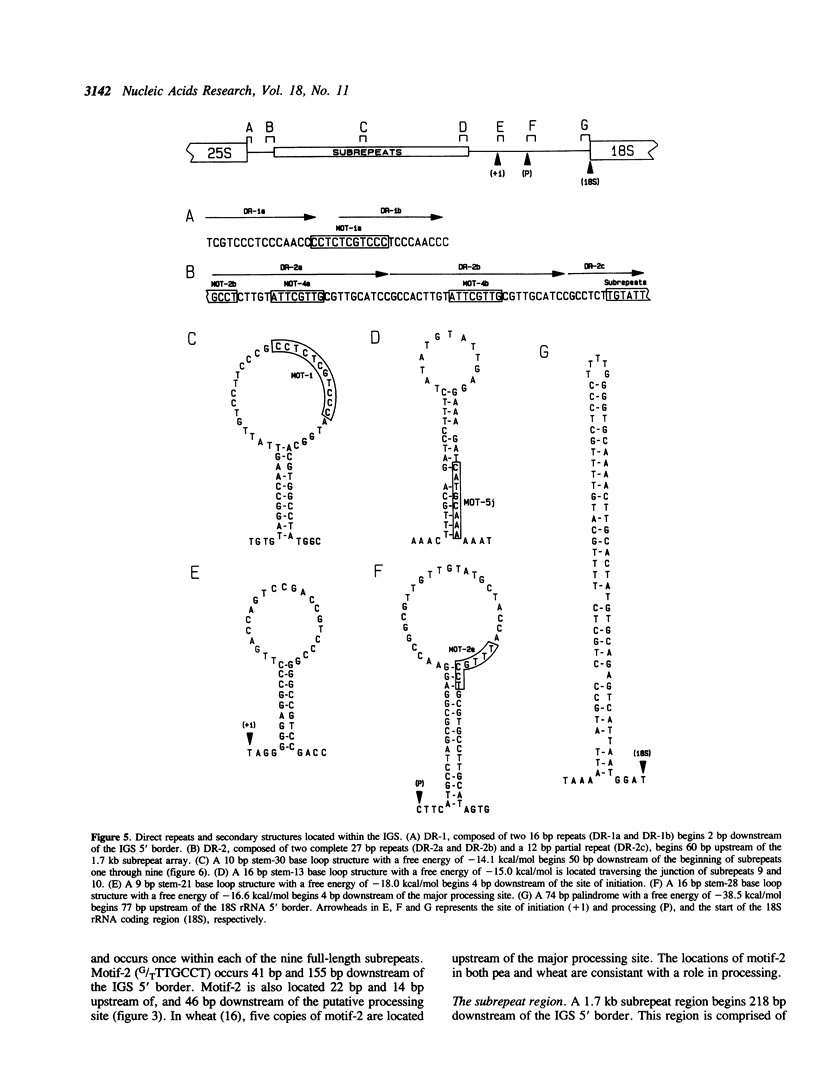
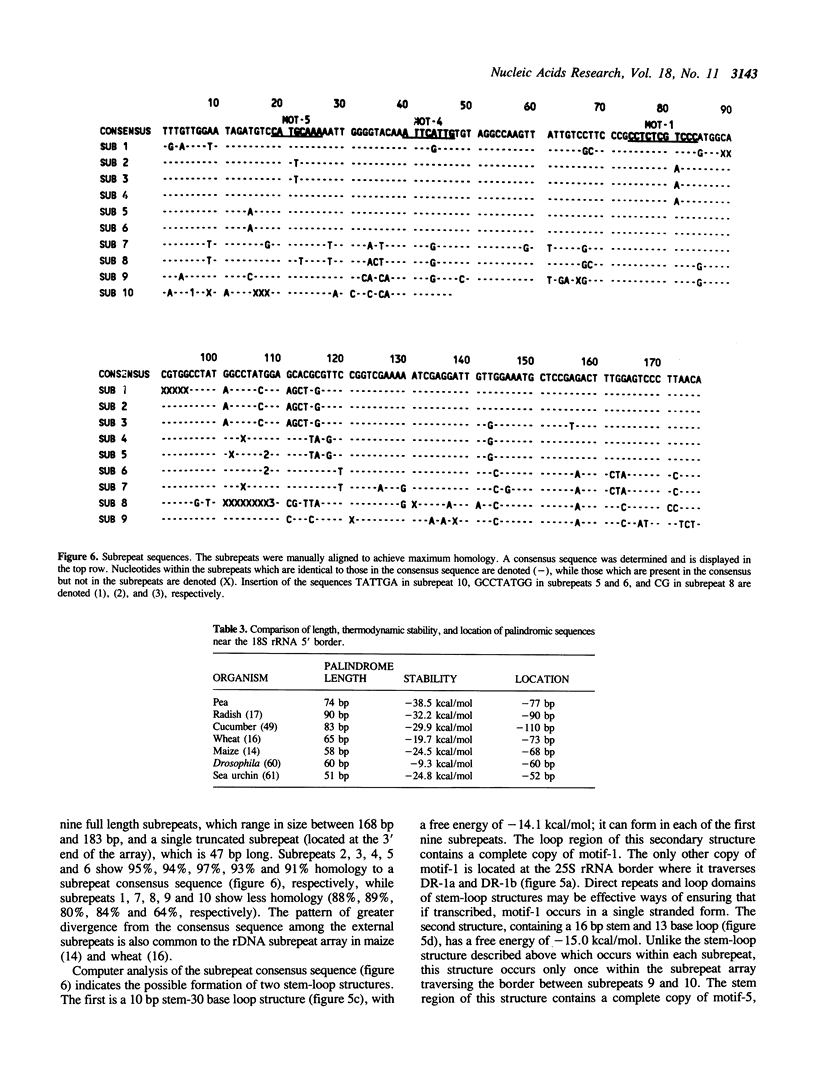
Abstract
The genomic clone, RRNpss1, representing the short ribosomal DNA length variant in Pisum sativum L. cv. Alaska, has been isolated and the 2859 bp intergenic spacer, along with the 25S rRNA 3' border and 18S rRNA 5' border, has been sequenced. The intergenic spacer contains nine tandem repeats, approximately 180 bp in length, which show greater than 80% sequence homology to each other. The RNA polymerase I transcription start site and a processing site, located 776 bp and 536 bp upstream of the 5' end of 18S rRNA, respectively, have been determined using S1 analysis. The region surrounding the +1 site shows strong homology between the positions -6 to +10 to the rDNA sites of initiation in radish, maize, and wheat. The sequence CATGCAAA is located 19 bp upstream of the site of initiation, and appears once within each subrepeat and twice more between the end of the subrepeat array and the site of initiation. A previously characterized HpaII site which shows developmental regulation of methylation is located 31 bp downstream of the site of initiation. Using RFLP linkage analysis, the short rDNA length variant of cv. Alaska is assigned to Chromosome 4 where it is genetically independent of the long rDNA length variant which is putatively assigned to Chromosome 7.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baerson S. R., Kaufman L. S. Increased rRNA gene activity during a specific window of early pea leaf development. Mol Cell Biol. 1990 Feb;10(2):842–845. doi: 10.1128/mcb.10.2.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. M., Platt T. Pol I transcription: which comes first, the end or the beginning? Cell. 1986 Dec 26;47(6):839–840. doi: 10.1016/0092-8674(86)90795-6. [DOI] [PubMed] [Google Scholar]
- Balzi E., Di Pietro A., Goffeau A., van Heerikhuizen H., Klootwijk J. The RNA polymerase I initiation site and the external transcribed spacer of the fission yeast Schizosaccharomyces pombe ribosomal RNA genes. Gene. 1985;39(2-3):165–172. doi: 10.1016/0378-1119(85)90310-5. [DOI] [PubMed] [Google Scholar]
- Barker R. F., Harberd N. P., Jarvis M. G., Flavell R. B. Structure and evolution of the intergenic region in a ribosomal DNA repeat unit of wheat. J Mol Biol. 1988 May 5;201(1):1–17. doi: 10.1016/0022-2836(88)90434-2. [DOI] [PubMed] [Google Scholar]
- Bateman E., Paule M. R. Promoter occlusion during ribosomal RNA transcription. Cell. 1988 Sep 23;54(7):985–992. doi: 10.1016/0092-8674(88)90113-4. [DOI] [PubMed] [Google Scholar]
