Abstract
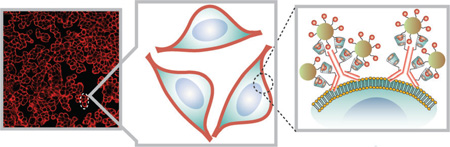
Be my guest: A supramolecular host–guest interaction is utilized for highly efficient bioorthogonal labeling of cellular targets. Antibodies labeled with a cyclodextrin host molecule bind to adamantane-labeled magnetofluorescent nanoparticles (see picture) and provide an amplifiable strategy for biomarker detection that can be adapted to different diagnostic techniques such as molecular profiling or magnetic cell sorting.
Keywords: antibodies, bioorthogonal labeling, host–guest systems, nanoparticles, supramolecular chemistry
Efficient nanoparticle (NP) labeling of cellular and other molecular targets is of fundamental importance for various biodiagnostic assays.[1–4] The ability to target nanoparticles to specific disease entities also has therapeutic and imaging implications.[5] To date, most labeling methods rely on the use of direct NP–ligand conjugates, wherein affinity ligands (e.g. antibodies, peptides) are covalently attached to the surface of NPs. Such a conjugation strategy, however, often requires time-consuming optimization processes to maximize the affinity, the ligand-to-NP ratio, and the colloidal stability of each new construct.[6] To overcome these issues, the use of synthetic reaction partners that are functionally orthogonal to biological systems has emerged as an appealing labeling platform both in vitro and in vivo.[7]
Supramolecular chemistry uses noncovalent interactions for the assembly of larger functional structures.[8] Noncovalent supramolecular interactions, such as those observed in host–guest binding pairs, allow for associations between recognition motifs that are specific and bioorthogonal and that do not require an additional catalyst.[9] Because the association between components of a noncovalent binding pair is typically diffusion-controlled, the reaction rate is much faster (ca. 109 m−1 s−1) than those of bioorthogonal covalent reactions (1–104 m−1 s−1).[10] Complexes formed through host–guest interactions are stable in biological systems and have thus been applied to many different biological applications.[11–14] The fast kinetics, specificity, stability, and bioorthogonal nature of these host–guest interactions prompted us to investigate this platform for cellular labeling with NPs. In particular, we hypothesized that this labeling strategy would enable us to design assay methods with: 1) stable and biocompatible components, 2) fast labeling for shorter assay time, 3) high signal-to-noise ratios, and 4) the capacity for signal amplification to detect scarce targets.
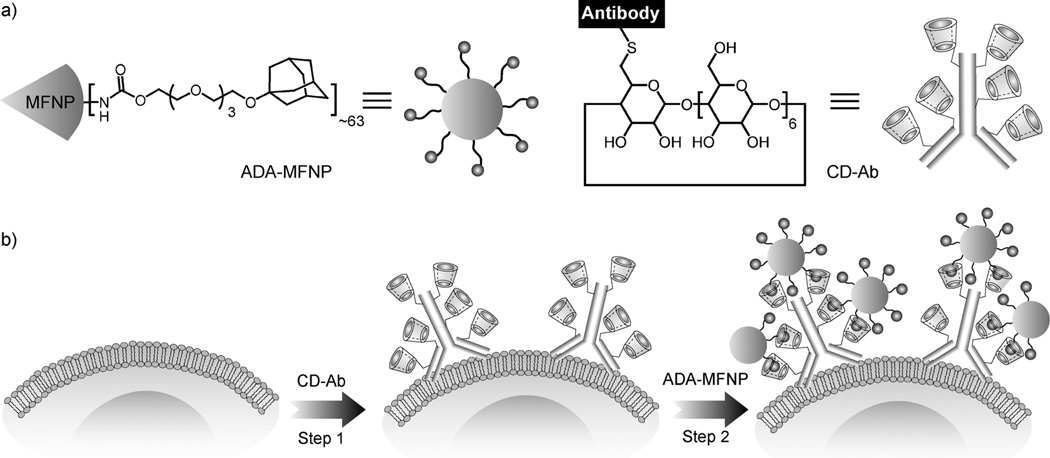
Herein, we present a modular labeling strategy, in which host–guest interactions between β-cyclodextrin (CD) and adamantane (ADA) are used as the coupling mechanism between NPs and antibodies (Scheme 1). This approach employs a two-step NP-labeling strategy, where CD-modified antibodies (CD–Abs) are used for primary target binding and subsequent noncovalent coupling with ADA-modified NPs. Using this approach, we were able to consistently achieve higher labeling efficiency than with either direct immunoconjugates or the noncovalent avidin–biotin system. Furthermore, we show that this supramolecular labeling strategy is easily adaptable to a variety of biodiagnostic assays, including molecular profiling, immunostaining, and magnetic cell sorting.
