonstrated the identification of SORBS3/vinexin-β, a multiple SH3 domain-containing protein involved in cell-cell adhesion and cytoskeletal function, as a RARγ corepressor. The interaction between the two proteins was shown to be mediated by the third SH3 domain (SH3.3) of vinexin-β and was uncoupled by phosphorylation of RARγ at two serine residues in its AF-1 domain.
In the first of the meeting’s short talks, Bruno Kieffer described recent work in the Rochette-Egly laboratory on the solution structure of the vinexin-β SH3.3 domain in complex with a proline-rich domain (PRM) of RARγ that mediates the interaction with vinexin-β. Analysis of the structure, in addition to biophysical interrogation of the effect of serine phosphorylation on the interaction surfaces of the two moieties, indicated that the phosphorylation of PRM modulated the conformations of the interacting partners by reducing their type II polyproline (PPII) character, an important conformation in disordered states of proteins. Kieffer concluded by suggesting that the interaction between the SH3.3 and PRM domains was modulated by a conformer selection mechanism.
Pre-genomic activation of kinase signaling cascades by retinoids was also the subject of a short talk by Aleksandr Piskunov (IGBMC, Strasbourg, France) of the Rochette-Egly laboratory. Members of the Gq family of heterotrimeric G proteins activate β-isoforms of phospholipase C which in turn catalyzes the hydrolysis of phosphatidylinositol phosphate to diacylglycerol and inositol triphosphate, resulting in protein kinase C activation and mobilization of cellular calcium.
Piskunov showed that RAR agonists induce the p38/MAPK/MSK1 pathway, and went on to describe an interaction between Gαq, a member of the Gq family, and RARα in lipid rafts, which are dynamic microdomains within the cell membrane lipid bilayer that assemble specific populations of transmembrane signaling proteins. Moreover, knockdown of either Gαq or p38 significantly attenuated RA induction of a reporter gene.
Recapitulating the phosphorylation of RARα serine 77 by kinase signaling cascades alluded to in Jean-Marc Egly’s talk, Piskunov demonstrated that mutation of these residues abrogated induction of the RARβ gene by RA, providing further evidence for the contribution of cellular kinase signaling pathways to retinoid signaling.
Continuing the theme of non-classical pathways of retinoid function, Noa Noy (Case Western Reserve University, Cleveland, USA) described a specific cell-surface RBP receptor, STRA6, a multitransmembrane domain and SH2-binding domain protein that is widely expressed in embryonic development and in adult organ systems, and previously characterized as a RA-induced gene. Mutations in STRA6 contribute to a spectrum of congenital malformations collectively known as Matthew Wood syndrome [Pasutto et al., 2007], and which include anophthalmia, congenital heart defects and mental retardation.
Noy pointed out that ectopic overexpression of RBP contributes to insulin resistance in animal models, a somewhat surprising observation given that RA suppresses obesity and improves insulin resistance. A clue to this apparent paradox is the crosstalk of STRA6 with the JAK/STAT pathway, a kinase cascade that mediates cellular response to peptide hormones including interleukins, interferons, leptin and insulin. RBP-retinol binding to STRA6 was shown to induce recruitment of JAK2 and STAT5 by STRA6, as well as triggering phosphorylation of all three proteins. Moreover, RBP-retinol induced STAT5, SOCS3 and PPARγ expression in adipocytes via a mechanism independent of RA.
In addition to its effects on transcription, Noy demonstrated that RBP-retinol abrogated activation of the insulin receptor in a STRA6-, STAT5- and SOCS3-dependent manner.
Finally, Noy showed that transient recruitment of cellular retinol binding protein I (CRBPI) to STRA6 upon retinol-RBP binding was required for retinol signaling, and that STRA6 signaling was abolished in CRBPI mutants. Her data sketch the outline of a negative feedback loop model in which retinol, signaling though RBP, induces SOCS3 and PPARγ via the JAK/STAT pathway, resulting in lipid accumulation (via PPARγ) and downregulation of glucose uptake through antagonism of insulin-insulin receptor signaling (via SOCS3).
The contribution of CRBP1 to retinoid signaling outlined in Noy’s talk is interesting in the context of data presented by Eduardo Farias (New York, USA), who demonstrated that CRBP-1 impaired c-myc-induced tumorigenesis. His data indicated opposing effects of RAR isotypes, such that RARα and RARβ drive cell cycle arrest and differentiation, whereas RARγ had a repressive effect on CRBP expression, which is silenced in a wide range of tumor types. He suggested that RARα/β agonists and RARγ antagonists could provide novel avenues for treatment of breast cancer.
