Abstract
Axon regeneration is a fundamental problem facing neuroscientists and clinicians. Failure of axon regeneration is caused by both extrinsic and intrinsic mechanisms. New techniques to exam gene expression such as Next Generation Sequencing of the Transcriptome (RNA-Seq) drastically increase our knowledge of both gene expression complexity (RNA isoforms) and gene expression regulation. By utilizing RNA-Seq, gene expression can now be defined at the level of isoforms, an essential step for understanding the mechanisms governing cell identity, growth and ultimately cellular responses to injury and disease.
Keywords: RNA-Seq, axon, regeneration, promoter, isoform, variant, untranslated region, coding DNA sequence, transcription start site
Adult central nervous system (CNS) neurons fail to regrow their axons and restore functional connections after an injury. When CNS axons are severed their distal portions undergo Wallarian degeneration--a process described by Santiago Ramon y Cajal nearly one hundred years ago1. Subsequently, the axonal endings proximal to cell bodies form dystrophic end bulbs that partially retract into highly dynamic structures2 that persist in the lesion site for weeks to months3 suggesting that severed axons retain motility but are inhibited in their attempts to regenerate. This view is supported by the finding that some CNS axons are able to extend long axons through permissive peripheral neuron grafts4,5. This and related findings led to the idea that the CNS environment present after an injury inhibits axon regeneration. Subsequently, major research efforts have focused on trying to understand the environmental influences that prohibit the axonal ends from growing across injury sites. These efforts identified key players that contribute to regenerative failure: immune cells including macrophages and microglia, reactive astrocytes which produce both physical and chemical barriers (reviewed in 6,7), and the by-products of myelin degradation8 (reviewed in 9). Since the identification of these extrinsic inhibitory influences, much work has focused on neutralizing or overcoming their effects. Unfortunately elimination of the various inhibitory factors does not result in major improvements in axonal regeneration10–13. Considering these findings, it is likely that the majority of neurons themselves are not in a state in which they can successfully regrow an axon, even when presented with favorable environmental conditions.
What evidence is there to suggest that adult CNS neurons need intrinsic modifications for axonal regeneration to succeed? First, there are substantial differences in the ability of embryonic versus adult CNS neurons to extend axons. A very simple observation is that culturing most adult CNS neurons is extremely difficult, if not impossible, whereas embryonic and early postnatal CNS neurons are easily cultured. This fundamental observation demonstrates that older CNS neurons are not capable of the plasticity and adaptability needed to survive in challenging conditions. In addition to this simple observation, there are clear differences in developmentally regulated gene expression changes that are associated with the growth properties of embryonic and early postnatal neurons suggesting differential gene expression changes contribute to the reduced axonal growth ability in mature CNS neurons14. Second, while axons from injured embryonic spinal cord can regenerate, if the same experimental lesion is performed in older spinal cords, regeneration fails15–17. Another piece of evidence stems from the observations that peripheral nervous system neurons, such as dorsal root ganglion (DRG) neurons, are capable of regenerating an axon18. DRG neurons exhibit robust growth in culture and grow axons into CNS white matter myelin tracts after injury19. Further, DRG neurons exhibit enhanced regeneration of both peripheral and central axons following injury so long there is a previous injury to the peripheral axon; this effect is known as a conditioning lesion18,20. If translation is pharmacologically blocked in DRG neurons, their ability to regenerate after injury is compromised21. These observations imply that failure of CNS neurons to regenerate axons is not solely due to the environment but that the pattern of neuronal gene expression is an important contributor to regenerative failure.
Several recent studies have identified genes important for axon regeneration such as the Krüppel-like transcription factors (KLFs) and cellular growth pathways involving mammalian target of rapamycin (mTOR) and the phosphatase and tensin homologue22–25 (PTEN). Since the importance and relevance of developmentally regulated transcription factors, such as the KLF family, and intrinsic growth pathways like mTOR and PTEN are nicely summarized in two recent reviews26,27, we will instead discuss efforts aimed at understanding how gene isoforms differ functionally and may be critical factors influencing the potential for axons to regenerate.
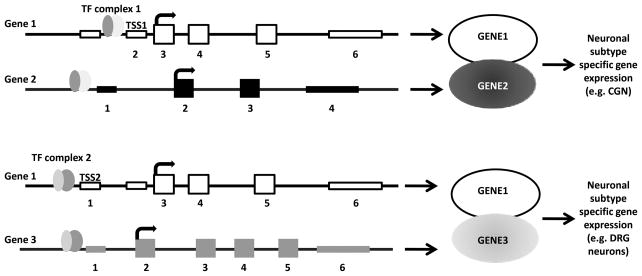
What are isoforms and why are they relevant to axon regeneration? Gene isoforms are messenger RNAs (mRNAs) that are produced from the same locus but are different in their transcription start sites (TSSs), protein coding DNA sequences (CDSs), and/or untranslated regions (UTRs), potentially altering gene function28,29. One mechanism of changing the expression of coregulated genes is through the use of alternative TSSs30,31. The idea that different binding partners confer different functional properties has been well studied in tissue specific gene regulation. For example, the same TF can direct gene expression in different tissues simply by binding with different TFs in each tissue32. This same idea may hold true for neuronal subtype specification and one way it could occur is through employment of alternative TSSs which direct different combinations of co-regulated genes (Figure 1). Alteration in CDSs can impart completely different protein functions, depending on what exons are ultimately expressed and what functional domains are included in the protein33. UTRs regulate the levels of primary transcript in numerous ways: transcript stability, folding, and turnover as well as translation efficiency34,35. In general, specific isoforms exhibit temporal or spatial differences in expression patterns, and it has been reported that alternatively expressed isoforms have different functions in the same cells. Exon usage has been shown to differ between tissues36 and it is likely that exon usage differs between neuronal subtypes and in different pathological states. Defining these isoforms is important for understanding neuronal gene expression in both health and disease. Indeed, some progress has been made to characterize known isoforms of regeneration associated genes (Table 1). However, a comprehensive understanding of the roles of these genes requires identification of the full isoform repertoire of each neuronal cell type under different conditions (i.e. injured vs. non-injured).
Figure 1. Example of how alternative TSSs can impart functional changes to isoforms from the same gene loci.