- Cassidy B. G., Yang-Yen H. F., Rothblum L. I. Additional RNA polymerase I initiation site within the nontranscribed spacer region of the rat rRNA gene. Mol Cell Biol. 1987 Jul;7(7):2388–2396. doi: 10.1128/mcb.7.7.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy B. G., Yang-Yen H. F., Rothblum L. I. Transcriptional role for the nontranscribed spacer of rat ribosomal DNA. Mol Cell Biol. 1986 Aug;6(8):2766–2773. doi: 10.1128/mcb.6.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E. S., Dover G. A. Multiple Pol I initiation sequences in rDNA spacers of Drosophila melanogaster. Nucleic Acids Res. 1982 Nov 11;10(21):7017–7026. doi: 10.1093/nar/10.21.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcasso-Tremousaygue D., Grellet F., Panabieres F., Ananiev E. D., Delseny M. Structural and transcriptional characterization of the external spacer of a ribosomal RNA nuclear gene from a higher plant. Eur J Biochem. 1988 Mar 15;172(3):767–776. doi: 10.1111/j.1432-1033.1988.tb13956.x. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Financsek I., Mizumoto K., Mishima Y., Muramatsu M. Human ribosomal RNA gene: nucleotide sequence of the transcription initiation region and comparison of three mammalian genes. Proc Natl Acad Sci U S A. 1982 May;79(10):3092–3096. doi: 10.1073/pnas.79.10.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Financsek I., Mizumoto K., Muramatsu M. Nucleotide sequence of the transcription initiation region of a rat ribosomal RNA gene. Gene. 1982 May;18(2):115–122. doi: 10.1016/0378-1119(82)90109-3. [DOI] [PubMed] [Google Scholar]
- Ganal M., Torres R., Hemleben V. Complex structure of the ribosomal DNA spacer of Cucumis sativus (cucumber). Mol Gen Genet. 1988 Jun;212(3):548–554. doi: 10.1007/BF00330863. [DOI] [PubMed] [Google Scholar]
- Grimaldi G., Di Nocera P. P. Transient expression of Drosophila melanogaster rDNA promoter into cultured Drosophila cells. Nucleic Acids Res. 1986 Aug 26;14(16):6417–6432. doi: 10.1093/nar/14.16.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Kuhn A., Bartsch I., Rosenbauer H. A transcription terminator located upstream of the mouse rDNA initiation site affects rRNA synthesis. Cell. 1986 Dec 26;47(6):901–911. doi: 10.1016/0092-8674(86)90805-6. [DOI] [PubMed] [Google Scholar]
- Grummt I. Mapping of a mouse ribosomal DNA promoter by in vitro transcription. Nucleic Acids Res. 1981 Nov 25;9(22):6093–6102. doi: 10.1093/nar/9.22.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S., Sollner-Webb B. A transcriptional terminator is a novel element of the promoter of the mouse ribosomal RNA gene. Cell. 1986 Dec 26;47(6):891–900. doi: 10.1016/0092-8674(86)90804-4. [DOI] [PubMed] [Google Scholar]
