Abstract
Numerous studies have indicated that each of the seven projections associated with the central pair of microtubules plays a distinct role in regulating eukaryotic ciliary / flagellar motility. Mutants which lack specific projections have distinct motility phenotypes. For example, Chlamydomonas pf6 mutants lack the C1a projection and have twitchy, non-beating flagella. The C1a projection is a complex of proteins including PF6, C1a-86, C1a-34, C1a-32, C1a-18 and calmodulin. To define functional domains within PF6 and to potentially assign functions to specific C1a components, we generated deletion constructs of the PF6 gene and tested for their ability to assemble and rescue motility upon transformation of mutant pf6 cells. Our results demonstrate that domains near the carboxyl-terminus of PF6 are essential for motility and/or assembly of the projection. The amino terminal half of PF6 is not required for C1a assembly; however, this region is important for stability of the C1a-34, C1a-32, and C1a-18 sub-complex and wild-type beat frequency. Analysis of double mutants lacking the amino terminus of PF6 and outer dynein arms reveal that C1a may play a role in modulating both inner and outer dynein arm activity.
Key Words/Phrases: Ciliary and flagellar motility, Chlamydomonas
Introduction
Motile cilia and flagella are found on diverse cell types; yet, for all eukaryotic cells which possess these organelles, movement is generated by dynein-driven sliding between adjacent doublet microtubules [Brokaw, 1989; Gibbons and Rowe, 1965; Satir, 1968; Summers and Gibbons, 1971]. The discovery that dynein generates force in a single direction led to a “switch point” model for ciliary beating in which dynein arms on opposite sides of the axoneme must alternate between active and inactive states [Sale and Satir, 1977; Satir, 1985; Satir and Matsuoka, 1989]. Switching between these states to generate simple oscillatory bends most likely involves structural cues generated from the basic microtubule sliding mechanism which couples the probability of dynein-microtubule interactions with the degree of flagellar bending [Lindemann, 1994; Lindemann, 2011; Sanchez et al., 2011]. However, the variety and complexity of ciliary waveforms and beat frequencies observed for different cell types as well as the ability to modulate these parameters suggests that additional control systems must be imposed upon this model to further regulate ciliary and flagellar beating. A significant number of studies have demonstrated that the central apparatus and radial spokes form part of a signaling network that modulates dynein activity to ultimately affect ciliary and flagellar movement [reviewed in Mitchell, 2009; Smith and Yang, 2004; Wirschell et al., 2009; Yang and Smith, 2009].
The central apparatus is composed of two singlet microtubules and at least seven biochemically and structurally distinct protein projections [recently reviewed in Mitchell, 2009]. Chlamydomonas mutants that lack the central apparatus are paralyzed, underscoring the importance of this structure in regulating motility [Witman et al., 1978]. In vitro analysis of protease-treated axonemes shows that microtubule sliding still occurs in the absence of the central pair, although the velocity of sliding is reduced [Smith, 2002b; Witman et al., 1978]. Addition of kinase inhibitors or free calcium to central pairless axonemes restores sliding velocity to wild-type levels, suggesting that the central apparatus is a part of a signal transduction network that alters dynein-driven microtubule sliding [Smith, 2002b; Smith, 2002a]. The finding that mutations in dynein arm components or components of the nexin-DRC (dynein regulatory complex) suppress paralysis in central pairless mutants further supports the hypothesis that dynein is a downstream effector of the central apparatus [Huang et al., 1982; Porter et al., 1994; Porter et al., 1992; Rupp et al., 1996].
Unlike mutants which fail to assemble the entire central apparatus, mutants that specifically lack one or more protein projections are not always paralyzed [Adams et al., 1981; Dutcher et al., 1984; Witman et al., 1978]. For example, destabilization of the entire C1 microtubule in pf16 mutants results in flagella that are paralyzed or form non-propagating bends [Dutcher et al., 1984]. Loss of the C1a projection in pf6 mutants results in twitchy flagella [Dutcher et al., 1984; Rupp et al. 2001]. Yet, loss of the C1b projection in cpc1 mutants results in flagella with a decreased beat frequency [Mitchell and Sale, 1999; Zhang and Mitchell 2004], and loss of the C1d projection results in uncoordinated beating [DiPetrillo and Smith, 2010; DiPetrillo and Smith 2011]. Although less is known about the C2 projections, knockdown of hydin expression, which destabilizes the C2b projection, results in flagella that arrest at specific switch points [Lechtreck et al., 2008; Lechtreck and Witman, 2007].The diverse effects caused by loss of specific projections suggest that each one makes a unique contribution to flagellar motility.
In this study, we focus on the function of the C1a projection. The C1a projection is a complex of proteins comprised of PF6, C1a-86, C1a-34, C1a-32, C1a-18, and the calcium binding protein calmodulin [Wargo et al., 2005]. The PF6 protein is large (2301 amino acids) and predicted to serve as a scaffold for the assembly of the smaller C1a components [Rupp et al., 2001]. Structural analyses of isolated axonemes following the induction of microtubule sliding have demonstrated that the C1 microtubule is oriented toward the site of active microtubule sliding in Chlamydomonas [Wargo and Smith, 2003]. While this orientation is retained in pf6 mutants, microtubule sliding patterns are disrupted in high calcium conditions [Wargo et al., 2004]. This observation combined with the motility defects observed for pf6 suggests that the C1a projection is important for coordinating dynein activity on specific doublets. The PF6 protein is highly conserved throughout eukaryotes, and a mammalian homolog, SPAG17, has been shown to localize to the central apparatus of murine sperm [Zhang et al., 2005]. Based on the finding that mutations in mouse models of other conserved central apparatus proteins such as SPAG6 (PF16) and SPAG16L (PF20) [Zhang et al., 2007], Hydin [Lechtreck et al., 2008] and Pcdp1 [Lee et al. 2008] cause phenotypes consistent with primary ciliary dyskinesia, mutations in SPAG17 may result in a similar phenotype.
Since PF6 mutants are paralyzed and fail to assemble all C1a complex members, specific functions for individual members of this complex remain unknown [Rupp et al., 2001; Wargo et al. 2005]. To define domains within the PF6 protein important for targeting and assembly of the C1a projection and potentially to determine the role of specific C1a components in regulating motility, we transformed pf6 cells with PF6 deletion constructs. We determined that the carboxyl-terminus of PF6 is necessary for assembly of the C1a projection and flagellar motility. We also identified domains important for assembly and stability of C1a sub-complex members, and as a consequence, we discovered a function for the C1a-34, C1a-32, and C1a-18 sub-complex in regulating motility.
Results
Generation of PF6 deletion constructs and recovery of motility
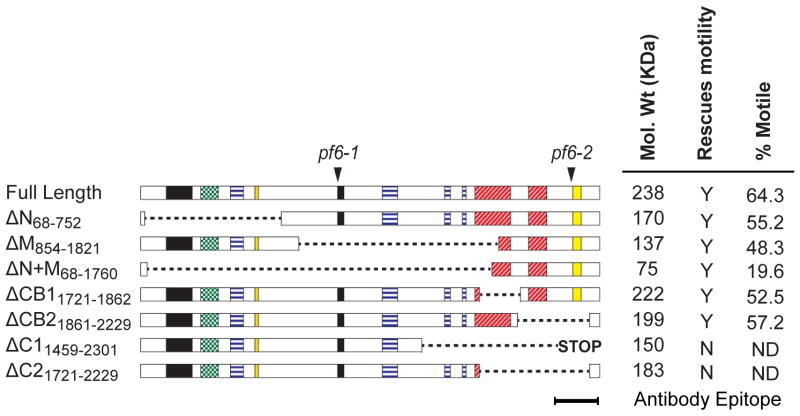
To define functional domains within the PF6 protein we obtained a genomic clone of the PF6 gene and generated deletion constructs using convenient restriction sites that preserved the translational reading frame. These constructs were then tested for their ability to rescue motility upon transformation of two different pf6 alleles, pf6-1 and pf6-2 (Figure 1 and Table I). We sequenced the entire PF6 gene in the original pf6-1 allele [Dutcher et al. 1984] and found a G to T transversion which results in a premature stop codonat amino acid 988. The pf6-2 allele was generated by insertional mutagenesis where a plasmid carrying NIT1 gene inserted into the 3’ end of the coding sequence [Rupp et al., 2001]. Evidently, a hybrid transcript is produced in this allele; however, the mutant protein presumably fails to assemble into pf6-2 axonemes. Since our antibody was generated against the C-terminus, which is lacking in this mutant, we cannot directly test for assembly of the truncated PF6 protein. For all constructs which rescued motility, motility was restored in both pf6 alleles with one exception. The exception was PF6ΔN (removes aa 68–752) which only restored motility in pf6-1. Apparently, the PF6ΔN construct behaves as if it is recessive to the pf6-2 allele. One possibility to explain this result is that a truncated chimeric protein may be expressed in pf6-2 that interferes with expression, trafficking, or assembly of the truncated PF6ΔN protein. The phenotypes for all other constructs were similar in both pf6 alleles; therefore, the following data reports transformants generated from the pf6-1 allele.
Figure 1. PF6 deletion constructs and recovery of motility.