Gene 1 has two TSSs (TSS1 and TSS2), one CDS (exons 3–5) and one 3′ UTR (exon 6). In this example the only difference in the two isoforms expressed from the Gene 1 locus is in the TSS. Transcription factor complex 1 (TF complex 1) simultaneously regulates Gene 1 and 2 by binding to cis-elements upstream of TSS1 and the TSS in Gene 2. A different TF complex (TF complex 2) regulates the expression of Gene 1 and Gene 3. In this example, GENE2 or GENE3 co expression with GENE1 results in different functional outcomes: specification of sensory neurons or cerebellar granular neurons (CGNs). Genomic DNA is indicated by the black line, UTRs are thinner boxes and CDS encoding exons are thick boxes.
Table 1. Regeneration associated gene isoforms.
The number of TSS, CDS, and 3′ UTR for each gene found in the UCSC Known Genes, Ensemble, and RefSeq mouse genome combined annotation. The relevant reference indicating a role for the gene in axon growth and regeneration is listed.
| Official Gene Symbol | Number of: | Reference | |||
|---|---|---|---|---|---|
| Isoforms | TSS | CDS | 3′UTR | ||
| Adcyap1 (Pacap) | 2 | 2 | 1 | 1 | Neuroscience, 151:63–73 |
| Atf3 | 1 | 1 | 1 | 1 | J Neurosci, 27:7911–7920 |
| Atrn (attractin) | 1 | 1 | 1 | 1 | FASEB J, 19:153–4 |
| Bex1 | 1 | 1 | 1 | 1 | J Neurochem, 115:910–920 |
| Cd44 | 7 | 1 | 7 | 2 | Neuron, 43:57–67 |
| Cdkn1a (p21) | 2 | 2 | 2 | 1 | J.Neurosci, 22:1303–1315 |
| Gal (galanin) | 1 | 1 | 1 | 1 | Neuron, 43:57–67 |
| Gap43 | 1 | 1 | 1 | 1 | Development, 128:1175–82 |
| IL-6 | 3 | 2 | 3 | 2 | J Neurosci, 24:4432–43; J Biol Chem, 283:416–26 |
| Il6st (Gp130) | 2 | 1 | 2 | 2 | Neuron, 64:617–623 |
| Itga1 (integrin alpha7) | 4 | 1 | 4 | 4 | Neuron, 43:57–67 |
| Itgb1 (integrin beta1) | 3 | 2 | 3 | 1 | Neuron, 43:57–67 |
| Jun | 1 | 1 | 1 | 1 | Neuron, 43:57–67 |
| Klf4 | 1 | 1 | 1 | 1 | Science, 326:298–301 |
| Klf6 | 1 | 1 | 1 | 1 | Science, 326:298–301 |
| Klf7 | 3 | 2 | 3 | 2 | Science, 326:298–301 |
| L1cam | 2 | 1 | 2 | 2 | PNAS, 102:14883–14888 |
| Lif | 2 | 2 | 2 | 1 | J Neurosci, 21:7161–70 |
| Mapk8ip1 (JIP1) | 3 | 2 | 2 | 1 | J Neurosci, 30:7804–7816. |
| Matn2 | 4 | 1 | 4 | 4 | J Cell Sci, 122:995–1004 |
| Mdk | 4 | 4 | 2 | 1 | J Neurosci Res, 87:2908–2915 |
| Mtap1a (Map1a) | 2 | 2 | 2 | 2 | FASEB J, 19:153–4 |
| Mtap1b | 3 | 2 | 3 | 3 | J Neurosci, 24:7204–7213 |
| Nosip | 2 | 1 | 2 | 1 | J Neuropathol Exp Neurol, 60:411–21 |
| Npr2 | 4 | 4 | 3 | 1 | J Neurosci Res, 86:3163–9 |
| Nrn1 (neurtitin) | 1 | 1 | 1 | 1 | FASEB J,19:153–4 |
| Pten | 2 | 1 | 2 | 2 | Science, 322:963–966; J Neurosci, 30:9306–15 |
| Ptprs (PtpSigma) | 7 | 2 | 6 | 4 | J Neurosci, 22:5481–91; Science, 326:592–596 |
| RhoA | 1 | 1 | 1 | 1 | J.Neurosci, 29:15266–76 |
| Rock2 | 4 | 3 | 4 | 4 | J.Neurosci, 29:15266–76 |
| Smad1 | 2 | 1 | 2 | 2 | J Neurosci, 29:7116–23 |
| Socs3 | 1 | 1 | 1 | 1 | Neuron, 64:617–623 |
| Sprr1a | 1 | 1 | 1 | 1 | J.Neurosci, 22:1303–1315 |
| Stat3 | 3 | 1 | 3 | 1 | J Neurosci, 26:9512–9 |
| Stk25 (Mst3b) | 1 | 1 | 1 | 1 | Nat Neurosci, 12:1407–14 |
| Tnfrsf12a (Fn14) | 1 | 1 | 1 | 1 | J Neurosci, 23:9675–86 |
| Tnfrsf19 (TROY) | 5 | 2 | 3 | 3 | Neuron, 45:353–359 |
| Trp53 (p53) | 2 | 1 | 1 | 2 | EMBO J, 25:4084–96 |
| Trpc4ap | 3 | 2 | 3 | 2 | J Biol Chem, 283:416–426 |
How are different isoforms expressed in specific cell types/conditions identified? First the appropriate comparison has to be made to identify relevant isoforms. To do this it is necessary to compare gene expression in regenerating versus non-regenerating neurons. Isoforms identified this way are commonly referred to as ‘regeneration associated’ genes (RAGs). Second, the sequence of all mRNA species within a cell needs to be defined. New technologies such as Next Generation Sequencing of the Transcriptome (RNA-Seq) make this possible29,37. RNA-Seq allows detection of all gene isoforms expressed from a locus, a tremendous advantages over traditional microarray approaches. The functional importance of isoform diversity in the CNS is starting to be explored38–40, but examples of the importance of specific isoforms to axon regeneration are limited even though the number of known isoforms produced from regeneration associated genes is high (Table 1). This knowledge gap is probably due to the technical challenges involved in studying the roles of individual isoforms in neurons before the use of RNA-Seq methodology. Not only was it difficult to identify all the mRNA isoforms expressed from a given gene locus in a given cell type, but the methods needed to determine the function of individual genes were and still are less than optimal. For example, overexpression studies rely on specific cDNAs, examining the function only of the protein coding region portion of the transcript, typically ignoring the function of 3′ or 5′ UTRs. Similarly troubling is the fact that loss of function studies typically disrupt the function of all or most isoforms. Here we highlight some examples of individual isoform function relevant to axon regeneration and outline some potential RNA-Seq analyses strategies to identify relevant isoforms and their regulation (Figure 2; Table 2).