- Ho K. C., Stafford D. W. Nucleotide sequence of the transcription initiation region for rRNA in the sea urchin Lytechinus variegatus. Gene. 1985;39(1):49–54. doi: 10.1016/0378-1119(85)90106-4. [DOI] [PubMed] [Google Scholar]
- Iida C. T., Kownin P., Paule M. R. Ribosomal RNA transcription: proteins and DNA sequences involved in preinitiation complex formation. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1668–1672. doi: 10.1073/pnas.82.6.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle J., Timmis J. N., Sinclair J. The Relationship between Satellite Deoxyribonucleic Acid, Ribosomal Ribonucleic Acid Gene Redundancy, and Genome Size in Plants. Plant Physiol. 1975 Mar;55(3):496–501. doi: 10.1104/pp.55.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. H., Learned R. M., Tjian R. Analysis of clustered point mutations in the human ribosomal RNA gene promoter by transient expression in vivo. Proc Natl Acad Sci U S A. 1988 Feb;85(3):669–673. doi: 10.1073/pnas.85.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L. S., Watson J. C., Thompson W. F. Light-regulated changes in DNase I hypersensitive sites in the rRNA genes of Pisum sativum. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1550–1554. doi: 10.1073/pnas.84.6.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto T., Nagamine M., Sasaki T., Takakusa N., Miwa T., Kominami R., Muramatsu M. Presence of a limited number of essential nucleotides in the promoter region of mouse ribosomal RNA gene. Nucleic Acids Res. 1985 May 24;13(10):3515–3532. doi: 10.1093/nar/13.10.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kownin P., Iida C. T., Brown-Shimer S., Paule M. R. The ribosomal RNA promoter of Acanthamoeba castellanii determined by transcription in a cell-free system. Nucleic Acids Res. 1985 Sep 11;13(17):6237–6248. doi: 10.1093/nar/13.17.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Grummt I. A novel promoter in the mouse rDNA spacer is active in vivo and in vitro. EMBO J. 1987 Nov;6(11):3487–3492. doi: 10.1002/j.1460-2075.1987.tb02673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Enhancer-like properties of the 60/81 bp elements in the ribosomal gene spacer of Xenopus laevis. Cell. 1984 May;37(1):285–289. doi: 10.1016/0092-8674(84)90324-6. [DOI] [PubMed] [Google Scholar]