Scheme 1.
a) Structure of the ADA-functionalized magnetofluorescent nanoparticles (ADA–MFNPs) and CD-modified antibodies (CD–Abs). b) Schematic depiction of the supramolecular labeling strategy. CD–Abs against the biomarker of interest were initially targeted to cells and then used as scaffolds for coupling ADA–MFNPs in live cells by host–guest complexation between CD and ADA.
We used MFNPs, which consist of an iron oxide core and a dextran shell modified with fluorochromes (VivoTag 680; VT680), as labeling agents.[15] ADA-functionalized MFNPs (ADA–MFNPs) were prepared by conjugating ADA poly-(ethylene glycol) succinimidyl ester to amine-modified MFNPs. Mono-thio-β-CD was anchored to maleimide-modified antibodies through Michael addition. CD–Abs were characterized using mass spectrometry (Figure S1 in the Supporting Information). Secondary antibody labeling was used to verify that the CD modifications did not affect the primary antibody binding to the cell surface markers (Figure S2 in the Supporting Information).
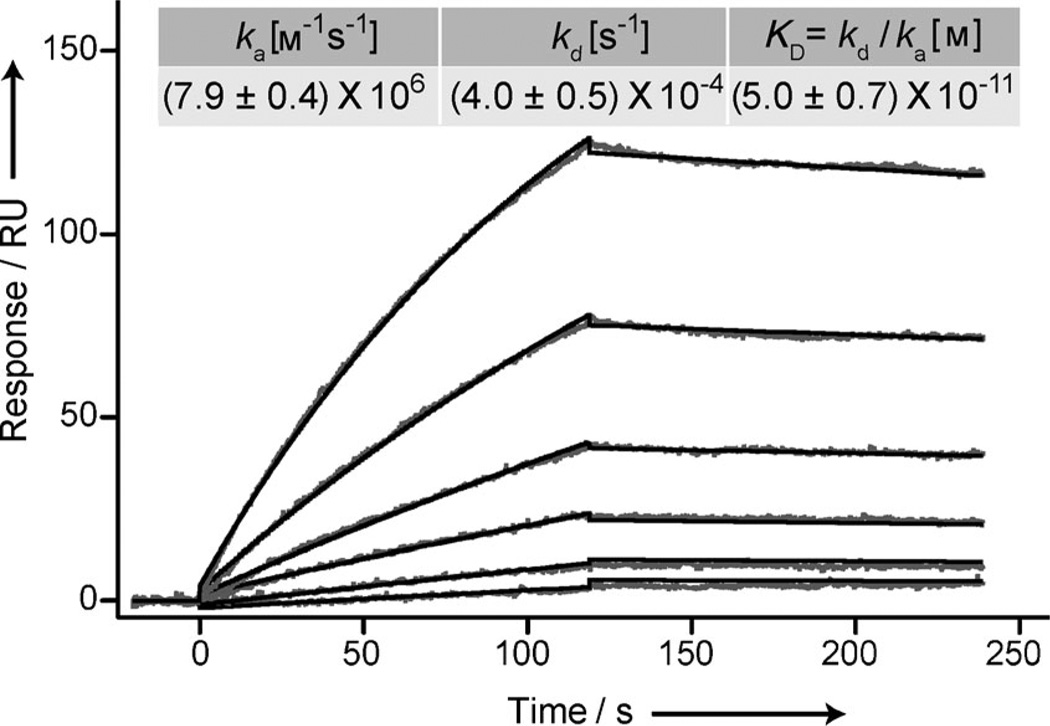
The affinity and binding kinetics between ADA–MFNPs and CD were characterized using surface plasmon resonance (SPR; Figure 1). We observed an exceptionally high association rate (ka = (7.9±0.4) × 106 m−1 s−1) as well as a slow off-rate (kd = (4.0±0.5) × 10−4 s−1). The observed high ka value is attributed to the fast and diffusion-controlled association between the noncovalent binding pair, whereas the slow off-rate and the resulting high binding affinity (KD = (5.0±0.7) × 10−11 m) indicate multivalent binding avidity of the NPs.[10, 16] The low KD value can also be attributed to the decrease in enthalpy owing to multivalent binding.[10] Furthermore, the CD–ADA complex maintained superb stability under harsh buffer conditions (Figure S3 in the Supporting Information).