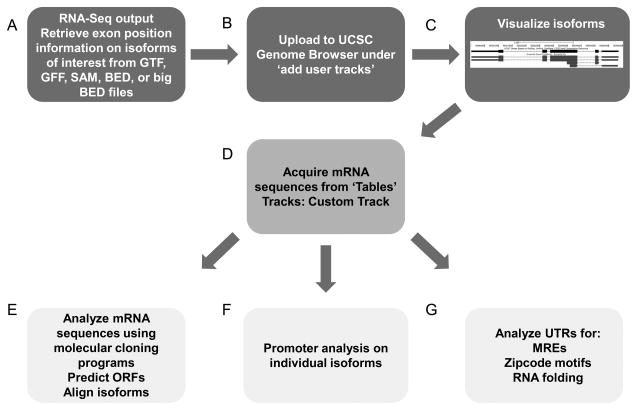
Figure 2. One approach to the downstream analysis of RNA-Seq data to identify and analyze mRNA isoforms.
Algorithms that assemble the small sequence fragments produced by Next Generation Sequencing will produce a file type (GFF, GTF, BED, big BED) that can be uploaded to the UCSC Genome Browser to visualize isoforms in the context of the reference genome (A, B, C). Once transcripts are visualized in the browser, the Tables tab will allow retrieval of the sequences in the user defined track, in this case, isoforms (C). After individual isoform sequences are in FASTA format they can be easily input into molecular cloning and analysis programs to predict open reading frames (ORFs) and align isoform sequences (E) as an example. TSS position information also facilitates isolation of the genomic promoter DNA sequence (F). UTR sequences can be uploaded to various programs for downstream analysis such as zipcode motifs, which target mRNAs within cells, and for microRNA Response Elements (MREs; G).
Table 2.
A list (not comprehensive) of frequently used programs for downstream RNA-Seq data analysis.
| Analysis | Software | Reference | PMID |
|---|---|---|---|
| Fragment/read mapping and transcript assembly | Cufflinks | Trapnell et al., 2010 | 20436464 |
| Tophat | Langmead et al., 2009 | 19261174 | |
| Bowtie | Trapnell et al., 2009 | 19289445 | |
| MapSplice | Wang et al., 2010 | 20802226 | |
| SOAP2 | Li et al., 2009 | 19497933 | |
| SpliceMap | Au et al., 2010 | 20371516 | |
|
| |||
| Promoter Analysis | MAPPER | Marinescu et al., 2004 | 15799782 |
| JASPAR | Bryne et al., 2008 | 18006571 | |
| oPOSSUM | Ho Sui et al., 2007 | 17576675 | |
| TRANSFAC | Matys et al., 2006 | 16381825 | |
|
| |||
| miRNA Response Element | TargetScan | Friedman et al., 2009 | 18955434 |
| MiRscan | Lim et al., 2003 | 12624257 | |
|
| |||
| zipcode motifs | REPFIND | Andken et al., 2007 | 17663765 |
Alternative transcription start sites
Cis-regulatory elements in the promoter contain sequences recognized by transcription factors and the basal transcription machinery; these are key elements and an important step in regulating isoform expression. Because of these cis-regulatory elements within the promoter, the location of the TSS is important for understanding the biogenesis of specific isoforms30,41. As mentioned, RNA-Seq enables the determination of individual isoform TSSs. Subsequently, the promoter DNA sequence can be isolated and then analyzed for cis-elements. Once this is accomplished for all the isoforms expressed in different cells types/states, such as a comparison between neurons capable or incapable of axon regeneration, it is possible to identify sets of isoforms undergoing similar patterns of gene expression regulation.
How can you predict the cis-elements in the promoters of individual isoforms? There are numerous freely available software programs capable of scanning nucleotide sequences and evaluating the frequency of transcription factor binding sites42–45. This enables the identification of candidate transcription factors regulating individual isoforms, which may be critically important if differential isoform expression arises from the TSSs, and not in the coding DNA sequence. Understanding gene expression regulation at the promoter level is not without precedent; using zebrafish RGCs as a model for developing and regenerating axons, two rat promoters for growth associated protein 43 (Gap43) were identified. One promoter is active when RGCs are growing to their targets in early development and a different promoter is active when they are regenerating46. While GAP43 is expressed in the developing and regenerating optic nerve, this study suggests that the genetic programs driving axon regeneration and axon path finding during development are different. Understanding the extent and importance of these differences is crucial to devising a plan to reactivate developmental gene programs that promote axon growth after injury.
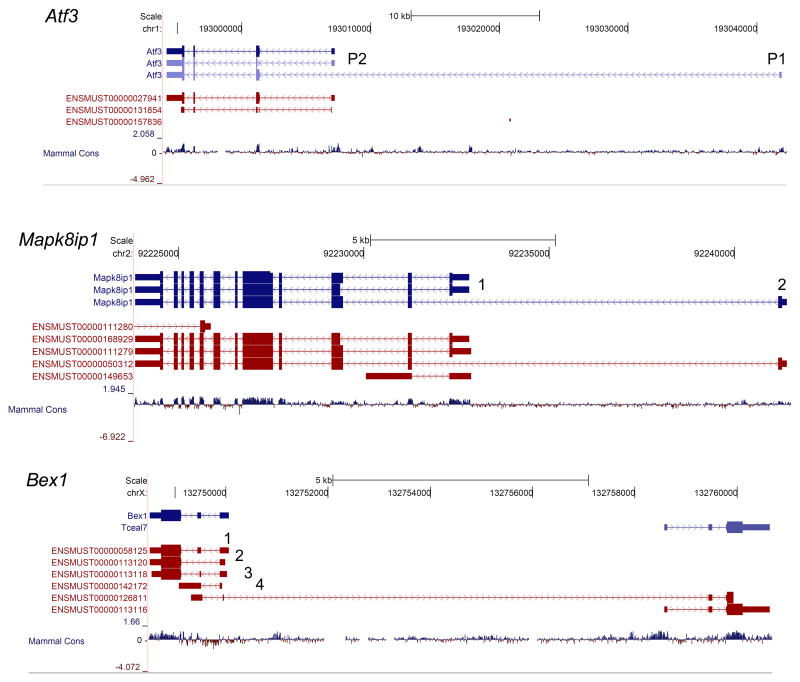
Do other regeneration associated genes utilize alternative TSSs? Activating transcription factor 3 (Atf3) is a known regeneration associated gene47,48 associated with numerous promoters. Atf3 expression increases after nerve injury and overexpression of a constitutively active form of Atf3 increases the rate of peripheral nerve regeneration47,49. In cancer studies an alternative TSS was previously characterized50. Each promoter is active under different conditions. One Atf3 isoform uses the P1 promoter (Figure 3) which is primarily activein response to stress and in numerous cancers50, whereas the conventional Atf3 promoter (P2) is typically activated in response to mitogenic stimuli51. While the CDS for these isoforms is the same, one way functional differences may occur is through differential promoter regulation resulting in functional differences resulting from changes in coregulated genes. Alternatively, these promoters may be driving varying levels of ATF3 expression. It is unclear which promoters are in use in regenerating DRG neurons and whether the same promoters are used during development when axons are first extending towards their targets.