Full length PF6 is 2301 amino acids long and contains coiled-coil (black), alanine-rich (green dotted), proline-rich (blue horizontal lines), and basic (red hatched) domains, as well as a sequence highly conserved in the PF6 mammalian homolog (yellow) [Zhang et al., 2005; Rupp et al., 2001]. Arrowheads indicate the location of lesions in pf6-1 and pf6-2 alleles. The removed amino acids in each construct are indicated by dotted lines. The endogenous stop in PF6ΔC1 was removed, but a downstream stop codon remains in-frame. The predicted molecular weight of each protein is indicated; rescue of motility refers to pf6-1 transformants; the percentage of motile cells was calculated using high-speed video microscopy (see Methods).
Table I.
Chlamydomonas strains used in this study.
| Strain | Parental | PF6 Defect | Phenotype | Reference |
|---|---|---|---|---|
|
nit1Δ A54e18 |
None (2301 aa) | Wild-type | (Nelson et al. 1994) | |
| pf6-1 | 137c | truncated (988 aa) | Twitchy | (Dutcher et al. 1984) |
| pf6-2 | A54e18 | carboxyl-terminal NIT1 insertion | Twitchy | (Rupp et al. 2001) |
| 54E8 | pf6-1 | Full Length transformant | Wild-type | This study |
| 18B6 | pf6-1 | Δ68–752 | Slow swimming | This study |
| 50A11 | pf6-1 | Δ854–1821 | Slow swimming | This study |
| 1H11 | pf6-1 | Δ68–1760 | Slow swimming | This study |
| 95H8 | pf6-1 | Δ1721–1862 | Wild-type | This study |
| 104H1 | pf6-1 | Δ1861–2229 | Wild-type | This study |
| None |
pf6-1 pf6-2 |
Δ1459–2301 | No rescue | This study |
| None |
pf6-1 pf6-2 |
Δ1721–2229 | No Rescue | This study |
| pf6pf28 | pf6-1 | truncated (988 aa) | Paralyzed(not twitchy) | This study |
| A10 | pf6pf28 | Full Length transformant | Slow swimming | This study |
| 3A3 | pf6pf28 | Δ68–752 | Slow swimming | This study |
| B12 | pf6pf28 | Δ854–1821 | Slow swimming | This study |
| BE10 | pf6pf28 | Δ68–1760 | Paralyzed | This study |
All strains used in this study are listed according to their original lab name, the construct transformed into the strain, where appropriate, the corresponding motility phenotype, and references for previously identified strains.
The PF6 gene encodes a large protein of 2301 amino acids with predicted coiled-coil domains as well as alanine-rich, proline-rich and basic domains (Figure 1). The first three constructs we generated were designed to delete approximately one third of the PF6 protein at the amino-terminus (PF6ΔN), the middle region (PF6ΔM, removes aa 854–1821), or the carboxyl-terminus (PF6ΔC1, removes aa1459–2301) (Figure 1). These constructs were co-transformed into pf6 cells with a plasmid carrying a selectable marker and successful transformants were screened for rescue of motility defects (see Methods). For all strains in which motility was restored, expression of the construct was confirmed by western blot (Figure 2). As a control for all experiments and subsequent analyses, pf6 cells were also transformed with the full-length wild-type PF6 gene.
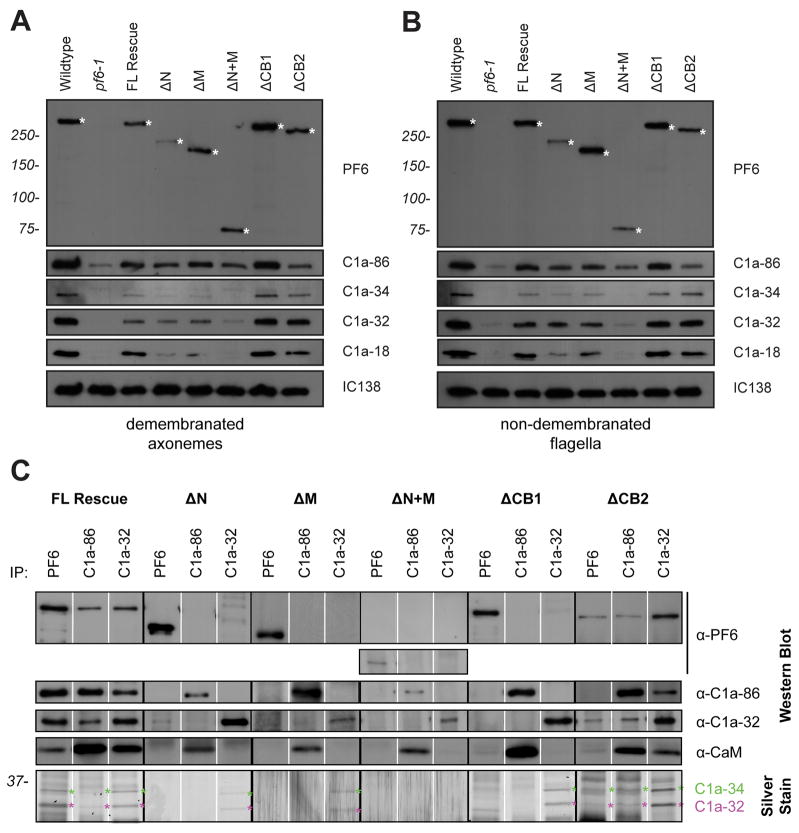
Figure 2. The assembly and stability of C1a components is defective in someΔPF6 transformants.
(A) Western blots of axonemes (5 μg per lane) isolated from deletion construct transformants with recovered motility, probed with antibodies generated against PF6, C1a-86, C1a-34, C1a-32 and C1a-18. Anti-IC138 is used as a loading control. The truncated PF6 protein is indicated by an asterisk. (B) Western blots of flagella (5 μg) isolated from deletion construct transformants with recovered motility, probed as described above. (C) Co-immunoprecipitation experiments performed using axonemal extracts and antibodies generated against PF6, C1a-86, and C1a-32. Precipitated proteins are identified by western blot. C1a-34 is visualized by gel silver staining. All C1a complex members co-precipitate from axonemal extracts isolated from cells expressing full length PF6, regardless of the antibody used. C1a components fail to co-immunoprecipitate from axonemal extracts isolated from PF6ΔN, PF6ΔM, PF6ΔN+M, or PF6ΔCB1. Anti-C1a-86 or anti-C1a-32 antibodies co-precipitate the C1a complex from PF6ΔCB2 extracts; however, anti-PF6 antibodies do not precipitate C1a-86.
Surprisingly, both PF6ΔN and PF6ΔM rescued motility (Figure 1). Twelve or more motile strains expressing each construct were isolated from a minimum of two independent transformation experiments. The co-transformation efficiency was approximately 3% in both cases, which is typical for co-transformation with two plasmids in Chlamydomonas [see for example Tam and Lefebvre 1993]. To quantify restoration of motility, we calculated the percentage of motile cells for rescued strains (Figure 1). While the percentages of motile cells for PF6ΔN (55.2%) and PF6ΔM (48.3%) are reduced compared to wild-type cells (83.5%), they are similar to that of pf6 cells transformed with the full length PF6 construct (64.3%). In contrast, in twelve independent transformation experiments using the carboxyl terminus deletion construct, PF6ΔC1, motility was not restored in a single strain among the 1200 transformants screened. In an effort to determine if the C1a projection failed to assemble or if the projection assembled and is not functional, 27 randomly-chosen non-motile strains transformed with PF6ΔC1 were screened by PCR for presence of the trans gene; in nine of these, the PF6 plasmid successfully integrated into the genome. Western blots of axonemes isolated from these nine strains indicated that neither C1a-86 nor C1a-32 was assembled onto the axoneme. The anti-PF6 antibodies were generated against the C-terminus of PF6 and could not be used to assess for assembly of PF6ΔC1. While it is possible that this approach fails to identify strains expressing the PF6ΔC1 construct, we suspect that the carboxyl-terminus of PF6 is required for assembly of C1a, consistent with the phenotype observed in both pf6 mutant alleles in which the carboxyl terminus of the PF6 protein is defective.
Based on these results, we tested whether the PF6 carboxyl terminus is sufficient to rescue motility. Transformation of pf6 with a construct lacking amino acids 68–1760 (PF6ΔN+M) restored motility; however, the percentage of motile cells was significantly reduced (19.6%) compared to other rescued strains (Figure 1). The reduced percentage of motile cells was not explained by reduced expression levels of the deletion construct. The truncated PF6 protein assembled into flagella of transformed pf6 cells at levels comparable to wild-type (Figure 2A).
To further define domains within the carboxyl terminus required for motility, we generated constructs PF6ΔCB1 (removes amino acids1721–1862) and PF6ΔCB2 (removes amino acids1861–2229), each of which removes one of the two basic domains in this region. Both constructs restored motility to pf6 cells. Transformation with PF6ΔC2 (removes amino acids 1721–2229), which lacks both basic domains but is a smaller deletion than PF6ΔC1, did not recover motility in six independent transformation experiments, screening ~1000 transformants for motility defects. To potentially assess for PF6 assembly in these strains, 22 randomly-chosen non-motile transformants were screened by PCR. The trans gene had successfully integrated into the genomes of 13 of the 22 strains, and western blots of axonemes isolated from these 13 strains indicated that PF6 failed to assemble onto the C1 tubule. (Note that sufficient antigen remained in the PF6ΔC2 protein for recognition by the anti-PF6 antibody.) While it is possible that we simply failed to identify strains expressing PF6ΔC2, this result suggests that at least one basic domain at the carboxyl-terminus is required for assembly of C1a. Assembly and stability of C1a components in PF6Δaa mutants
Previous studies have shown that PF6 is one component of a complex that includes C1a-86, C1a-34, C1a-32, C1a-18, and CaM and is required for assembly of the C1a central pair projection [Wargo et al., 2005]. To determine if the PF6 complex components assemble in PF6Δaa mutants, we isolated axonemes from motile strains and probed western blots with antibodies generated against each component (Figure 2). For all rescued strains, a protein of the corresponding predicted molecular weight is recognized by the anti-PF6 antibody. We observed some variability between preparations of axonemes with respect to the amount of PF6 protein present on blots, particularly with PF6ΔN. TheC1a-86 protein is also present in axonemes isolated from all rescued strains, although protein levels appear slightly reduced in PF6ΔN, PF6ΔM, and PF6ΔN+M.