Developmental biology of retinoid signaling
The functions of RA and RARs in developmental biology is a complex interplay of factors ranging from tissue-selective distribution of RA and its biosynthetic precursors, expression of RAR isoforms, and the functional interactions of RARs and RXRs with transcriptional coregulators to effect co-ordinated programs of gene activation and repression. Retinol dehydrogenases (RDH) comprise a family of enzymes that generate all-trans retinaldehyde from all-trans retinol during the initial step of retinoic acid biosynthesis. Downstream of the generation of retinaldehyde substrates by RDH enzymes is the conversion of these to retinoic acid by tissue-specific enzymes of the retinaldehyde dehydrogenase (Raldh) family, which is a key component in the establishment of morphogenetic gradients of retinoic acid which play demonstrably crucial roles in early somitogenesis and neurogenesis.
Tissue-specificity of RA biosynthesis: retinol dehydrogenases (RDHs) and retinaldehyde dehydrogenases (RALDHs)
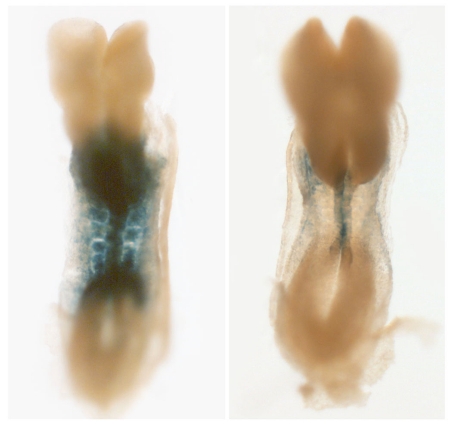
Pascal Dollé (IGBMC, Strasbourg, France) discussed Cre-Lox conditional knockout model-based studies on the role of one member of the Rdh family of enzymes, Rdh10, in early embryogenesis, and the insights these afford into tissue-specific principles in retinoid signaling. While Rdh10 null mutants are early embryonic lethal (E10.5-E12.5), retinoid signaling as reported by a RARE-Lacz reporter transgene, is significantly compromised (Figure 6), but not fully abolished – a reflection of the existence of other members of this enzyme family possibly acting during tumorigenesis. Rdh10-/- mutant embryos display a number of developmental defects, including abnormal patterning in the posterior hindbrain and compaction of the cervical somitic region. Defects in Rdh10-/- mutants were also the subject of a short talk by Gregg Duester (La Jolla, USA), who noted impaired forelimb development as well as defects in early somitic development related to ectopic Fgf8 expression in the trunk at early stages of development. Reiterating the tissue-specific nature of Raldh action, Duester noted that while RA signaling was compromised in the somitic mesoderm, it was intact in the neuroectoderm.
Figure 6. Highly diminished RARE-lacZ reporter transgene activity in an early mouse embryo with a targeted Rdh10 gene disruption.
See text for details.
Dollé next used rescue experiments involving RA or retinaldehyde transplacental supplementation of pregnant mice to determine whether Rdh10 activity was required for generating tissue-specific distributions of substrates for embryonic RALDH. He found that RA rescue did not result in viable Rdh10-/- mutant embryos, and that these mutants displayed organogenesis defects characteristic of RA deficiency in the heart (reduced myocardial compact zone, epicardial detachment), large vessels (pulmonary artery atresia) and lungs (absence of left lung, hypoplastic right lung). In contrast, retinaldehyde supplementation rescued most developmental defects and resulted in viable, fertile mutants, suggesting that by failing to recapitulate the precise tissue-specific gradients of RA required during development, RA supplementation is unable to rescue mutants lacking Rdh10.
Dollé concluded with data implicating a member of the Raldh family, Raldh3, in development of the inner ear vestibular system. Raldh3 is expressed in discrete areas of the inner ear, including the spiral ganglia of the cochlea and specific regions of the vestibular sensory epithelium, such as the utricle and saccule. Reflecting the role of these structures in hearing and balance, Raldh-/- mutant mice exhibit an abnormal spontaneous circling behavior and motor skill deficiencies, as well as impaired development of inner ear vestibular structures. In particular, there is an absence in these mice of otoconia, mineralized structures in the utricle and saccule that provide mass loads for detection of the gravity and linear acceleration. Dollé concluded by speculating that RA-based therapeutics might be profitably used to offset the otoconial degeneration that contributes to a variety of senescent diseases.