Figure 3. UCSC Genome Browser tracks for Atf3, Mapk8ip1, and Bex1.
Mouse isoforms in the UCSC known genes database are listed at the top (in blue) and isoforms in the Ensemble database are below (in dark red). Bars indicate exons and the CDS is indicated with a thicker bar. The arrows on the lines connecting the boxes indicate strand. Arrows pointing to the right indicate ( + ) strand, arrows pointing left indicate (−) strand. For example, Atf3 is in the (−) strand and so the 5′ end of the transcript is on the right. The mammalian conservation at the DNA level is shown in the bottom track. For Atf3, two TSSs have been identified and the associated P1 and P2 promoter are labeled. Two TSSs for Mapk8ip1 have been identified and are labeled. Bex1 produces four isoforms which have different CDS.
The use of alternative TSSs is not limited to Atf3. Numerous regeneration associated genes have known alternative TSSs annotated in the UCSC Genome Browser (Figure 1; Table 1). Some examples are themitogen-activated protein kinase 8 interacting protein 1 (Mapk8ip1/Jip1), protein tyrosine phosphatase receptor type S (Ptprs), and Rho-associated coiled-coil containing protein kinase 252–55 (Rock2). MAPK8IP1/JIP1, a scaffolding protein for the c-Jun N-terminal kinases (Jnks), is a newly identified regeneration associated gene with multiple TSSs53 (Figure 3). Mapk8ip1 null DRG neurons exhibit delayed neurite extension and reduced neurite length, possibly due to a reduction in JNK phosphorylation53. Which isoforms are expressed after peripheral nerve injury is currently unknown. If Gap43 serves as an example, it will be interesting to determine if these alternative promoters are active in a particular combination during development and then in another combination during axon regeneration. Identifying the set of promoters in use during development and in regeneration is necessary if the goal is to define what pathways need to be reactivated to elicit an optimal intrinsic regeneration response in injured neurons.
Alternative protein coding DNA sequences (CDS)
Isoforms harboring changes in the CDS have been the most thoroughly characterized because they commonly give rise to proteins with different functional properties33. In neurons, some well characterized genes which produce isoforms with changes in CDS are members of the immunoglobulin superfamily, molecules which have well defined roles in facilitating axon growth and guidance. Three genes in this family, neural cell adhesion molecule 1 (Ncam1), L1 cell adhesion molecule (L1cam), and deleted in colorectal cancer (Dcc), all undergo alternative splicing giving rise to isoforms that exhibit cell type and developmental stage specific expression in the nervous system. For example, Ncam1 has isoforms that produce proteins of 180kD, 140kD, or 120kD in size56. These isoforms show preferential expression in different cell types and contribute to different aspects of neuronal phenotype such as neuronal process branching and synaptic maturation (reviewed in57). In the injured spinal cord of rats treated with a monoclonal antibody to Nogo-A (IN-1), Ncam expression increased. This result suggests that Ncam could be a target to enhance axon regeneration58; however which Ncam isoforms were upregulated remains unknown. L1cam is another regeneration associated gene which undergoes alternative splicing yielding two alternative isoforms with different functions59–61. Alternative splicing that includes both exons 2 and 27 defines the predominantly neuronal, long L1CAM isoform59,62. Specifically exon 27 is involved in L1CAM targeting to neuronal growth cones and exon 2 is used in determining ligand specificity63,64. The L1CAM isoform without exons 2 and 27 is considered the short form and is predominantly expressed in glial and nonneuronal cells62,65. NCAM and L1CAM isoforms are examples of how the same gene can produce proteins with functional and cell type dependant expression differences, but isoforms with CDS changes may also define specific populations of neurons. For example, a recent study which profiled the transcriptome of developing cortical layers using RNA-Seq identified over 1500 layer specific isoforms66. Interestingly, numerous regeneration associated genes were found to have layer specific isoforms (Mapk8ip1, Ptprs, Rock2, Bex1, and microtubule associated protein 1 a: Mtap1a) indicating that these genes have the potential to produce differentially regulated isoforms, and thus differ in their function as well. The brain expressed X-linked (Bex1) gene locus produces four different isoforms, each having different CDSs (Figure 3). Understanding the possible alterations in function and/or expression pattern is relevant to axon regeneration because BEX1 interacts with the p75 neurotrophin receptor (p75NTR), is expressed in a variety of neuronal tissues during development, and regulates neuronal differentiation67. Recently, it has been shown that BEX1 is localized to neuronal processes, its expression substantially increases in both motor and dorsal root ganglion neurons after a peripheral nerve injury, and that Bex1 null mice undergo delayed peripheral nerve regeneration52.
The KLFs are zinc finger transcription factors, most of which are expressed in the mammalian CNS. The first study demonstrating the importance of two members of the Krüppel-like family of transcription factors (KLFs), KLF6 and KLF7, for optic nerve regeneration was performed in zebrafish68. Interestingly, several members of this TF family have opposing expression patterns throughout development. This pattern correlates with the age of the neuron and its ability to grow an axon: KLF4 and -9 are highly expressed in adult neurons which show reduced axon regeneration potential, overexpression of KLF4 or -9 suppresses neurite outgrowth in vitro, and conditional knockout of KLF4 in RGC neurons results in enhanced axon regeneration after optic nerve crush22,69. Conversely, KLF6 and -7 are highly expressed in embryonic neurons but downregulated in the adult, and overexpression in RGC, hippocampal, or cortical neurons in vitro increases neurite outgrowth22,69.