- Learned R. M., Smale S. T., Haltiner M. M., Tjian R. Regulation of human ribosomal RNA transcription. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3558–3562. doi: 10.1073/pnas.80.12.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L., Zengel J. M. Ribosomal genes in Escherichia coli. Annu Rev Genet. 1986;20:297–326. doi: 10.1146/annurev.ge.20.120186.001501. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Loenen W. A., Brammar W. J. A bacteriophage lambda vector for cloning large DNA fragments made with several restriction enzymes. Gene. 1980 Aug;10(3):249–259. doi: 10.1016/0378-1119(80)90054-2. [DOI] [PubMed] [Google Scholar]
- Long E. O., Rebbert M. L., Dawid I. B. Nucleotide sequence of the initiation site for ribosomal RNA transcription in Drosophila melanogaster: comparison of genes with and without insertions. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1513–1517. doi: 10.1073/pnas.78.3.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen M. D., Hunter B., Phillips R. L., Rubenstein I. The structure of the maize ribosomal DNA spacer region. Nucleic Acids Res. 1986 Jun 25;14(12):4953–4968. doi: 10.1093/nar/14.12.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A termination site for Xenopus RNA polymerase I also acts as an element of an adjacent promoter. Cell. 1986 Dec 26;47(6):913–920. doi: 10.1016/0092-8674(86)90806-8. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Tower J., Sollner-Webb B. A complex control region of the mouse rRNA gene directs accurate initiation by RNA polymerase I. Mol Cell Biol. 1985 Mar;5(3):554–562. doi: 10.1128/mcb.5.3.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. Transcription of cloned Xenopus laevis ribosomal DNA microinjected into Xenopus oocytes, and the identification of an RNA polymerase I promoter. Cell. 1982 Oct;30(3):835–842. doi: 10.1016/0092-8674(82)90288-4. [DOI] [PubMed] [Google Scholar]
- Murray M. G., Thompson W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980 Oct 10;8(19):4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard C. S., Reeder R. H. Sequence elements essential for function of the Xenopus laevis ribosomal DNA enhancers. Mol Cell Biol. 1988 Oct;8(10):4282–4288. doi: 10.1128/mcb.8.10.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polans N. O., Weeden N. F., Thompson W. F. Inheritance, organization, and mapping of rbcS and cab multigene families in pea. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5083–5087. doi: 10.1073/pnas.82.15.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R. H. Enhancers and ribosomal gene spacers. Cell. 1984 Sep;38(2):349–351. doi: 10.1016/0092-8674(84)90489-6. [DOI] [PubMed] [Google Scholar]
- Reeder R. H., Roan J. G., Dunaway M. Spacer regulation of Xenopus ribosomal gene transcription: competition in oocytes. Cell. 1983 Dec;35(2 Pt 1):449–456. doi: 10.1016/0092-8674(83)90178-2. [DOI] [PubMed] [Google Scholar]