Retinoid signaling in somitic development and neurogenesis
In vertebrate species, the vertebral column, skeletal muscle and dermis are derived from embryonic structures called somites, which are formed by the periodic, bilaterally symmetrical process of mesodermal segmentation. The fibroblast growth factor (FGF) pathway establishes a traveling posterior to anterior gradient of activity in the presomitic mesoderm such that the segmentation transcriptional program, involving genes such as Mesp2, is triggered only in cells exposed to sufficiently low levels of FGF. Opposed to the FGF gradient is a RA gradient, and the antagonistic effects of signaling pathways regulated by these ligands establishes a determination front for segmentation. Raldh2/Aldh1a2 catalyzes the synthesis of RA from retinaldehyde in somitic and rostral presomitic mesoderm; its inhibition by disulfiram in the chick, or by gene targeting in the mouse, results in left-right asymmetry of somitogenesis [Vermot et al., 2005; Vermot and Pourquie, 2005]. Olivier Pourquié (IGBMC, Strasbourg, France) drew attention to defects in somitogenesis, including delayed somite formation on the right side, in a mouse model containing a mutation in Rere/Atrophin2, previously characterized as a coregulator for members of the COUP-TF subfamily of NRs [Wang and Tsai, 2008]. In support of a role for Atrophin 2 in retinoid signaling, developmental defects in the Rere/Atrophin2 mutant were strikingly phenocopied in the Raldh2 mutants. Pourquié proceeded to show data from a RARE-LacZ mouse crossed into the Atrophin 2 mutant that Atrophin 2 was required for the in vivo response to RA, and that Atrophin 2 potentiated the transcriptional response to RA in F9 cells. Supporting biochemical and in vivo data demonstrated that a complex comprising Atrophin 2, p300 and COUP-TFII/Nr2f2 was recruited to RA-responsive genes, and that knockdown of COUP-TFII and Atrophin 2 abrogated RA signaling. Pourquié demonstrated that RARs were expressed in the pre-somitic mesoderm in mouse E8.5 embryos, and that asymmetric expression of COUP-TFII overlapped with the RA response domain in the right pre-somitic mesoderm. Finally, Pourquié discussed multi-dimensional protein identification technology (MUDPIT) analysis of Atrophin 2-associated proteins which demonstrated the existence in the Atrophin 2 interaction network of HDAC1, HDAC2, p300 and a protein named STRAP (serine/threonine kinase receptor-associated protein), mutations in which result in somite desynchronization defects.
Kate Storey (University of Dundee, Dundee, UK) continued the developmental theme with a discussion of neurogenesis, which has several parallels with somitogenesis in the context of retinoid signaling, notably the functional antagonism between retinoid- and FGF-regulated pathways. During neurogenesis, retinoid signaling, driven in part by Wnt promotion of retinoid biosynthesis represses FGF action, resolves mesodermal and neural cell fates and promotes commitment to the neural cell lineage. Genes involved in neurogenesis whose expression is subject to mutually opposed regulation by FGF (repression) and retinoids (induction (activation) include neurogenin (Ngn)1 and Ngn2, NeuroM and Crabp1, although these represent only a small fraction of the major re-organization of transcriptomes which takes place along the neural axis during neurogenesis. The FGF/retinoid antagonistic paradigm so clearly characterized in this context is not a general rule however, and Storey went on to discuss how retinoid signaling during embryonic stem (ES) cell differentiation can, under certain conditions, promote FGF/Erk signaling, at least transiently. Specifically, RA initially induces a rapid increase in Erk1/2 signaling via induction of Fgf8, which ChIP data from the laboratory of Gregg Duester (La Jolla, USA) implicate as the result of direct binding of RAR to the Fgf8 promoter. As differentiation progresses however, there is a net repression of ERk1/2 signaling, most likely via repression of Fgf4.