In PF6ΔN, PF6ΔM, and PF6ΔN+M, levels of C1a-34, and C1a-18 are reduced in the axoneme compared to pf6 cells rescued with the full length wild-type PF6 (Figure 2A). Because we observed some variability with respect to the amount of PF6 protein present on western blots, we performed densitometric analyses from multiple western blots from four different preparations of isolated axonemes (Supplemental Figure S1). After normalizing to the amount of PF6 protein present, these three strains still showed reduced assembly of C1a-34 and C1a18 relative to PF6. This reduction is most dramatic in PF6ΔN+M axonemes where these two proteins as well as C1a-32 are virtually absent. To determine if these components fail to assemble into flagella or if the complex is destabilized during preparation of axonemes, we probed western blots of isolated flagella (Figure 2B). In flagella isolated from strains PF6ΔN and PF6ΔM, C1a-34, and C1a-18 are reduced in abundance compared to wild-type, although not as reduced in flagella compared to isolated axonemes. C1a-32 appears to assemble into flagella at wild-type levels. Therefore, in these two strains both the assembly and stability of these components appears to be compromised. In strain PF6ΔN+M, all three components are severely reduced in isolated flagella. We have shown previously that C1a-34, C1a-32, and C1a-18 comprise a stable sub-complex [Wargo et al., 2005]. Evidently, the assembly and stability of this sub-complex is dependent upon interactions with the amino terminal half of PF6.
In wild-type cells, the entire C1 complex is extracted from the axoneme with high salt buffers and precipitated with antibodies generated against individual C1a components [Wargo et al., 2005]. To further assess the stability of interactions between complex components in pf6 transformants, we performed the same immunoprecipitation experiments using axonemal extracts obtained from rescued strains. These interactions are maintained in extracts isolated from pf6 mutants transformed with the full-length PF6 construct (Figure 2C); all antibodies precipitate all complex members. However, in PF6ΔN, PF6ΔM, PF6ΔN+M, and PF6ΔCB1, the complete complex does not co-precipitate with each antibody. Anti-PF6 antibodies only precipitate PF6 in these strains with a small amount of C1a-32 co-precipitating in PF6ΔN; anti-C1a-86 antibodies co-precipitate C1a-86 and CaM; and anti-C1a-32 antibodies co-precipitate C1a-34 and C1a-32, except in PF6ΔN+M where only C1a-32 is visualized by western blot. C1a-18 is difficult to visualize since it migrates very close to the antibody light chains. This result indicates that while C1a sub-complexes assemble, interactions between the sub-complexes and truncated PF6 are negatively affected.
In immunoprecipitation experiments using axonemal extracts isolated from PF6ΔCB2, all complex components co-immunoprecipitate using antibodies generated against either C1a-86 or C1a-32. However, anti-PF6 antibodies precipitate PF6, C1a-34 and C1a-32 but not C1a-86 or CaM. Assembly of C1a-86 is slightly reduced in this strain. In addition, much of the antigen used to generate the PF6 antibody is lacking from this construct, indicating that this result may be due to inefficient immunoprecipitation using this particular antibody.
Structural analyses of axonemes isolated from PF6Δaa mutants
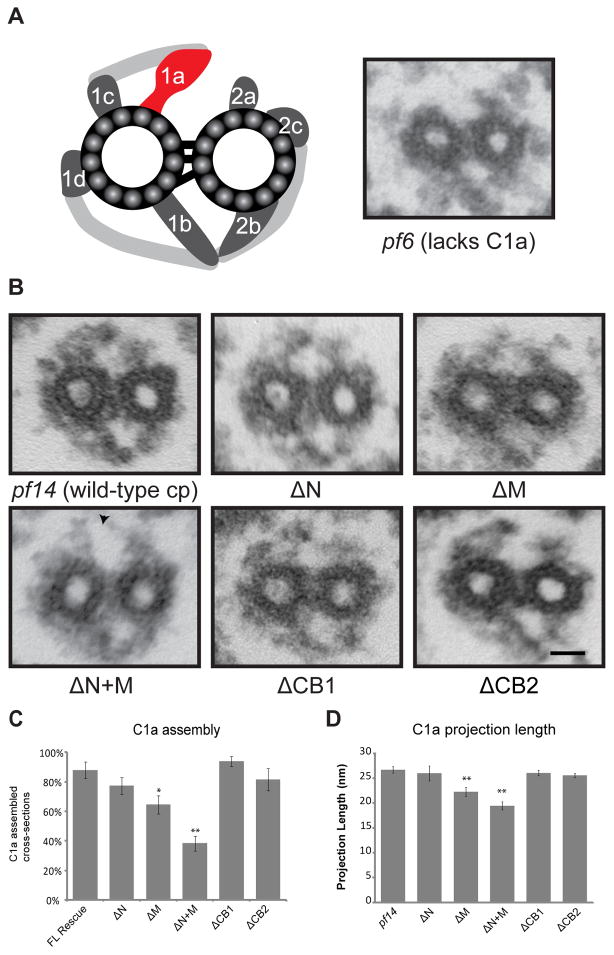
To verify the structural integrity of the C1a projection, we prepared axonemes from PF6Δaa mutants for thin section transmission electron microscopy (Figure 3). To easily distinguish central pair projections from the radial spoke heads, we generated double mutants of all PF6Δaa transformants using the pf14 strain which lacks the radial spokes. For all strains, the results were the same for both single and pf14 double mutants. TheC1a projection is visible in transverse sections of all strains (Figure 3B); however, in PF6ΔN, PF6ΔM, and most notably, PF6ΔN+M we observed a significant percentage of transverse sections in which no C1a projection is assembled (Figure 3C). This observation correlates with the strains exhibiting significant defects in the stability of the C1a-34, C1a-32, and C1a-18 sub-complex.
Figure 3. Assembly of the C1a projection is defective in some PF6Δaa transformants.
(A) Left panel: Illustration of Chlamydomonas central apparatus indicating the location of C1a projection [adapted from Mitchell and Sale, 1999]. Right panel: Transverse section of pf6 mutant axoneme; the C1a projection fails to assemble. (B) Representative images of transverse sections of the central apparatus from pf14 and pf14PF6Δaa transformants. The C1a projection is restored in all PF6Δaa transformants, however the percentage of transverse sections in which the C1a projection assembles varies depending on the PF6Δaa construct transformed (see (C)). A gap is observed between the C1a and C2a projections in PF6ΔN+M (arrow). Scale bar = 25 nm. (C) Percentage of transverse sections with visible C1a projection. PF6ΔM and PF6ΔN+M have a significant percentage of transverse sections lacking the projection. (*p<0.01, **p<0.001, Z test.) N ≥ 25 images. (D) C1a projection length. PF6ΔM and PF6ΔN+M have significantly shorter C1a projections (**p < 0.001, Student’s t test.) N ≥ 6 images; transverse sections lacking C1a were not included in the average. Error bars indicate SEM.
In transverse sections in which the C1a projection is clearly visible, we assessed defects in C1a structure by measuring the length of the C1a projection. In pf14 cells, the C1a projection has a length of 26.70 ± 0.67 nm. Projections in PF6ΔN, PF6ΔCB1, and PF6ΔCB2 are not significantly different in length from those of pf14 (p=0.18). The projections in both PF6ΔM and PF6ΔN+M are significantly shorter in length (22.27 ± 0.91 nm and 19.44 ± 0.79 nm, respectively (p<0.001, Student’s t test)) compared to C1a in pf14 (Figure 3D, see also Supplemental Figure 2 for additional transverse sections). The PF6 constructs in both of these strains lack amino acids in the middle region of the PF6 protein. In addition, PF6ΔN+M lacks the C1a-34, C1a-32, and C1a-18 sub-complex.
Functional analysis of PF6Δaa mutants
For those strains in which the PF6 deletion construct restored motility, we used high-speed video microscopy to further characterize flagellar motility. Representative examples of swimming cells for all strains are shown in Figure 4 and supplemental movies. The waveforms of transformed strains appear nearly wild-type. As a general method to quantitatively compare waveforms, we calculated the radius of curvature for the principle bend of the recovery stroke. No significant differences were measured among the transformed strains compared to wild-type (data not shown).