Is there reason to believe that Klfs give rise to multiple KLF isoforms? A study examining zinc-finger transcription factors, which includes Klfs, found that over 50% of these genes produce multiple isoforms70. Indeed, new isoforms for Klf13 were identified with changes in the number of zinc finger domains, which is likely to affect DNA binding ability and thus Klf13 function70. The expression and identity of Klf6 and Klf7 isoforms in the CNS is unknown. It is known that Klf7 expression is high in developing TrkA+ DRG neurons71, but only Klf6 has been reported to be upregulated after a sciatic nerve injury72. Currently, we do not know if different isoforms of Klf6 or -7 exist, but it is plausible that different isoforms would have different functions in the context of axon regeneration.
Alternative 5′ and 3′ Untranslated Regions
What are untranslated regions (UTRs) and what is their function? UTRs are regions of mRNAs that do not code for protein. They are interesting because they confer regulatory information to the primary transcript. UTRs can regulate mRNA stability, translation rate, harbor sequences that mediate subcellular targeting of mRNAs (zipcode sequences) and/or those that govern global gene expression such as microRNA (miRNA) Response Elements (MREs)34,35,73,74.
Alternative splicing and polymorphisms in the 3′UTR have been shown to affect isoform function through regulation by miRNAs, which can modulate translation via regulation of the primary transcript75. MiRNAs typically downregulate transcript expression by triggering degradation or halting translation. For example, the homeobox (Hox) genes are responsible for patterning the anterior/posterior body axis and their expression along the body axis is tightly controlled during development. In Drosophila, several isoforms of the Hox gene family differ in the lengths of their 3′UTRs, imparting differences in the MREs within those transcripts76. Thus these isoforms undergo developmental regulation and could have specific function in setting up anteroposterior body axis76. In addition to development, different sequences in the UTRs of isoforms may affect pathological states. For example, polymorphisms within the 3′UTR of amyloid precursor protein (App) are associated with altered miRNA regulation of the primary transcript and could be linked to Alzheimer’s disease progression77. MiRNA activity in neurons can regulate activity dependent dendritic spine formation78,79. While these are examples of individual isoform regulation, miRNAs have the potential to simultaneously regulate hundreds of transcripts, also making them global regulators of gene expression75. A role for miRNAs in axon regeneration is just starting to be explored80 but understanding which miRNAs regulate genes that are prohibitive to axon growth could provide an important new mechanism to globally alter gene expression in ways that would activate intrinsic axon growth pathways in injured neurons.
One of the most well characterized examples of a UTR effecting function and mRNA targeting is the alternative transcripts produced from the brain derived neurotrophic factor (Bdnf) gene. Bdnf produces two isoforms that differ in the lengths of their 3′ UTRs while coding for the same protein81. The isoform with the shorter 3′ UTR is sequestered in the neuronal cell body while the transcript with the longer 3′ UTR is targeted to dendrites. This differential targeting enables activity dependent translation in the dendrites where BDNF has an essential role in regulating dendritic arborizations and long term potentiation81. The glucocorticoid receptor (GR; gene name: nuclear receptor subfamily 3, group C, member 1: Nr3c1) is another example of a gene with enormous diversity in both the 5′ and 3′ UTRs. Because GR is ubiquitously expressed, this diversity is thought to enable tissue specific isoform expression (reviewed in82,83).
Of the isoform regulatory mechanisms conferred by UTRs, one most relevant to axon regeneration may be targeting of mRNAs to the axon via zipcode motifs. For example, L1cam and beta-1-integrin (Itgb1)promote axonal growth in CNS injury models. However, gene therapy strategies using them might fail because the transcript in question does not target appropriately within the neuron to promote axon growth60,84. How are transcripts targeted to specific subcellular locations in neurons? Transcripts are targeted by ‘zipcodes’ which are sequences found in the 3′ UTR that enable mRNA trafficking proteins, such as zipcode binding protein 1 (Zbp1) and poly(rC) binding protein 1 (Pcbp1 aka: heterogeneous nuclear ribonucleoproteinE1; hnRNP), which bind and shuttle the transcript to the area of the cell where it will be translated73,85. One of the first characterized zipcode motifs and mRNA binding proteins involved in neuronal process transcript targeting was demonstrated in the cytoskeletal protein component beta actin (Actb)86,87. Actb targeting relies on a relatively short nucleotide motif (~40–55 base pairs in length) and disruption of transcript targeting by mutating the nucleotide motif alters cytoskeletal organization87. Identifying additional zipcodes is challenging because methods have relied primarily on alignment, examination of conserved sequences, and mutation or deletion analysis. This is where using RNA-Seq becomes advantageous because if the targeting of individual isoforms is to be understood, then the sequence identity has to be recovered88. Predicting motifs that can target a transcript to the axon will allow for future gene therapy approaches to appropriately target growth promoting transcripts to the growth cone of injured axons and perhaps facilitate meaningful regeneration.
In summary, to fully understand neuronal specific gene expression and the mechanisms that regulate differential isoform expression between neuronal subtypes, we need to understand the full repertoire of transcripts expressed from each gene; a new technology, RNA-Seq, makes this possible. With this technology we now have the ability to precisely define the isoforms expressed within specific neuronal populations, examine their sequences for changes in the 5′ or 3′ UTRs which may alter TSS, transcript expression regulation, and examine changes in the CDS which may affect functional domains in the protein. Since these alterations in mRNAs generated from the same gene loci may affect function, they are necessary to consider during the elucidation of cell type specific isoform function. Applying RNA-Seq to the question of axon regeneration will undoubtedly increase our ability to define the isoforms relevant to axonal growth and our ability to understand the regulation of ‘regenerating’ versus ‘non-regenerating’ neurons.
Acknowledgments
This work was supported by National Institutes of Health grants HD057632 and NS059866, U.S. Army (W81XWH-05-1-0061), the Walter G. Ross foundation and the Buoniconti Foundation. We thank Dr. Murray Blackmore for helpful discussions, Dr. Dario Motti, Dr. Kunie Sakurai and Samuel Beckerman for helpful edits of the manuscript.