Norbert Ghyselinck (IGBMC, Strasbourg, France) concluded the developmental strand with a discussion of specific and overlapping roles of RARs during male germ cell differentiation. Dietary vitamin A deficiency induces testis degeneration via arrest of spermatogonia proliferation, which can be reversed by vitamin A or RA supplementation, indicating that germ cell differentiation depends on RA signals. Ghyselinck showed that RARγ is expressed at specific stages during spermatogenesis, specifically in single cell spermatogonia, as well as in aligned A spermatogonia in later stages (Figure 7). Cre-lox-mediated ablation of RARγ or RARα individually in Sertoli cells is sufficient to induce testis degeneration through loss of germ cells, although the specific developmental deficit in each mutant appears to occur at different points during spermatogenesis. RARα and RARγ appear to be functionally redundant to some extent in this context since expression of Stra8, a RA-inducible germ cell marker, which is undetectable in RARγ-deficient mice, can be restored by administration of the RARα agonist BMS753. Observing that ablation of RXRα, RXRβ and RXRγ in spermatogonia results in a loss of germ cells similar to that in RARγ and RARα, Ghyselinck proposed a model in which RARα and RARγ function cell-autonomously as heterodimers with RXRs.
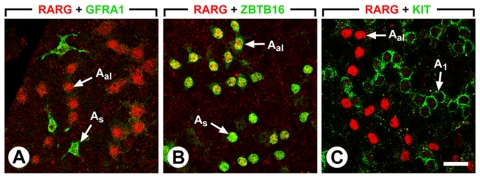
Figure 7. RARγ is expressed in Aal and A1 spermatogonia.
External views of seminiferous tubules of wild-type, sexually mature, mice that were double-immunostained with anti-RARγ (red nuclear signal) and anti-GFRA1 (green membrane signal) antibodies in panel A, with anti-RARγ (red nuclear signal) and ZBTB16 (green nuclear signal) antibodies in panel B, and with anti-RARγ (red nuclear signal) and anti-KIT (green membrane signal) antibodies in panel C. Legend: As, Aal, and A1 indicate A single, A aligned and A1 spermatogonia, respectively. Bar: 50 µm.
Retinoid signaling and disease
Consistent with their promotion of exit from the cell cycle and progression towards differentiated cell fates, retinoids have anti-proliferative and apoptotic effects and have found application in a number of inflammatory conditions including skin disorders, as well as diseases such as breast cancer and leukemia. Moreover, the functional integrity of RARs is often subverted in gene translocation events commonly encountered in leukemias, resulting in ectopic repression of transcriptional programs involved in myeloid cell differentiation [Duong and Rochette-Egly, 2011]. Harnessing retinoid signaling pathways has met with mixed success as a cancer therapeutic strategy, providing an impetus for more detailed models of retinoid action in cancer cells and the exploration of alternative strategies, including epigenomics, for intervention in retinoid pathways opposing tumor progression.
Breast cancer
Sylvie Mader (Montreal, Canada) described recent efforts in her laboratory focused on RALDHs as therapeutic leverage points in breast cancer. RALDH3 is the predominant RALDH family member in normal breast tissue, as well as immortalized and breast cancer epithelial cell lines. Mader noted that the majority of breast cancer cell lines tested have low RA synthetic capacity, which correlates with low levels of RALDH3 expression at both the mRNA and protein levels, a pattern recapitulated in luminal breast tumors. In contrast, RALDH3 was highly expressed in normal breast tissue and mediated retinoic acid biosynthesis in human immortalized mammary gland 184B5 cells. Given these observations, Mader next asked whether ectopic RALDH expression might inhibit growth and proliferation of breast cancer cell lines and luminal tumors. RALDH3 was capable of inducing G0/G1 arrest in tumorigenic SKBR3 cells, inhibited colony formation in MCF-7 breast cancer cells and retarded tumor growth in MCF-7 xenograft models. At the transcriptional level, Mader demonstrated a remarkable overlap between patterns of gene expression in luminal cells exposed to retinoic acid or expressing RALDH3, resulting in the activation of luminal-specific transcription factors and of anti-proliferative and pro-apoptotic gene networks. The 17β-estradiol/ER proliferative paradigm is well established in models of epithelial breast cancer, and Mader’s closing remarks illuminated the dynamic antagonism that exists between this signaling conduit and that of retinoids. She observed that RALDH3 expression is blocked by 17β-estradiol in an ERα-dependent manner, while this effect is reversed by anti-estrogens such as Fulvestrant and Tamoxifen.