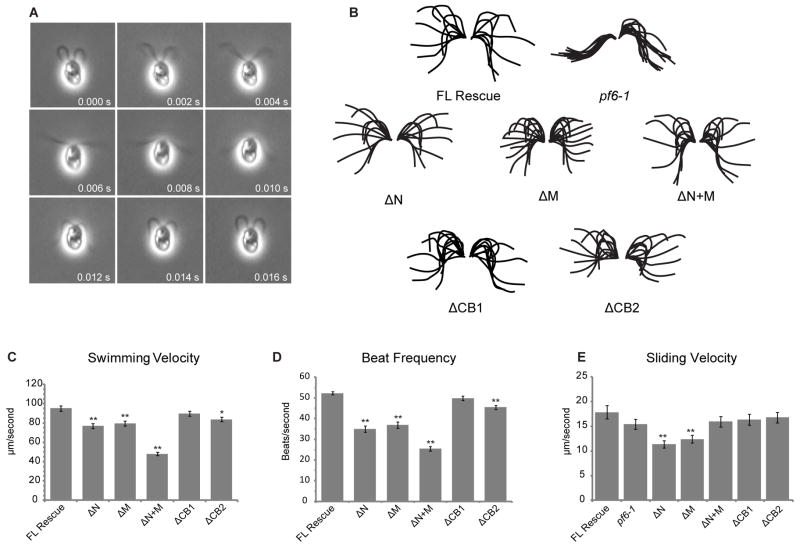
Figure 4. Swimming velocity, beat frequency, and microtubule sliding velocity are reduced in some PF6Δaa strains.
(A) Montage of images obtained from high speed video recording of cell transformed with full-length PF6, demonstrating a complete beat cycle. Videos of all other transformants are included in supplemental data. (B) Tracings of complete beat cycles from pf6-1 and each of the PF6 transformants. pf6 is paralyzed, however, waveforms of all PF6Δaa transformants are similar to flagella containing full-length PF6. Each flagellar trace represents one frame of video separated by 2 ms. (C) Mean swimming velocity of strains containing truncated PF6 protein. Velocity is significantly decreased in PF6ΔN, PF6ΔM, and PF6ΔN+M (**p < 0.001, Student’s t test). A small but significant decrease is also observed in PF6ΔCB2 (*p < 0.01, Student’s t test). (D) Asymmetric beat frequency is significantly decreased in PF6ΔN, PF6ΔM, PF6ΔN+M, and PF6ΔCB2 (**p < 0.001, Student’s t test). (E) Mean microtubule sliding velocities for PF6 strains. Microtubule sliding velocities were obtained using an in vitro microtubule sliding assay and isolated axonemes as indicated in Methods. Velocity is significantly decreased in PF6ΔN and PF6ΔM (**p < 0.001, Student’s t test). N ≥ 50 for all swimming velocity, beat frequency, and sliding velocity measurements. Total N is derived from three independent experiments. Error bars indicate SEM. Data is presented in tabular form in Table 2.
We also compared mean swimming velocity and mean beat frequency for all strains (Figure 4 and Table 2). For pf6 transformed with the wild-type PF6, cells swim with a mean velocity of 95.14 ± 2.63 μm/s and a mean beat frequency of 52.29 ± 0.77 Hz (all errors are given as SEM). However, strains with significantly reduced assembly / stability of the C1a-34, C1a-32, and C1a-18 sub-complex exhibited obvious motility defects. Strains expressing PF6ΔN, which lacks the amino-terminus, have a significantly reduced swimming velocity of 77.06 ± 2.57 μm/s (p< 0.001, Student’s t test) and beat frequency of 34.97 ± 1.61 Hz (p< 0.001, Student’s t test) compared to strains transformed with the full length PF6 construct. Strains expressing PF6ΔM also have a significant reduction in swimming velocity (79.61 ± 2.44 μm/s, p< 0.001, Student’s t test) and beat frequency (36.98 ± 1.51 Hz, p < 0.001, Student’s t test). The most significant swimming defects were observed when only the carboxyl-terminus of PF6 remained; for the 19.6% of PF6ΔN+M cells that swim, the mean velocity was 48.06 ± 1.74 μm/s with a flagellar beat frequency of 25.53 ± 1.08 Hz. Removal of the first basic domain within the carboxyl-terminus in PF6ΔCB1 did not significantly reduce either swimming velocity or beat frequency. A small but significant reduction was observed in both parameters in PF6ΔCB2 transformants which lack the second basic domain (83.77 ± 2.24 μm/s and 45.43 ± 1.05 Hz, p < 0.01, Student’s t test). Combined, these data suggest that the C1a projection is involved in modulating flagellar beat frequency and that the C1a-34, C1a-32, and C1a-18 sub-complex may be involved in this modulation.
Table II.
Analysis of motility and microtubule sliding velocities.
| Construct (pf6-1 background) | Swimming Velocity (μm/sec) | Asymmetric Beat Frequency (beats/sec) | Symmetric Beat Frequency (beats/sec) | Microtubule Sliding Velocity (μm/sec) |
|---|---|---|---|---|
| Full Length PF6 | 95.14 ± 2.63 | 52.29 ± 0.77 | 68.68 ±1.70 | 17.80 ± 1.35 |
| pf6-1 | ND | ND | ND | 15.36 ± 1.03 |
| PF6ΔN (Δ68–752) | * 77.06 ± 2.57 | * 34.97 ± 1.61 | 67.49 ± 1.65 | * 11.29± 0.71 |
| PF6ΔM (Δ854–1821) | * 79.61 ± 2.44 | * 36.98 ± 1.51 | 67.12 ± 1.12 | * 12.34± 0.77 |
| PF6ΔN+M (Δ68–1760) | * 48.06 ± 1.74 | * 25.53 ± 1.08 | 62.59± 0.86 | 15.89 ± 1.04 |
| PF6ΔCB1 (Δ1721–1862) | 89.92 ± 2.63 | 49.77 ± 1.06 | 70.49 ± 1.39 | 16.29 ± 1.14 |
| PF6ΔCB2 (Δ1861–2229) | 83.77±2.24 | * 45.43 ± 1.05 | 68.15 ± 1.57 | 16.73 ± 1.09 |
The listed strains were analyzed for cell swimming velocity, asymmetric beat frequency, and symmetric beat frequency. Microtubule sliding velocity was measured for axonemes isolated from the listed strains using an in vitro assay.
p ≤ 0.001
We also tested whether pf6 transformants exhibited the photoaccumulation and photoshock responses (see Methods for assay; data not shown). Both of these behaviors are regulated by changes in intraflagellar calcium concentration. Cells transformed with full length PF6, P F 6ΔCB1, and PF6ΔCB2 rapidly photoaccumulate. PF6ΔN and PF6ΔMcells also photoaccumulate, but at a slower rate which can be accounted for by their slower swimming velocities. PF6ΔN+M photoaccumulate, but very slowly compared to all other strains. This result can be explained by the observation that only 19.6% were observed to swim. Photoshock occurs in Chlamydomonas upon exposure to intense light and results in a conversion of the forward-swimming, asymmetric waveform to a symmetric waveform, subsequently reversing the swimming direction of the cell. All of the strains exhibited a normal photoshock response (data not shown) and beat frequencies for the symmetric waveform were not significantly different from wild-type (Table II).
Microtubule sliding velocities are reduced when PF6 is truncated
To determine if the reduced beat frequencies and swimming velocities are due to altered dynein-driven microtubule sliding, we measured sliding velocities in isolated axonemes induced to slide in the presence of ATP and protease to disrupt inter-doublet linkages (Figure 4 and Table 2). We previously demonstrated that axonemes isolated from pf6 mutants have microtubule sliding velocities that are not significantly different from that of wild-type [Smith, 2002b]. Axonemes isolated from pf6 mutants rescued with full length PF6 have mean sliding velocities of 17.80 ± 1.35 μm/s (Figure 4). This velocity is not significantly different from pf6-1 mutants (15.36 ± 1.03 μm/s p = 0.15, Student’s t test). Removal of domains in PF6ΔN and PF6ΔM results in a significant decrease in mean sliding velocity to 11.29 ± 0.71 μm/sec and 12.34 ± 0.77 μm/sec, respectively (p < 0.001, Student’s t test). These two mutants have defects in the assembly/stability of C1a-34, C1a-32, and C1a-18 sub-complex. No significant decrease (p> 0.26) in mean sliding velocity was observed in PF6ΔN+M (15.89 ± 1.04 μm/s), PF6ΔCB1 (16.29 ± 1.14 μm/s), or PF6ΔCB2 (16.73 ± 1.09 μm/sec).
Both inner and outer arm dyneins are affected by PF6 deletions
The most obvious motility defect observed in strains PF6ΔN, PF6ΔM, and PF6ΔN+M was a reduction in the asymmetric beat frequency. Comparative studies of inner and outer dynein arm mutants have indicated that high beat frequency is largely determined by the activity of the outer dynein arms whereas the inner dynein arms primarily modulate waveform [recently reviewed in King and Kamiya, 2009; see also Brokaw and Kamiya 1987]. Based on previous studies of dynein arm mutants, we hypothesized that the reduction in beat frequency for strains PF6ΔN, PF6ΔM, and PF6ΔN+M may result from mis-regulation of outer dynein arm activity. If our hypothesis is correct, we predicted that double mutants with truncations in PF6 and lacking the outer dynein arms would have beat frequencies equal to those in mutants lacking only the outer dynein arms. If the inner arms are the target of this regulation, then double mutants will have beat frequencies slower than the outer dynein armless mutant.