References
- 1.Ramon y Cajal S. Degeneration and regeneration of the nervous system. London: Oxford University Press; 1928. [Google Scholar]
- 2.Tom VJ, Steinmetz MP, Miller JH, Doller CM, Silver J. Studies on the development and behavior of the dystrophic growth cone, the hallmark of regeneration failure, in an in vitro model of the glial scar and after spinal cord injury. J Neurosci. 2004;24(29):6531–6539. doi: 10.1523/JNEUROSCI.0994-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Raisman G. Sprouts from cut corticospinal axons persist in the presence of astrocytic scarring in long-term lesions of the adult rat spinal cord. Exp Neurol. 1995;134(1):102–111. doi: 10.1006/exnr.1995.1041. [DOI] [PubMed] [Google Scholar]
- 4.Richardson PM, McGuinness UM, Aguayo AJ. Axons from CNS neurons regenerate into PNS grafts. Nature. 1980;284(5753):264–265. doi: 10.1038/284264a0. [DOI] [PubMed] [Google Scholar]
- 5.David S, Aguayo AJ. Axonal elongation into peripheral nervous system “Bridges” after central nervous system injury in adult rats. Science. 1981;214(4523):931–933. doi: 10.1126/science.6171034. [DOI] [PubMed] [Google Scholar]
- 6.Yiu G, He Z. Glial inhibition of CNS axon regeneration. Nat Rev Neurosci. 2006;7(8):617–627. doi: 10.1038/nrn1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitch MT, Silver J. CNS injury, glial scars, and inflammation: Inhibitory extracellular matrices and regeneration failure. Exp Neurol. 2008;209(2):294–301. doi: 10.1016/j.expneurol.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry M. Post-injury myelin-breakdown products inhibit axonal growth: An hypothesis to explain the failure of axonal regeneration in the mammalian central nervous system. Bibl Anat. 1982;23(23):1–11. [PubMed] [Google Scholar]
- 9.Filbin MT. Myelin-associated inhibitors of axonal regeneration in the adult mammalian CNS. Nat Rev Neurosci. 2003;4(9):703–713. doi: 10.1038/nrn1195. [DOI] [PubMed] [Google Scholar]
- 10.Horn KP, Busch SA, Hawthorne AL, van Rooijen N, Silver J. Another barrier to regeneration in the CNS: Activated macrophages induce extensive retraction of dystrophic axons through direct physical interactions. J Neurosci. 2008;28(38):9330–9341. doi: 10.1523/JNEUROSCI.2488-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tom VJ, Sandrow-Feinberg HR, Miller K, et al. Combining peripheral nerve grafts and chondroitinase promotes functional axonal regeneration in the chronically injured spinal cord. J Neurosci. 2009;29(47):14881–14890. doi: 10.1523/JNEUROSCI.3641-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kigerl KA, Gensel JC, Ankeny DP, et al. Identification of two distinct macrophage subsets with divergent effects causing either neurotoxicity or regeneration in the injured mouse spinal cord. J Neurosci. 2009;29(43):13435–13444. doi: 10.1523/JNEUROSCI.3257-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JK, Geoffroy CG, Chan AF, et al. Assessing spinal axon regeneration and sprouting in nogo-, MAG-, and OMgp-deficient mice. Neuron. 2010;66(5):663–670. doi: 10.1016/j.neuron.2010.05.002. Available: www.refworks.com via the Internet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg JL, Klassen MP, Hua Y, Barres BA. Amacrine-signaled loss of intrinsic axon growth ability by retinal ganglion cells. Science. 2002;296(5574):1860–1864. doi: 10.1126/science.1068428. [DOI] [PubMed] [Google Scholar]
- 15.Bregman BS, Goldberger ME. Anatomical plasticity and sparing of function after spinalcord damage in neonatal cats. Science. 1982;217(4559):553–555. doi: 10.1126/science.7089581. [DOI] [PubMed] [Google Scholar]
- 16.Saunders NR, Balkwill P, Knott G, Habgood MD, Mollgard K, et al. Growth of axons through a lesion in the intact CNS of fetal rat maintained in long-term culture. Proc Biol Sci. 1992;250(1329):171–180. doi: 10.1098/rspb.1992.0146. [DOI] [PubMed] [Google Scholar]
- 17.Treherne JM, Woodward SK, Varga ZM, Ritchie JM, Nicholls JG. Restoration of conduction and growth of axons through injured spinal cord of neonatal opossum in culture. Proc Natl Acad Sci U S A. 1992;89(1):431–434. doi: 10.1073/pnas.89.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McQuarrie IG, Grafstein B. Axon outgrowth enhanced by a previous nerve injury. Arch Neurol. 1973;29(1):53–55. doi: 10.1001/archneur.1973.00490250071008. [DOI] [PubMed] [Google Scholar]
- 19.Davies SJ, Fitch MT, Memberg SP, et al. Regeneration of adult axons in white matter tracts of the central nervous system. Nature. 1997;390(6661):680–683. doi: 10.1038/37776. [DOI] [PubMed] [Google Scholar]
- 20.McQuarrie IG, Grafstein B, Gershon MD. Axonal regeneration in the rat sciatic nerve: Effect of a conditioning lesion and of dbcAMP. Brain Res. 1977;132(3):443–453. doi: 10.1016/0006-8993(77)90193-7. [DOI] [PubMed] [Google Scholar]
- 21.Smith DS, Skene JH. A transcription-dependent switch controls competence of adult neurons for distinct modes of axon growth. J Neurosci. 1997;17(2):646–658. doi: 10.1523/JNEUROSCI.17-02-00646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore DL, Blackmore MG, Hu Y, et al. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326(5950):298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park KK, Liu K, Hu Y, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322(5903):963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christie KJ, Webber CA, Martinez JA, Singh B, Zochodne DW. PTEN inhibition to facilitate intrinsic regenerative outgrowth of adult peripheral axons. J Neurosci. 2010;30(27):9306–9315. doi: 10.1523/JNEUROSCI.6271-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu K, Lu Y, Lee JK, et al. PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat Neurosci. 2010;13(9):1075–1081. doi: 10.1038/nn.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu K, Tedeschi A, Park KK, He Z. Neuronal intrinsic mechanisms of axon regeneration. Annu Rev Neurosci. 2011;34:131–152. doi: 10.1146/annurev-neuro-061010-113723. [DOI] [PubMed] [Google Scholar]