A number of models exist for the development of resistance by breast tumors to treatment with Tamoxifen, one of which invokes the role of a cancer stem cell population in the generation of novel chemotherapeutically-resistant cell lineages in tumors. Maria del Mar Vivanco (Bilbao, Spain) described recent work in her laboratory which sheds light on the role of tumor stem cells using a Tamoxifen-resistant MCF-7 cell line. These cells resemble progenitor cells in their expression of high levels of Sox2, a member of the SRY-related HMG-box family of transcription factors required for the maintenance of cancer stem cells. Ectopic overexpression of Sox2 was shown to confer increased resistance of cells to the anti-proliferative effects of Tamoxifen, and clinically, Sox2 was more highly expressed in recurrent tumors than in primary tumors.
While most of the speakers reiterated the pro-differentiative, antiproliferative effects of retinoid signaling, Vincent Giguère (Montreal, Canada) sounded a dissonant note by discussing data casting RA and RARβ as cancer promoters. RARβ null mice exhibit delayed mammary gland development and tumorigenesis is delayed in a MMTV-ErbB2 model in a RARβ null background. Giguère cited downregulation of the Cxc12/SDF-1 pathway in RARβ null mice as a possible mechanistic basis for RARβ’s promotion of tumorigenesis. While provocative, Giguère’s results did not exclude the possibility that there is developmental overcompensation for germline loss of RARβ from other RAR isoforms, a possibility given the overlapping functions of members of this subfamily, and a conditional mammary gland knockout of RARβ would more clearly address the nature of its role in breast cancer.
Leukemias
Hematopoiesis is the continuous formation of all blood cell types from hematopoietic stem cells residing in the bone marrow. Cell fates during hematopoiesis are a function of both extracellular signaling networks mediated by soluble growth factors and their receptors, as well as specific transcriptomic fingerprints triggered in individual cell populations in response to these signaling events. In myeloid leukemia, the mechanisms that normally promote differentiation into the diverse myeloid lineages are disrupted, leading to pathological accumulation of immature hematopoietic cell types.
Henk Stunnenberg (Nijmegen, The Netherlands) opened with an overview of the European Union-funded BLUEPRINT of Hematopoietic Epigenomes, a team science endeavor that aims to generate genomic DNA sequence, epigenomic and transcriptomic datasets from about 60 distinct types of primary hematopoietic cells and blood cancer subtypes. Oncofusions involving AML and PML proteins are well characterized in leukemias: the fusion protein, promyelocytic leukemia (PML)-retinoic acid receptor-α (RARA), initiates acute promyelocytic leukemia (APL) through both a block to differentiation and increased self-renewal of leukemic progenitor cells. Similarly, the fusion of AML1 and the transcriptional corepressor ETO (RUNX1T1) through a t(8;21) translocation is a frequent genetic lesion in acute myeloid leukemia (AML). Stunnenberg posed the questions: where do these oncofusion proteins bind, do they alter the epigenomic environment where they bind, and do common mechanisms exist by which they achieve these ends? Stunnenberg described a comparison of AML1 and AML1/ETO sites in SKNO/Kasumi cells and AML patient blasts, and demonstrated a redistribution of targets from gene promoters in the case of AML1 towards introns and intergenic regions for AML1/ETO. Stunnenberg pointed out that AML1/ETO bound preferentially to multimerized sites, and speculated that this was due to the oligomerization properties of AML1, previously shown to be required for the interaction of ETO with transcriptional corepressors. Members of the erythroblast transformation-specific (ETS) family of transcription factors are key regulators of embryonic development, cell proliferation, differentiation, angiogenesis, inflammation, and apoptosis. Stunnenberg described an overlap between binding sites for ERG, a member of the ETS family, and AML1-ETO in U937 cells, and highlighted the fact that elevated expression of ERG in AML was associated with poor prognosis in the disease. Addressing the epigenomic consequences of AML1-ETO, Stunnenberg demonstrated that AML1/ETO binding in U937 cells was associated with local hypoacetylation, consistent with the association of ETO with corepressors. Drawing parallels between the AML1/ETO and PML/RARα paradigms, he described extensive overlap between the binding sites of the two fusion proteins. He concluded by speculating that a common mechanism for disease progression was the recruitment of the oncofusion proteins to promoters of ETS family-regulated genes encoding proteins required for myeloid differentiation, and the subsequent epigenomic silencing of these genes via a mechanism involving association of corepressors with the fusion proteins.