To test this, we generated double mutants which lack both the C1a projection and outer dynein arms (pf6-1x pf28); the double mutants are completely paralyzed with no visible twitchy movements. Motility in pf6pf28 was rescued by transformation with PF6ΔN and PF6ΔM; western blots of axonemes confirm that the truncated PF6 is expressed (Figure 5). The PF6ΔN+M construct did not rescue motility upon transformation of pf6pf28; this result may not be surprising given the low percentage of motile cells observed in pf6-1 transformants. The mean beat frequency of pf6pf28 transformed with full length PF6 is 19.14 ± 0.34 Hz, which is not significantly different from the mean beat frequency of pf28 mutants (19.60 ± 0.25 Hz). In contrast, pf6pf28 cells transformed with either the PF6ΔNor PF6ΔM construct had beat frequencies significantly slower (14.34 ± 0.33 Hz and 10.39 ± 0.23 Hz, respectively, p< 0.001, Student’s t test) than that of pf28 (Figure 5B). Since this reduction in beat frequency occurs in the absence of the outer arm dyneins, the simplest explanation is that the truncations in PF6ΔN or PF6ΔM result in reduced ability of the inner dynein arms to generate force required for wild-type beat frequencies.
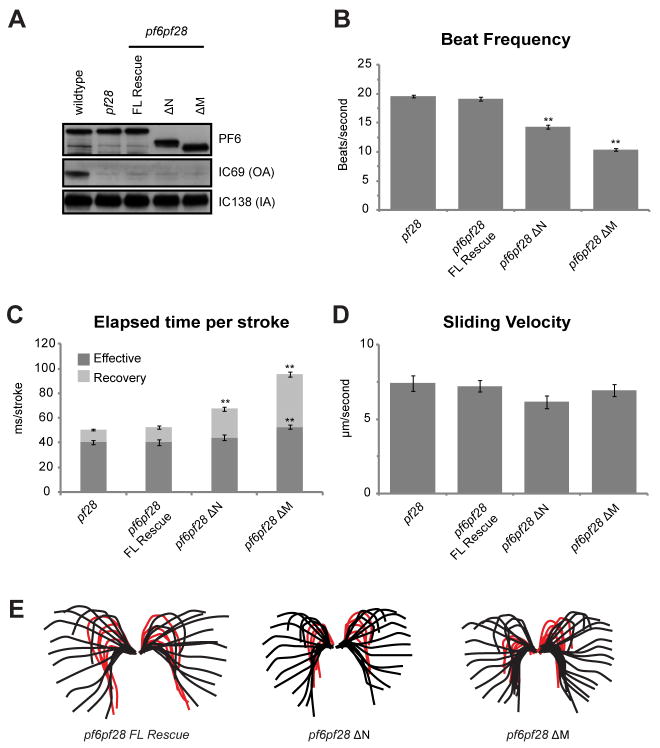
Figure 5. Analysis of motility defects in PF6 Δaa transformants in the absence of the outer dynein arms.
(A) Western blots of axonemes (10 μg per lane) isolated from transformed pf6pf28 strains, probed with antibodies generated against PF6, IC69, and IC138. IC69 is an outer arm dynein intermediate chain. IC138 is an inner dynein arm intermediate chain. (B) Beat frequencies are significantly rescued in pf6pf28 strains expressing PF6ΔN or PF6ΔM (**p < 0.001, Student’s t test). (C) Quantification of stroke timing indicates that the recovery stroke is significantly longer in PF6ΔN and PF6ΔM mutants (**p < 0.001, Student’s t test). (D) Mean microtubule sliding velocities are not significantly different in pf6pf28 strains expressing PF6ΔN or PF6ΔM. (E) Waveform tracings indicate that waveform shape is similar in all mutants. The beginning of the effective stoke was designated at the point when the proximal end of the flagellum first began to reverse direction moving downward. By this criterion the effective stroke is initiated before the recovery stroke has reached the end of the flagellum. The same criterion was used to establish the beginning of the recovery stroke – the point at which the proximal portion of the flagellum begins to reverse direction. As is true for the effective stroke, the recovery stroke begins before the tip of the flagellum has completed the effective stroke. Each individual flagellar trace represents one frame of video separated by 2 ms. Red tracings indicate the recovery stroke. N ≥ 50 for all beat frequency and sliding velocity measurements. N ≥ 40 strokes and 10 cells for stroke time measurements. Total N is derived from three independent experiments. Error bars indicate SEM. Note that because the recovery stroke is delayed for some mutants, red tracings may overlap and, therefore, appear less numerous.
In additional analyses of beating flagella in the rescued double mutants, we did not observe any obvious defects in waveform (see tracings, Figure 5E). However, the length of time for the completion of the recovery stroke is significantly slower for the pf6pf28 cells transformed with PF6ΔN and PF6ΔM (Figure 5D, E). For pf6p f28 cells transformed with the full length PF6 construct, the recovery stroke took 12.34 ± 1.29 ms from start to completion. For pf6pf28 cells transformed with PF6ΔN and PF6ΔM , the time for completion of the recovery stroke significantly increased to 23.34 ± 1.51 ms and 42.64 ± 2.10 ms, respectively (p < 0.001, Student’s t test). In contrast, the amount of time required for the effective stroke was not increased in PF6ΔN strains (43.99 ± 2.23 ms compared to 40.33 ± 2.4 ms in strains rescued with wild-type PF6, p=0.28). A small but significant increase in the time required for the effective stroke was observed in pf6pf28 strains rescued with PF6ΔM strains (52.60 ± 1.74 ms, p < 0.001). These results suggest that modulation of inner dynein arm activity to produce the recovery stroke is defective in the absence of C1a complex members and the primary cause of decreased beat frequency.
To determine if the decreased sliding velocity for axonemes isolated from ΔPF6 transformants was due to decreased activity of outer versus inner dynein arms, we also measured microtubule sliding velocities for axonemes isolated from pf6pf28 cells transformed with PF6ΔN and PF6ΔM. The mean sliding velocity of axonemes isolated from pf6pf28 mutants rescued with wild-type PF6 are not significantly different from that of pf28 (p = 0.75). Similarly, the mean sliding velocities of pf6pf28 cells transformed with either PF6ΔN or PF6ΔM were not significantly different from that of pf28 (6.16 ± 0.44 μm/s (p = 0.065) and 6.94 ± 0.40 μm/sec (p = 0.46), respectively). This result indicates that the reduction in sliding velocity observed in pf6 mutants transformed with these same constructs is most likely a result of impaired outer dynein arm activity.
Discussion
Previous studies have shown that the C1a projection of the central apparatus is required for wild-type flagellar motility; loss of this projection in pf6 mutants results in twitchy flagella which fail to propel the cell forward [Dutcher et al., 1984; Rupp et al., 2001]. The C1a projection is comprised of six different proteins including PF6, C1a-86, C1a-34, C1a-32, C1a-18 and CaM [Wargo et al., 2005]. In the absence of PF6, all subunits fail to assemble, resulting in the loss of the C1a projection. This observation indicates that PF6 is required for assembly of the C1a complex but does not reveal the role of specific subunits in regulating flagellar motility. By generating mutants expressing PF6 protein lacking specific domains we have shown that: at least one basic domain at the carboxyl-terminus of PF6 is required for assembly of PF6; the amino and middle regions of PF6 are required for the assembly and stability of the C1a-34, C1a-32, C1a-18 sub-complex; stable interactions with C1a-34, C1a-32, and C1a-18 are required for wild-type beat frequency; and the C1a projection plays a role in modulating the activity of both the inner and outer dynein arms.
A minimum of one C-terminal basic domain of PF6 is required for C1a projection assembly
Based on the size of the PF6 protein as well as the pf6 mutant phenotype, PF6 is proposed to act as a scaffold for assembly of subunits comprising the C1a projection [Rupp et al., 2001; Wargo et al., 2005]. This model predicts that the PF6 protein has domains required for microtubule binding (directly or indirectly) as well as for associating with complex members. Our analyses of mutants expressing PF6 deletion constructs indicate that the carboxyl terminus is both necessary and sufficient for assembly of PF6 onto the C1 microtubule. This conclusion is based on several observations. First, the carboxyl-terminus of PF6 is disrupted in both pf6 alleles. Second, transformants with constructs lacking the carboxyl terminus (PF6ΔC1 and PF6ΔC2) failed to rescue motility in both pf6 alleles. And finally, motility is recovered in pf6 cells transformed with a PF6 construct which only expresses amino acids 1761–2301, although assembly of the entire PF6 complex is defective.
Our studies show that at least one C-terminal basic domain is required for PF6 motility. Smaller deletions within the carboxyl-terminus (PF6ΔCB1 and PF6ΔCB2) rescued motility, while removal of both basic domains did not (PF6ΔC2). The deletion in PF6ΔCB2 also removes a conserved ASM domain, which is a putative microtubule binding domain found in several central apparatus proteins, including PF6, hydin, and FAP74 [Ponting, 2006]. Since PF6 retains the ability to assemble and rescue motility in the absence of this domain, this domain is not required for assembly onto the C1 central microtubule. We previously reported that the C1a components extracted from isolated axonemes failed to bind microtubules assembled in vitro [Wargo et al., 2005]. We have also expressed the PF6 carboxyl-terminus in bacteria and used purified expressed protein in an in vitro microtubule binding assay; the truncated protein fails to bind microtubules (unpublished results). Based on this result, we suspect that additional adaptor proteins are required for PF6 assembly onto the C1 microtubule.