Figure 1.
SPR sensorgram to determine the binding kinetics of ADA–MFNP to CD immobilized on a gold-coated surface (see the Supporting Information for details). An increase in the SPR signal after ADA–MFNP injection (t = 0) into the flow-through device confirmed NP binding. ADA–MFNP binding was characterized by running multiple cycles and measuring binding at varying concentrations during the sample injection step (1:2 dilution series: 500, 250, 125, 62.5, 31.2, and 15.6 ng MFNP mL−1). The resulting binding curves were double-reference subtracted and fitted to a one-to-one binding model. The rate constants listed in the inset are calculated from five separate measurements.
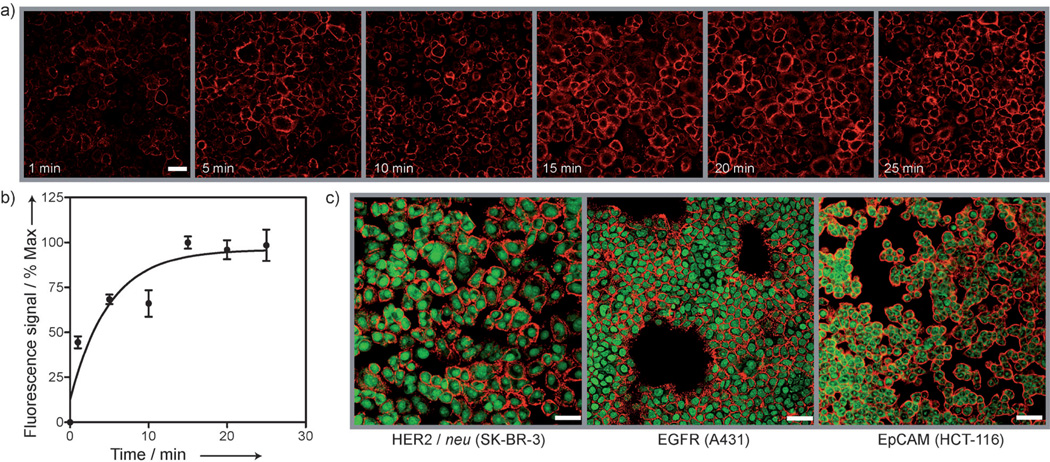
We evaluated the effectiveness of the CD–ADA method for cellular labeling. Live mammalian SK-BR-3 cells were targeted through a two-step labeling method (Scheme 1), wherein cells were first incubated with CD–Abs (CD–HER2/neu) for primary target binding, and subsequently coupled to ADA–MFNPs. Time-lapse fluorescence images taken during the ADA–MFNP incubation (Figure 2a) confirmed fast particle binding with the fluorescence signal reaching saturation in less than 15 min (Figure 2b). The CD–ADA method was then applied to label various surface receptors in live cells. SK-BR-3, A431, and HCT-116 cells were labeled with ADA–MFNPs by targeting their overexpressed markers, HER2/neu, EGFR (epidermal growth factor receptor), and EpCAM (epithelial cell adhesion molecule), respectively. A strong fluorescence signal was detected on the targeted cell surface (Figure 2 c), thus indicating NP localization to membrane-associated biomarkers. Cross-section transmission electron microscopy (TEM) further confirmed the presence of the NPs on the cell membrane (Figure S4 in the Supporting Information). Notably, the CD–ADA labeling was highly selective with very low nonspecific binding; a negligible fluorescence signal was detected when primary targeting with CD–Abs was omitted (Figure S5 in the Supporting Information). In addition, the CD–ADA system retained its stability and reactivity when cell labeling was performed in different buffers, in biological media, and at different temperatures (Figures S6 and S7 in the Supporting Information). Negligible release of the ADA–MFNP labels was observed from the competition experiment with nonfluorescent ADA–NP or adamantyl amine, presumably owing to the slow off-rate as a result of multivalent binding avidity of the ADA–MFNPs.