- 27.Moore DL, Goldberg JL. Multiple transcription factor families regulate axon growth and regeneration. Dev Neurobiol. 2011 doi: 10.1002/dneu.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Gerstein M, Snyder M. RNA-seq: A revolutionary toolfor transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu JQ, Habegger L, Noisa P, et al. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc Natl Acad Sci U S A. 2010;107(11):5254–5259. doi: 10.1073/pnas.0914114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 31.Hittelman AB, Burakov D, Iniguez-Lluhi JA, Freedman LP, Garabedian MJ. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 1999;18(19):5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu X, Lin J, Zack DJ, Qian J. Computational analysis of tissue-specific combinatorial gene regulation: Predicting interaction between transcription factors in human tissues. Nucleic Acids Res. 2006;34(17):4925–4936. doi: 10.1093/nar/gkl595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breitbart RE, Andreadis A, Nadal-Ginard B. Alternative splicing: A ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- 34.van der Velden AW, Thomas AA. The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int J Biochem Cell Biol. 1999;31(1):87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- 35.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A. 2007;104(23):9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 38.Polo-Parada L, Bose CM, Plattner F, Landmesser LT. Distinct roles of different neural cell adhesion molecule (NCAM) isoforms in synaptic maturation revealed by analysis of NCAM 180 kDa isoform-deficient mice. J Neurosci. 2004;24(8):1852–1864. doi: 10.1523/JNEUROSCI.4406-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Bates R, Yin DM, et al. Specific regulation of NRG1 isoform expression by neuronal activity. J Neurosci. 2011;31(23):8491–8501. doi: 10.1523/JNEUROSCI.5317-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanaykina N, Abelson K, King D, et al. In vitro and in vivo effects on neural crest stem cell differentiation by conditional activation of Runx1 short isoform and its effect on neuropathic pain behavior Ups. J Med Sci. 2010;115(1):56–64. doi: 10.3109/03009730903572065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roni V, Carpio R, Wissinger B. Mapping of transcription start sites of human retina expressed genes. BMC Genomics. 2007;8:42. doi: 10.1186/1471-2164-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen QK, Hertz GZ, Stormo GD. MATRIX SEARCH 1.0: A computer program that scans DNAsequences for transcriptional elements using a database of weight matrices. Comput Appl Biosci. 1995;11(5):563–566. doi: 10.1093/bioinformatics/11.5.563. [DOI] [PubMed] [Google Scholar]
- 43.Kel AE, Gossling E, Reuter I, et al. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31(13):3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marinescu VD, Kohane IS, Riva A. MAPPER: A search engine for the computational identification of putative transcription factor binding sites in multiple genomes. BMC Bioinformatics. 2005;6:79. doi: 10.1186/1471-2105-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marstrand TT, Frellsen J, Moltke I, et al. Asap: A framework for over-representation statistics for transcription factor binding sites. PLoS One. 2008;3(2):e1623. doi: 10.1371/journal.pone.0001623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Udvadia AJ, Koster RW, Skene JH. GAP-43 promoter elements in transgenic zebrafish reveal a difference in signals for axon growth during CNS development and regeneration. Development. 2001;128(7):1175–1182. doi: 10.1242/dev.128.7.1175. [DOI] [PubMed] [Google Scholar]
- 47.Seijffers R, Mills CD, Woolf CJ. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J Neurosci. 2007;27(30):7911–7920. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saul KE, Koke JR, Garcia DM. Activating transcription factor 3 (ATF3) expression in the neural retina and optic nerve of zebrafish during optic nerve regeneration. Comp Biochem Physiol A-Mol Integr Physiol. 2010;155(2):172–182. doi: 10.1016/j.cbpa.2009.10.042. [DOI] [PubMed] [Google Scholar]
- 49.Tsujino H, Kondo E, Fukuoka T, et al. Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: A novel neuronal marker of nerve injury. Mol Cell Neurosci. 2000;15(2):170–182. doi: 10.1006/mcne.1999.0814. [DOI] [PubMed] [Google Scholar]
- 50.Miyazaki K, Inoue S, Yamada K, et al. Differential usage of alternate promoters of the human stress response gene ATF3 in stress response and cancer cells. Nucleic Acids Res. 2009;37(5):1438–1451. doi: 10.1093/nar/gkn1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyer VR, Eisen MB, Ross DT, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283(5398):83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 52.Khazaei MR, Halfter H, Karimzadeh F, et al. Bex1 is involved in the regeneration of axons after injury. J Neurochem. 2010;115(4):910–920. doi: 10.1111/j.1471-4159.2010.06960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barnat M, Enslen H, Propst F, et al. Distinct roles of c-jun N-terminal kinase isoforms in neurite initiation and elongation during axonal regeneration. J Neurosci. 2010;30(23):7804–7816. doi: 10.1523/JNEUROSCI.0372-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Y, Tenney AP, Busch SA, et al. PTPsigma is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science. 2009;326(5952):592–596. doi: 10.1126/science.1178310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duffy P, Schmandke A, Schmandke A, et al. Rho-associated kinase II (ROCKII) limits axonal growth after trauma within the adult mouse spinal cord. J Neurosci. 2009;29(48):15266–15276. doi: 10.1523/JNEUROSCI.4650-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cunningham BA, Hemperly JJ, Murray BA, et al. Neural cell adhesion molecule: Structure, immunoglobulin-like domains, cell surface modulation, and alternative RNA splicing. Science. 1987;236(4803):799–806. doi: 10.1126/science.3576199. [DOI] [PubMed] [Google Scholar]
- 57.Brennaman LH, Maness PF. NCAM in neuropsychiatric and neurodegenerative disorders. Adv Exp Med Biol. 2010;663:299–317. doi: 10.1007/978-1-4419-1170-4_19. [DOI] [PubMed] [Google Scholar]