Stable assembly of the C1a-34, C1a-32, C1a-18 sub-complex is required for wild-type beat frequency
Truncation of either the amino terminus or middle region of the PF6 protein results in reduced assembly and stability of the C1a-34, C1a-32, and C1a-18 sub-complex; the subcomplex completely fails to assemble in a mutant which only retains the carboxyl terminus of PF6. Therefore, the amino terminal two-thirds of the PF6 protein must form a binding site for this sub-complex. Strain PF6ΔCB1 shows wild-type assembly of this sub-complex, yet immunoprecipitation experiments indicate that there is reduced association of PF6 complex members. Most likely the lack of amino acids 1721–1862 in PF6ΔCB1 slightly changes the conformation of PF6 so that assembly of the complex is unaffected but that the stability of the associations between complex members is slightly reduced.
Interestingly, strains with significant defects in assembly/stability of C1a-34, C1a-32, and C1a-18 swim with a reduced velocity and beat frequency. Therefore, stable association of wild-type levels of the C1a-34, C1a-32, and C1a-18 sub-complex with PF6 is important for producing wild-type beat frequency. The reduction in beat frequency correlated with a decrease in dynein-driven microtubule sliding velocities in PF6ΔN and PF6ΔM transformants. In contrast, no reduction in microtubule sliding velocity was observed in the PF6ΔN+M transformant in which a majority of the population fails to assemble a visible C1a projection. This result is similar to that observed for the pf6 mutant which lacks the C1a projection and do not have decreased sliding velocity compared to wild-type axonemes [Smith, 2002b]. These combined results suggest that in wild-type axonemes, the amino / middle region of PF6 protein and/or the C1a-34, C1a-32, and C1a-18 subcomplex contributes to modulating dynein-driven microtubule sliding.
It is important to note that the C1a-86 subunit, the CaM interacting protein, assembled in all strains with restored motility and retained its ability to interact with CaM. One prediction is that restoration of motility requires assembly of C1a-86. For the transformants in which the PF6Δaa failed to rescue motility, it appeared that the entire PF6 complex did not assemble into axonemes. Therefore, we were unable to test whether C1a-86 is specifically required for motility. In future studies we will test our prediction by reducing the expression of C1a-86 using RNAi methods newly developed for Chlamydomonas [DiPetrillo and Smith, 2010; Dymek et al., 2011; Molnar et al., 2009; Zhao et al., 2009].
The C1a projection affects the activity of both the inner and outer dynein arms
Our results indicate that both the inner and outer dynein arms are the targets of regulation by the C1a projection. This conclusion is supported by the observation that in double mutants lacking both PF6 and the outer dynein arms, microtubule sliding velocities of PF6ΔN and PF6ΔM transformants do not significantly differ from that of the single outer dynein armless mutant. Therefore, the reduction in sliding velocity observed in the pf6 single mutants transformed with these constructs is evidently dependent on the presence of the outer dynein arms. Further evidence for the C1a projection modulating outer dynein arm activity comes from the discovery of sup-pf-1, a suppressor mutation which alters the structure of outer arm dynein β heavy chain and restores motility to pf6 mutants [Huang et al., 1982; Porter et al., 1994].
In contrast, pf6pf28 cells lacking the outer arm dyneins and transformed with PF6ΔN and PF6ΔM have a reduced beat frequency compared to the pf6pf28 transformed with full length PF6. This reduced beat frequency is largely the result of a slowed recovery stroke. These results indicate that in wild-type axonemes, the amino / middle region of PF6 protein and/or the C1a-34, C1a-32, and C1a-18 subcomplex also affects microtubule sliding driven by the inner dynein arms to produce an effective recovery stroke.
Results from previous studies of Chlamydomonas mutants have suggested that the activity of the outer dynein arms is primarily controlled by the mechanical state of the axoneme whereas the activity of the inner dynein arms is regulated by the central pair/radial spoke control system [Hayashibe et al., 1997]. The activities of the two motor systems are predicted to be coupled by the mechanical state of the axoneme, potentially via newly described structural connections between the two rows of dynein arms [Heuser et al., 2009; Nicastro et al., 2005; Nicastro et al., 2006]. Based on the structural organization of the axoneme, it is likely that C1a modulation of outer dynein arm activity is mediated by these connections to the inner dynein arms as predicted by Kikushima [2009].
Conclusions
The C1a projection is clearly required for wild-type motility. Loss of the C1a projection in pf6 mutants causes total paralysis. Remarkably, deletion of the amino two-thirds of the PF6 protein did not affect PF6 assembly. However, only a small percentage of cells recovered motility and beat frequencies are significantly reduced. The question is, given the location of the central apparatus relative to the dynein arms, what is the mechanism by which the C1a projection regulates dynein arm activity? One explanation for this observation is that the amino terminus of PF6 or the C1a-34, C1a-32, and C1a-18 complex, interacts with the radial spoke heads. Numerous studies supported a model in which interactions between the central pair projections and radial spokes provide regulatory cues to modulate dynein-driven microtubule sliding [see for example Mitchell, 2003; Omoto and Kung, 1979; Omoto and Kung, 1980; Satir, 1982; Satir, 1989; Warner and Satir, 1974]. Future studies will be aimed at identifying key central apparatus components which interact with the radial spoke heads.
Materials and Methods
Strains and cell culture
Strains of Chlamydomonas reinhardtii used in this study are listed in Table 1. Strain A54-e18 (nit1-1, ac17, sr1, mt+) has wild-type motility and was obtained from Paul Lefebvre (University of Minnesota, St. Paul, MN). The strains pf6-1 (CC-1029) and pf6-2 (CC-3926) were obtained from the Chlamydomonas Genetics Center (University of MN, St. Paul MN). The double mutant pf6pf28 was isolated from nonparental ditype tetrads from pf6 and pf28 (CC-1877) and is paralyzed. Double mutants with pf14(CC-613) were generated by mating and isolation of nonparental ditype tetrads. All strains were grown in TAP media [Gorman and Levine, 1965] under constant light conditions.
Generation of PF6 deletion constructs
All of the constructs used in this paper were generated from plasmid p5B9A/S containing a genomic clone of PF6 [Rupp et al., 2001], obtained from Mary Porter (University of Minnesota, St. Paul, MN). Digestion of the plasmid with NheI, NsiI, or AatII, followed by circular ligation of the vector containing fragment produced constructs PF6Δ68–752 (PF6ΔN), PF6Δ1459–2301 (PF6ΔC1), and PF6Δ1861–2229 (PF6ΔCB2), respectively. PF6Δ854–1821 (PF6ΔM) was generated by double digestion with PshAI and BsrGI, followed by blunting with mung bean nuclease (NEB, Ipswich, MA) and religation. PF6Δ68–1760 (PF6ΔN+M) was generated by double digestion with NheI and BsiWI, followed by blunting with mung bean nuclease (NEB) and religation. For PF6Δ1721–1862 (PF6ΔCB1) and PF6Δ1721–2229(PF6ΔC2), a6.6 kb BstEII/SalI fragment was blunted with Klenow and subcloned into the EcoRV site of pBluescript. The resulting construct was digested with AfeI/AatII, blunted with Klenow, and religated together; partial digestion of the two AatII sites resulted in two constructs. A 2.1 kb fragment was digested from one construct with MauBI/SnaBI and replaced into the same sites on the p5B9A/S plasmid to generate PF6ΔCB1. For PF6ΔC2, a 1.3 kb fragment was digested from the appropriate construct with NsiI and replaced into the same sites in p5B9A/S.
Transformation
Transformation of Chlamydomonas strains with PF6 deletion constructs was carried out using the glass bead method [Kindle, 1990; Kindle et al., 1989]. Briefly, autolysin treated cells were combined with 1 μg of PF6 construct plasmid DNA, 1 μg of PCR amplified AphVIII DNA (confers resistance to paromomycin [Sizova et al., 2001; Sizova et al., 1996]), and 5% PEG. Glass beads were added to the tube, which was vortexed for 45 seconds before diluting with TAP media. After growth overnight in constant light, cells were plated on TAP agar containing 10 μg/ml (pf6-2) or 15 μg/ml (pf6; pf6pf28) paromomycin. Selected cells were picked into 96-well dishes and screened by light microscopy for restoration of motility.
Axoneme isolation, extraction, and immunoprecipitation
Flagella were isolated, demembranated, and extracted as described in Dymek and Smith [2007], except that isolated axonemes were extracted only in 0.5 M KI buffer (10 mM Hepes, pH 7.4, 5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, 30 mM NaCl, and 0.5 M KI) at 6 mg/ml for 30 minutes on ice, followed by dialysis into cold NaLow buffer (10 mM Hepes, pH 7.4, 5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, and 30 mM NaCl). For immunoprecipitations, 50–75 μg of affinity-purified PF6, C1a-86, or C1a-32 antibody was allowed to preincubate with ~50 μl protein-A beads (Invitrogen) and 500 ul ice-cold TBST150 (150 mM NaCl, 50 mM Tris-HCl, and 0.5 mM EDTA, pH 7.5) for 1 hour. Beads were washed 2 times with TBST150 before adding 500 μg of dialyzed extract and TBST150. This was allowed to mix at 4°C for 2 hours. The beads were subsequently washed four more times in TBST150 and fixed for gels in 60 μl TBST150 and 40 μl of 5X SDS-PAGE sample buffer.