In addition to the well-characterized functions of transcription factors and coregulators, there is an increasing recognition of miRNAs as key players in orchestrating specific transcriptional endpoints during hematopoiesis. Clara Nervi (University of Rome, Rome, Italy) focused on the role of one such miRNA, miR-223, in normal hematopoietic development as well as in the deficits in differentiation characteristic of the progression of leukemic cells, and the interface between this miRNA and retinoid signaling. Administration of RA in HL60 cells results in the association of miR-223 with the miRNA processing ribonuclease Dicer1, a key step in the processing of miRNAs. Nervi showed that ectopic expression of miR-223 potentiated granulocytic differentiation of ALP-NB4 cells, and that the AML1/ETO oncofusion targeted miR-223 for heterochromatic silencing.
Further evidence for the functional interaction of miR-223 and AML1/ETO in hematopoietic pathologies was provided by the similar myeloproliferative disorders in miR-223 knockout and AM1/ETO transgenic mice. Nervi provided evidence that one of the targets of miR-223 in this context was NFIA: treatment of HL60 cells with RA resulted in silencing of the NFIA promoter accompanied by recruitment of miR-223. Moreover, NFIA was downregulated in HL60 cells following overexpression of miR-223. Progressing to the clinical setting, Nervi demonstrated that expression of miR-223 in primary blasts from AML patients restored myeloid differentiation.
Failure of tumors to respond to retinoid therapy is a common clinical observation, suggesting that establishing an environment more receptive to retinoid signaling might be a profitable approach in treatment of the disease. In this context, Christine Chomienne (Hôpital St Louis, Paris, France) discussed epigenetic strategies to enhance RAR promoter permissiveness as a therapeutic strategy in leukemogenesis. She demonstrated that non-responsiveness of thyroid cancer cells to RA was associated with reduced histone acetylation and increased DNA methylation of the RARβ promoter. The combination of RA with a histone deacetylase inhibitor resulted in increased expression of RARβ in these cells. In the clinical setting, the combination of RA with histone deacetylases such as valproic acid in non-APL leukemias was associated with complete remission in older patients, albeit with a modest response rate.
Hugues deThé (Hôpital St Louis, Paris, France) concluded the meeting with a discussion of data on the molecular mechanisms of RA and arsenic trioxide (As2O3) therapy in APL, a combination therapy that has become standard of care for the disease, with an emphasis on the PML-RARα oncofusion. Both RA and As2O3 degrade PML-RARα, and synergize in the treatment of APL in mice, radically extending their life span. This synergy arises from the fact that they engage parallel proteasome-dependent PML/RARα degradation pathways, As2O3 targeting the PML moiety of the oncofusion for sumoylation and RA targeting the RAR portion of the molecule in a manner evoking the targeting of ERα for degradation by Fulvestrant. deThé highlighted the role of a CC motif in the B2 box region of PML, pointing out that amino acid variability in this region of the molecule, secondary to mis-sense mutations in the PML gene, was a critical determinant of the responsiveness to As2O3 treatment. deThé invoked a derepression model for PML-RARα-targeted therapy, suggesting that loss of PML-RAR was the motive force behind the reversion to a default myeloid differentiation pathway.
Conclusions
This brief, focused meeting reflected the ongoing efforts of a highly active clinical and translational global research community and the contributions that it continues to make not only in the context of retinoid signaling, but to the broader picture of nuclear receptor signaling. It reiterated the myriad critical junctures along the axis of retinoid signaling – retinoic acid biosynthesis, the interaction between ligand and receptor, or the action of RAR coregulators, has been shown to have profound consequences both in early embryonic development and in a variety of human pathologies. In addition to these better characterized components, compelling evidence was made for non-coding RNAs, DNA looping and excision repair mechanisms, as well as epigenomic programming of promoters, as emerging mechanistic principles not only in retinoid signaling, but in the wider context of signal transduction by the NR superfamily. Finally, the field has shown itself to be highly active in the translation of mechanistic principles of retinoid signaling into strategies that are showing promise in the treatment of myriad neoplastic and inflammatory conditions, and one can justifiably anticipate further exciting advances in the mechanism, biology and pathology of retinoid signaling in the coming years.
Acknowledgments
The author thanks the European Molecular Biology Organization (EMBO) and the meeting organizers, Cécile Rochette-Egly, Christine Chomienne and Vincent Laudet for their kind invitation to write a review of the meeting.