- Saiga H., Mizumoto K., Matsui T., Higashinakagawa T. Determination of the transcription initiation site of Tetrahymena pyriformis rDNA using in vitro capping of 35S pre-rRNA. Nucleic Acids Res. 1982 Jul 24;10(14):4223–4236. doi: 10.1093/nar/10.14.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaak J., Mao J., Söll D. The 5.8S RNA gene sequence and the ribosomal repeat of Schizosaccharomyces pombe. Nucleic Acids Res. 1982 May 11;10(9):2851–2864. doi: 10.1093/nar/10.9.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A., La Volpe A., Boncinelli E. Nucleotide sequence of a complete ribosomal spacer of D. melanogaster. Nucleic Acids Res. 1985 Feb 25;13(4):1089–1101. doi: 10.1093/nar/13.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner J. A., Ohrlein A., Grummt I. In vitro mutagenesis and transcriptional analysis of a mouse ribosomal promoter element. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2137–2141. doi: 10.1073/pnas.81.7.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Wilkinson J. A., Roan J., Reeder R. H. Nested control regions promote Xenopus ribosomal RNA synthesis by RNA polymerase I. Cell. 1983 Nov;35(1):199–206. doi: 10.1016/0092-8674(83)90222-2. [DOI] [PubMed] [Google Scholar]
- Suiter K. A., Wendel J. F., Case J. S. LINKAGE-1: a PASCAL computer program for the detection and analysis of genetic linkage. J Hered. 1983 May-Jun;74(3):203–204. doi: 10.1093/oxfordjournals.jhered.a109766. [DOI] [PubMed] [Google Scholar]
- Toloczyki C., Feix G. Occurrence of 9 homologous repeat units in the external spacer region of a nuclear maize rRNA gene unit. Nucleic Acids Res. 1986 Jun 25;14(12):4969–4986. doi: 10.1093/nar/14.12.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler B. M., Giles N. H. Structure of a Neurospora RNA polymerase I promoter defined by transcription in vitro with homologous extracts. Nucleic Acids Res. 1985 Jun 25;13(12):4311–4332. doi: 10.1093/nar/13.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano Y., Kominami R., Mishima Y., Muramatsu M. The nucleotide sequence of the putative transcription initiation site of a cloned ribosomal RNA gene of the mouse. Nucleic Acids Res. 1980 Dec 20;8(24):6043–6058. doi: 10.1093/nar/8.24.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. C., Kaufman L. S., Thompson W. F. Developmental regulation of cytosine methylation in the nuclear ribosomal RNA genes of Pisum sativum. J Mol Biol. 1987 Jan 5;193(1):15–26. doi: 10.1016/0022-2836(87)90622-x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]