Gel Electrophoresis, Western Blotting, and Silver-Stained Gels
Gel electrophoresis, Western blotting, and silver-staining were performed as described in Wargo et al. [2005], utilizing antibodies generated in that study. Antibodies were affinity purified and used at the following dilutions: anti-PF6, 1:1000; anti-C1a-86 (PF6-IP1), 1:5000; anti-C1a-34 (PF6-IP2), 1:2000; anti-C1a-32 (PF6-IP3), 1:2000; anti-C1a-18 (PF6-IP4), 1:100; anti-CaM, 1:7000. The IC138 antibodies were provided by Winfield Sale (Emory University, Atlanta, GA) and used at dilution of 1:20,000 [Hendrickson et al., 2004]. The IC69 antibodies were provided by George Witman (University of Massachusetts Medical School, Worcester, MA) and used at dilution of 1:10,000. Axonemal western blots were analyzed by densitometry using ImageJ software (http://imagej.nih.gov/ij/, 1997–2011).
Electron Microscopy
Axonemes from mutants were prepared for thin-section electron microscopy by fixation in 1% gluteraldehyde and 1% tannic acid in 0.1 M sodium cacodylate, postfixation in 1% osmium tetraoxide, and dehydration in ethanol. The resulting pellets were embedded in LX112 resin, sectioned, mounted on Formvar-coated, carbon-stabilized copper grids, and stained with uranyl acetate and Reynolds lead citrate. Grids were examined at 100 kV in a JEM 1010 transmission electron microscope with a side-mount 2Kx2K AMT camera. Central pair images were viewed and analyzed in ImageJ software.
Motility Analysis
High speed video recordings of Chlamydomonas motility were produced on an Axioskop microscope (Zeiss) utilizing a PCO.1200hs (PCO.imaging) video camera and CamWare software. Percent motility and swimming velocity information was collected in phase-contrast at 60 frames per second with a 10X .30 NA objective lens. For percent motility, video was replayed frame-by-frame, and motile cells were counted. At this magnification, both paralyzed cells and cells attached to the microscope slide were considered “non-motile.” Swimming velocity was calculated in ImageJ software by measuring the distance travelled by a motile cell during a set time frame. Beat frequency information was collected at 500 frames per second with a 40X .65 NA objective lens. Video was replayed frame-by-frame and beats counted manually for a set time frame. All recording took place in the presence of a red filter, except for “photoshock” experiments. All data are presented as mean ± SEM. The Student’s t test was used to determine the significance between means.
For waveform traces and calculating the time of effective and recovery strokes (Figure 5c), the beginning of the effective stoke was designated at the point when the proximal end of the flagellum first began to reverse direction moving downward. By this criterion the effective stroke is initiated before the recovery stroke has reached the end of the flagellum. The same criterion was used to establish the beginning of the recovery stroke – the point at which the proximal portion of the flagellum begins to reverse direction. As is true for the effective stroke, the recovery stroke begins before the tip of the flagellum has completed the effective stroke. Importantly, the waveforms of all strains were characterized in exactly the same way.
Photoaccumulation ability was determined by putting a liquid culture of cells into a Petri dish that was covered by black plastic, leaving one edge exposed to light. Time for accumulation of cells at one end was compared to wild-type strains and compensated for reductions in swimming velocity. Photoshock ability was determined by removing the red filter while recording video and visualizing waveform switching.
Sliding assay
Microtubule sliding assays were performed on isolated axonemes as described previously [Dymek and Smith, 2007; Okagaki and Kamiya, 1986; Smith, 2002a; Smith, 2002b]. Briefly, axonemal sliding was induced with Klow buffer (10 mM Hepes, pH 7.4, 5 mM MgSO4, 1 mM DTT, 0.5 mM EGTA, and 50 mM CH3CO2K) containing 1 mM ATP and 2μg/ml type VIII protease (Sigma-Aldrich). Experiments were observed on an Axioskop 2 microscope (Zeiss) with dark-field optics utilizing a silicon intensified target camera (VE-1000 SIT, Dage-MTI, Inc., Michigan City, IN) interfaced with a PC via an IMAQ PCI-1450 image acquisition device (National Instruments). A script written in LabView 7.1 software (National Instruments) was used to facilitate data collection. ImageJ software was utilized for calibration and measurement of data. All data are presented as mean ± SEM. The Student’s t test was used to determine the significance between means.
Supplementary Material
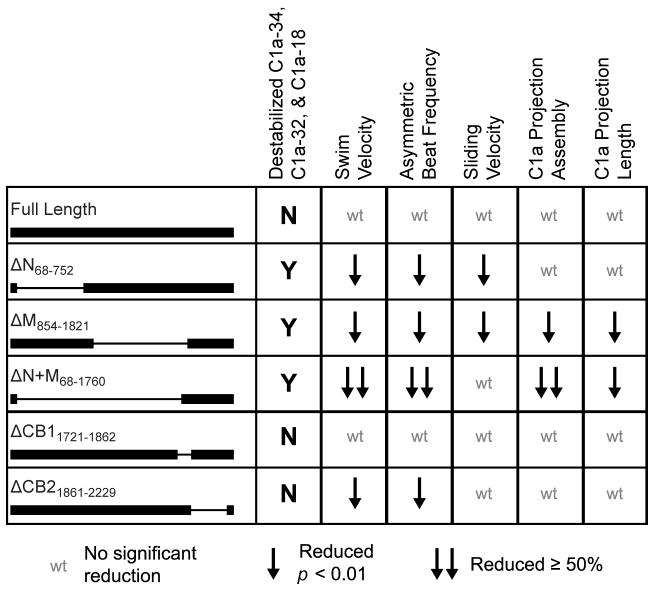
Figure 6. Summary of structural and functional defects observed in PF6Δaa transformants.
Motility and complex assembly defects are observed in PF6ΔN, PF6ΔM, and PF6ΔN+M strains which lack domains at the amino terminus of PF6. Single downward arrow indicates significant reduction (p < 0.01, Student’s t test). Double downward arrows indicate reduction that is greater than 50% of wild-type and highly significant (p < 0.001, Student’s t test).
Acknowledgments
The authors thank Erin Dymek and Christen DiPetrillo for critical reading of and suggestions for improving the manuscript. We thank Louisa Howard for her technical assistance in preparing samples for electron microscopy. This work is supported by NIH GM66919 (EFS).
References
- Adams GM, Huang B, Piperno G, Luck DJ. Central-pair microtubular complex of Chlamydomonas flagella: polypeptide composition as revealed by analysis of mutants. J Cell Biol. 1981;91(1):69–76. doi: 10.1083/jcb.91.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw CJ. Direct measurements of sliding between outer doublet microtubules in swimming sperm flagella. Science. 1989;243(4898):1593–6. doi: 10.1126/science.2928796. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ, Kamiya R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskeleton. 1987;8(1):68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- DiPetrillo CG, Smith EF. Pcdp1 is a central apparatus protein that binds Ca(2+)-calmodulin and regulates ciliary motility. J Cell Biol. 2010;189(3):601–12. doi: 10.1083/jcb.200912009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPetrillo CG, Smith EF. The Pcdp1 complex coordinates the activity of dynein isoforms to produce wild-type ciliary motility. Mol Biol Cell. 2011;22(23):4527–4538. doi: 10.1091/mbc.E11-08-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK, Huang B, Luck DJ. Genetic dissection of the central pair microtubules of the flagella of Chlamydomonas reinhardtii. J Cell Biol. 1984;98(1):229–36. doi: 10.1083/jcb.98.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Heuser T, Nicastro D, Smith EF. The CSC is required for complete radial spoke assembly and wild-type ciliary motility. Mol Biol Cell. 2011;22(14):2520–31. doi: 10.1091/mbc.E11-03-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Smith EF. A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. J Cell Biol. 2007;179(3):515–26. doi: 10.1083/jcb.200703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR, Rowe AJ. Dynein: a protein with adenosine triphosphatase activity from cilia. Science. 1965;149:424–425. doi: 10.1126/science.149.3682.424. [DOI] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1965;54(6):1665–9. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashibe K, Shingyoji C, Kamiya R. Induction of temporary beating in paralyzed flagella of Chlamydomonas mutants by application of external force. Cell Motil Cytoskeleton. 1997;37(3):232–9. doi: 10.1002/(SICI)1097-0169(1997)37:3<232::AID-CM5>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol. 2009;187(6):921–33. doi: 10.1083/jcb.200908067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Ramanis Z, Luck DJ. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for Flagellar function. Cell. 1982;28(1):115–24. doi: 10.1016/0092-8674(82)90381-6. [DOI] [PubMed] [Google Scholar]
- Kikushima K. Central pair apparatus enhances outer-arm dynein activities through regulation of inner-arm dyneins. Cell Motil Cytoskeleton. 2009;66(5):272–80. doi: 10.1002/cm.20355. [DOI] [PubMed] [Google Scholar]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1990;87(3):1228–32. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL, Schnell RA, Fernandez E, Lefebvre PA. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J Cell Biol. 1989;109(6 Pt 1):2589–601. doi: 10.1083/jcb.109.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Kamiya R. Axonemal Dyneins: Assembly, Structure, and Force Generation. In: Witman GB, editor. The Chlamydomonas Sourcebook. Elsevier; 2009. pp. 131–191. [Google Scholar]