References
- Aagaard M. M., Siersbaek R., Mandrup S. Molecular basis for gene-specific transactivation by nuclear receptors. Biochim Biophys Acta. 2011;1812:824–35. doi: 10.1016/j.bbadis.2010.12.018. [DOI] [PubMed] [Google Scholar]
- Chambon P. A decade of molecular biology of retinoic acid receptors. Faseb J. 1996;10:940–54. [PubMed] [Google Scholar]
- Delacroix L., Moutier E., Altobelli G., Legras S., Poch O., Choukrallah M. A., Bertin I., Jost B., Davidson I. Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol Cell Biol. 2010;30:231–44. doi: 10.1128/MCB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong V., Rochette-Egly C. The molecular physiology of nuclear retinoic acid receptors. From health to disease. Biochim Biophys Acta. 2011;1812:1023–31. doi: 10.1016/j.bbadis.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Glass C. K., Rosenfeld M. G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- Heery D. M., Kalkhoven E., Hoare S., Parker M. G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–6. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Hu X., Lazar M. A. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–6. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- Korzus E., Torchia J., Rose D. W., Xu L., Kurokawa R., McInerney E. M., Mullen T. M., Glass C. K., Rosenfeld M. G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–7. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- Lalevee S., Anno Y. N., Chatagnon A., Samarut E., Poch O., Laudet V., Benoit G., Lecompte O., Rochette-Egly C. Genome-wide in silico identification of new conserved and functional retinoic acid receptor response elements (direct repeats separated by 5 bp) J Biol Chem. 2011;286:33322–34. doi: 10.1074/jbc.M111.263681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Hener P., Zhang Z., Ganti K. P., Metzger D., Chambon P. Induction of thymic stromal lymphopoietin expression in keratinocytes is necessary for generating an atopic dermatitis upon application of the active vitamin D3 analogue MC903 on mouse skin. J Invest Dermatol. 2009;129:498–502. doi: 10.1038/jid.2008.232. [DOI] [PubMed] [Google Scholar]
- Li M., Hener P., Zhang Z., Kato S., Metzger D., Chambon P. Topical vitamin D3 and low-calcemic analogs induce thymic stromal lymphopoietin in mouse keratinocytes and trigger an atopic dermatitis. Proc Natl Acad Sci U S A. 2006;103:11736–41. doi: 10.1073/pnas.0604575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark M., Ghyselinck N. B., Chambon P. Function of retinoic acid receptors during embryonic development. Nucl Recept Signal. 2009;7:e002. doi: 10.1621/nrs.07002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J. H., Rao N. A., Stunnenberg H. G. Genome-wide interplay of nuclear receptors with the epigenome. Biochim Biophys Acta. 2011;1812:818–23. doi: 10.1016/j.bbadis.2010.10.005. [DOI] [PubMed] [Google Scholar]
- McKenna N. J., O'Malley B. W. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–74. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Pasutto F., Sticht H., Hammersen G., Gillessen-Kaesbach G., Fitzpatrick D. R., Nurnberg G., Brasch F., Schirmer-Zimmermann H., Tolmie J. L., Chitayat D., Houge G., Fernandez-Martinez L., Keating S., Mortier G., Hennekam R. C., von der Wense A., Slavotinek A., Meinecke P., Bitoun P., Becker C., Nurnberg P., Reis A., Rauch A. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am J Hum Genet. 2007;80:550–60. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette-Egly C., Germain P. Dynamic and combinatorial control of gene expression by nuclear retinoic acid receptors (RARs) Nucl Recept Signal. 2009;7:e005. doi: 10.1621/nrs.07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Ganti K. P., Mukherji A., Ye T., Hua G., Metzger D., Li M., Chambon P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–41. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- Tsai M. J., O'Malley B. W. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–86. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- Vermot J., Gallego Llamas J., Fraulob V., Niederreither K., Chambon P., Dolle P. Retinoic acid controls the bilateral symmetry of somite formation in the mouse embryo. Science. 2005a;308:563–6. doi: 10.1126/science.1108363. [DOI] [PubMed] [Google Scholar]
- Vermot J., Pourquie O. Retinoic acid coordinates somitogenesis and left-right patterning in vertebrate embryos. Nature. 2005b;435:215–20. doi: 10.1038/nature03488. [DOI] [PubMed] [Google Scholar]
- Wang L., Tsai C. C. Atrophin proteins: an overview of a new class of nuclear receptor corepressors. Nucl Recept Signal. 2008;6:e009. doi: 10.1621/nrs.06009. [DOI] [PMC free article] [PubMed] [Google Scholar]