- 58.Bareyre FM, Haudenschild B, Schwab ME. Long-lasting sprouting and gene expression changes induced by the monoclonal antibody IN-1 in the adult spinal cord. J Neurosci. 2002;22(16):7097–7110. doi: 10.1523/JNEUROSCI.22-16-07097.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacob J, Haspel J, Kane-Goldsmith N, Grumet M. L1 mediated homophilic binding and neurite outgrowth are modulated by alternative splicing of exon 2. J Neurobiol. 2002;51(3):177–189. doi: 10.1002/neu.10052. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Bo X, Schoepfer R, et al. Growth-associated protein GAP-43 and L1 act synergistically to promote regenerative growth of purkinje cell axons in vivo. Proc Natl Acad Sci U S A. 2005;102(41):14883–14888. doi: 10.1073/pnas.0505164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miura M, Kobayashi M, Asou H, Uyemura K. Molecular cloning of cDNA encoding the rat neural cell adhesion molecule L1. two L1 isoforms in the cytoplasmic region are produced by differential splicing. FEBS Lett. 1991;289(1):91–95. doi: 10.1016/0014-5793(91)80915-p. [DOI] [PubMed] [Google Scholar]
- 62.Takeda Y, Asou H, Murakami Y, et al. A nonneuronal isoform of cell adhesion molecule L1: Tissue-specific expression and functional analysis. J Neurochem. 1996;66(6):2338–2349. doi: 10.1046/j.1471-4159.1996.66062338.x. [DOI] [PubMed] [Google Scholar]
- 63.Kamiguchi H, Lemmon V. A neuronal form of the cell adhesion molecule L1 contains a tyrosine-based signal required for sorting to the axonal growth cone. J Neurosci. 1998;18(10):3749–3756. doi: 10.1523/JNEUROSCI.18-10-03749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Angelis E, Brummendorf T, Cheng L, Lemmon V, Kenwrick S. Alternative use of a mini exon of the L1 gene affects L1 binding to neural ligands. J Biol Chem. 2001;276(35):32738–32742. doi: 10.1074/jbc.M105156200. [DOI] [PubMed] [Google Scholar]
- 65.Itoh K, Sakurai Y, Asou H, Umeda M. Differential expression of alternatively spliced neural cell adhesion molecule L1 isoforms during oligodendrocyte maturation. J Neurosci Res. 2000;60(5):579–586. doi: 10.1002/(SICI)1097-4547(20000601)60:5<579::AID-JNR2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 66.Belgard TG, Marques AC, Oliver PL, et al. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71(4):605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vilar M, Murillo-Carretero M, Mira H, et al. Bex1, a novel interactor of the p75 neurotrophin receptor, links neurotrophin signaling to the cell cycle. EMBO J. 2006;25(6):1219–1230. doi: 10.1038/sj.emboj.7601017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Veldman MB, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312(2):596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 69.Blackmore MG, Moore DL, Smith RP, et al. High content screening of cortical neurons identifies novel regulators of axon growth. Mol Cell Neurosci. 2010;44(1):43–54. doi: 10.1016/j.mcn.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ravasi T, Huber T, Zavolan M, et al. Systematic characterization of the zinc-finger-containing proteins in the mouse transcriptome. Genome Res. 2003;13(6B):1430–1442. doi: 10.1101/gr.949803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lei L, Ma L, Nef S, Thai T, Parada LF. mKlf7, a potential transcriptional regulator of TrkA nerve growth factor receptor expression in sensory and sympathetic neurons. Development. 2001;128(7):1147–1158. doi: 10.1242/dev.128.7.1147. [DOI] [PubMed] [Google Scholar]
- 72.Nilsson A, Moller K, Dahlin L, Lundborg G, Kanje M. Early changes in gene expression in the dorsal root ganglia after transection of the sciatic nerve; effects of amphiregulin and PAI-1 on regeneration. Brain Res Mol Brain Res. 2005;136(1–2):65–74. doi: 10.1016/j.molbrainres.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 73.Singer RH. RNA zipcodes for cytoplasmic addresses. Curr Biol. 1993;3(10):719–721. doi: 10.1016/0960-9822(93)90079-4. [DOI] [PubMed] [Google Scholar]
- 74.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 76.Thomsen S, Azzam G, Kaschula R, Williams LS, Alonso CR. Developmental RNA processing of 3′UTRs in hox mRNAs as a context-dependent mechanism modulating visibility to microRNAs. Development. 2010;137(17):2951–2960. doi: 10.1242/dev.047324. [DOI] [PubMed] [Google Scholar]
- 77.Delay C, Calon F, Mathews P, Hebert SS. Alzheimer-specific variants in the 3′UTR of amyloid precursor protein affect microRNA function. Mol Neurodegener. 2011;6(1):70. doi: 10.1186/1750-1326-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schratt GM, Tuebing F, Nigh EA, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 79.Fiore R, Khudayberdiev S, Christensen M, et al. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28(6):697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu B, Zhou S, Wang Y, et al. Profile of MicroRNAs following rat sciatic nerve injury by deep sequencing: Implication for mechanisms of nerve regeneration. PLoS One. 2011;6(9):e24612. doi: 10.1371/journal.pone.0024612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.An JJ, Gharami K, Liao GY, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134(1):175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu NZ, Cidlowski JA. Glucocorticoid receptor isoforms generate transcription specificity. Trends Cell Biol. 2006;16(6):301–307. doi: 10.1016/j.tcb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 83.Revollo JR, Cidlowski JA. Mechanisms generating diversity in glucocorticoid receptor signaling. Ann N Y Acad Sci. 2009;1179:167–178. doi: 10.1111/j.1749-6632.2009.04986.x. [DOI] [PubMed] [Google Scholar]
- 84.Tan CL, Kwok JC, Patani R, et al. Integrin activation promotes axon growth on inhibitory chondroitin sulfate proteoglycans by enhancing integrin signaling. J Neurosci. 2011;31(17):6289–6295. doi: 10.1523/JNEUROSCI.0008-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Snee M, Kidd GJ, Munro TP, Smith R. RNA trafficking and stabilization elements associate with multiple brain proteins. J Cell Sci. 2002;115(Pt 23):4661–4669. doi: 10.1242/jcs.00137. [DOI] [PubMed] [Google Scholar]
- 86.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17(4):2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127(2):441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andken BB, Lim I, Benson G, et al. 3′-UTR SIRF: A database for identifying clusters of short interspersed repeats in 3′ untranslated regions. BMC Bioinformatics. 2007;8:274. doi: 10.1186/1471-2105-8-274. [DOI] [PMC free article] [PubMed] [Google Scholar]