- Lechtreck KF, Delmotte P, Robinson ML, Sanderson MJ, Witman GB. Mutations in Hydin impair ciliary motility in mice. J Cell Biol. 2008;180(3):633–43. doi: 10.1083/jcb.200710162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Witman GB. Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J Cell Biol. 2007;176(4):473–82. doi: 10.1083/jcb.200611115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L, Campagna DR, Pinkus JL, Mulhern H, Wyatt TA, Sisson JH, Pavlik JA, Pinkus GS, Fleming MD. Primary ciliary dyskinesia in mice lacking the novel ciliary protein Pcdp1. Mol Cell Biol. 2008;28(3):949–57. doi: 10.1128/MCB.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann CB. A model of flagellar and ciliary functioning which uses the forces transverse to the axoneme as the regulator of dynein activation. Cell Motil Cytoskeleton. 1994;29(2):141–54. doi: 10.1002/cm.970290206. [DOI] [PubMed] [Google Scholar]
- Lindemann CB. Experimental evidence for the geometric clutch hypothesis. Curr Top Dev Biol. 2011;95:1–31. doi: 10.1016/B978-0-12-385065-2.00001-3. [DOI] [PubMed] [Google Scholar]
- Mitchell DR. Orientation of the central pair complex during flagellar bend formation in Chlamydomonas. Cell Motil Cytoskeleton. 2003;56(2):120–9. doi: 10.1002/cm.10142. [DOI] [PubMed] [Google Scholar]
- Mitchell DR. The flagellar central pair apparatus. In: Witman GB, editor. The Chlamydomoans Sourcebook. 2. Elsevier; 2009. pp. 235–252. [Google Scholar]
- Mitchell DR, Sale WS. Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J Cell Biol. 1999;144(2):293–304. doi: 10.1083/jcb.144.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009;58(1):165–174. doi: 10.1111/j.1365-313X.2008.03767.x. [DOI] [PubMed] [Google Scholar]
- Nelson JA, Savereide PB, Lefebvre PA. The CRY1 gene in Chlamydomonas reinhardtii: structure and use as a dominant selectable marker for nuclear transformation. Mol Cell Biol. 1994;14(6):4011–9. doi: 10.1128/mcb.14.6.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, McIntosh JR, Baumeister W. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc Natl Acad Sci USA. 2005;102(44):15889–94. doi: 10.1073/pnas.0508274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313(5789):944–8. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Okagaki T, Kamiya R. Microtubule sliding in mutant Chlamydomonas axonemes devoid of outer or inner dynein arms. J Cell Biol. 1986;103(5):1895–902. doi: 10.1083/jcb.103.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto CK, Kung C. The pair of central tubules rotates during ciliary beat in Paramecium. Nature. 1979;279(5713):532–4. doi: 10.1038/279532a0. [DOI] [PubMed] [Google Scholar]
- Omoto CK, Kung C. Rotation and twist of the central-pair microtubules in the cilia of Paramecium. J Cell Biol. 1980;87(1):33–46. doi: 10.1083/jcb.87.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics. 2006;22(9):1031–5. doi: 10.1093/bioinformatics/btl022. [DOI] [PubMed] [Google Scholar]
- Porter ME, Knott JA, Gardner LC, Mitchell DR, Dutcher SK. Mutations in the SUP-PF-1 locus of Chlamydomonas reinhardtii identify a regulatory domain in the beta-dynein heavy chain. J Cell Biol. 1994;126(6):1495–507. doi: 10.1083/jcb.126.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Power J, Dutcher SK. Extragenic suppressors of paralyzed flagellar mutations in Chlamydomonas reinhardtii identify loci that alter the inner dynein arms. J Cell Biol. 1992;118(5):1163–76. doi: 10.1083/jcb.118.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp G, O'Toole E, Gardner LC, Mitchell BF, Porter ME. The sup-pf-2 mutations of Chlamydomonas alter the activity of the outer dynein arms by modification of the gamma-dynein heavy chain. J Cell Biol. 1996;135(6 Pt 2):1853–65. doi: 10.1083/jcb.135.6.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp G, O'Toole E, Porter ME. The Chlamydomonas PF6 locus encodes a large alanine/proline-rich polypeptide that is required for assembly of a central pair projection and regulates flagellar motility. Mol Biol Cell. 2001;12(3):739–51. doi: 10.1091/mbc.12.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale WS, Satir P. Direction of active sliding of microtubules in Tetrahymena cilia. Proc Natl Acad Sci U S A. 1977;74(5):2045–9. doi: 10.1073/pnas.74.5.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez T, Welch D, Nicastro D, Dogic Z. Cilia-like beating of active microtubule bundles. Science. 2011;333(6041):456–9. doi: 10.1126/science.1203963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P. Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J Cell Biol. 1968;39(1):77–94. doi: 10.1083/jcb.39.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P. Mechanisms and controls of microtubule sliding in cilia. Symp Soc Exp Biol. 1982;35:179–201. [PubMed] [Google Scholar]
- Satir P. In: Switching mechanisms in the control of ciliary motility. Satir B, editor. New York: Alan R. Liss, Inc; 1985. p. 46. [Google Scholar]
- Satir P. The role of axonemal components in ciliary motility. Comp Biochem Physiol A. 1989;94(2):351–7. doi: 10.1016/0300-9629(89)90558-6. [DOI] [PubMed] [Google Scholar]
- Satir P, Matsuoka T. Splitting the ciliary axoneme: implications for a “switch-point” model of dynein arm activity in ciliary motion. Cell Motil Cytoskeleton. 1989;14(3):345–58. doi: 10.1002/cm.970140305. [DOI] [PubMed] [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene. 2001;277(1–2):221–9. doi: 10.1016/s0378-1119(01)00616-3. [DOI] [PubMed] [Google Scholar]
- Sizova IA, Lapina TV, Frolova ON, Alexandrova NN, Akopiants KE, Danilenko VN. Stable nuclear transformation of Chlamydomonas reinhardtii with a Streptomyces rimosus gene as the selective marker. Gene. 1996;181(1–2):13–8. doi: 10.1016/s0378-1119(96)00384-8. [DOI] [PubMed] [Google Scholar]
- Smith EF. Regulation of flagellar dynein by calcium and a role for an axonemal calmodulin and calmodulin-dependent kinase. Mol Biol Cell. 2002a;13(9):3303–13. doi: 10.1091/mbc.E02-04-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF. Regulation of flagellar dynein by the axonemal central apparatus. Cell Motil Cytoskeleton. 2002b;52(1):33–42. doi: 10.1002/cm.10031. [DOI] [PubMed] [Google Scholar]
- Smith EF, Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton. 2004;57(1):8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers KE, Gibbons IR. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc Natl Acad Sci U S A. 1971;68(12):3092–6. doi: 10.1073/pnas.68.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam LW, Lefebvre PA. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics. 1993;135(2):375–84. doi: 10.1093/genetics/135.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo MJ, Dymek EE, Smith EF. Calmodulin and PF6 are components of a complex that localizes to the C1 microtubule of the flagellar central apparatus. J Cell Sci. 2005;118(Pt 20):4655–65. doi: 10.1242/jcs.02585. [DOI] [PubMed] [Google Scholar]
- Wargo MJ, McPeek MA, Smith EF. Analysis of microtubule sliding patterns in Chlamydomonas flagellar axonemes reveals dynein activity on specific doublet microtubules. J Cell Sci. 2004;117(Pt 12):2533–44. doi: 10.1242/jcs.01082. [DOI] [PubMed] [Google Scholar]
- Wargo MJ, Smith EF. Asymmetry of the central apparatus defines the location of active microtubule sliding in Chlamydomonas flagella. Proc Natl Acad Sci U S A. 2003;100(1):137–42. doi: 10.1073/pnas.0135800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner FD, Satir P. The structural basis of ciliary bend formation. Radial spoke positional changes accompanying microtubule sliding. J Cell Biol. 1974;63(1):35–63. doi: 10.1083/jcb.63.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M, Nicastro D, Porter ME, Sale WS. The regulation of axonemal bending. In: Witman GB, editor. The Chlamydomonas Sourcebook. 2. Elsevier; 2009. pp. 253–282. [Google Scholar]
- Witman GB, Plummer J, Sander G. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J Cell Biol. 1978;76(3):729–47. doi: 10.1083/jcb.76.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Smith EF. The Flagellar Radial Spokes. In: Witman GB, editor. The Chlamydomonas Sourcebook. 2. Elsevier; 2009. pp. 209–234. [Google Scholar]
- Zhang H, Mitchell DR. Cpc1, a Chlamydomonas central pair protein with an adenylate kinase domain. J Cell Sci. 2004;117(Pt 18):4179–88. doi: 10.1242/jcs.01297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jones BH, Tang W, Moss SB, Wei Z, Ho C, Pollack M, Horowitz E, Bennett J, Baker ME, et al. Dissecting the axoneme interactome: the mammalian orthologue of Chlamydomonas PF6 interacts with sperm-associated antigen 6, the mammalian orthologue of Chlamydomonas PF16. Mol Cell Proteomics. 2005;4(7):914–23. doi: 10.1074/mcp.M400177-MCP200. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Tang W, Zhou R, Shen X, Wei Z, Patel AM, Povlishock JT, Bennett J, Strauss JF., 3rd Accelerated mortality from hydrocephalus and pneumonia in mice with a combined deficiency of SPAG6 and SPAG16L reveals a functional interrelationship between the two central apparatus proteins. Cell Motil Cytoskeleton. 2007;64(5):360–76. doi: 10.1002/cm.20189. [DOI] [PubMed] [Google Scholar]
- Zhao T, Wang W, Bai X, Qi Y. Gene silencing by artificial microRNAs in Chlamydomonas. Plant J. 2009;58(1):157–164. doi: 10.1111/j.1365-313X.2008.03758.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.