Figure 2.
a) Kinetics of labeling in live cells. Images show the increased intensity of the fluorescence signal (VT680 fluorescence from ADA–MFNP) from the cell surface as a function of nanoparticle incubation time. b) Fluorescence intensity, plotted against nanoparticle incubation time, reached saturation within 15 min. c) Confocal microscopy images of SK-BR-3, A431, and HCT-116 cells targeted with CD-modified antibodies against HER2/neu, EGFR, and EpCAM, respectively, and coupled with ADA–MFNP (red fluorescence). Cell nuclei were stained using TO-PRO-1 (green). Scale bars 50 µm.
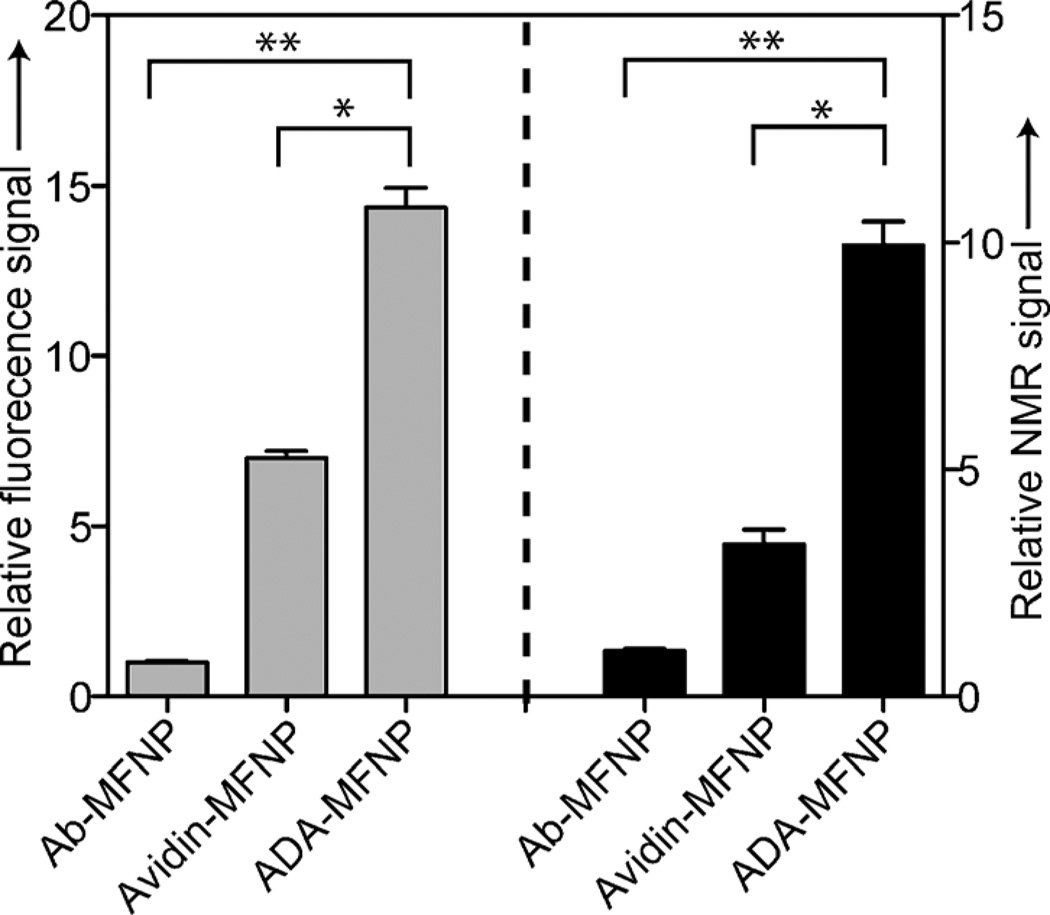
We compared the performance of CD–ADA labeling with those of other conventional methods: direct labeling with antibody-MFNP conjugates and two-step noncovalent labeling using the avidin–biotin system. The CD–ADA supramolecular method consistently showed higher NP loading to the targeted cells. For example, the relative increase in fluorescence signal was 15-fold compared to the direct labeling method (Figure 3). More interestingly, the CD–ADA method showed two times higher fluorescence intensity than the avidin–biotin method (Figure 3), although both methods had a similar antibody loading (CD–Ab or biotin–Ab) on cells (Figure S8 in the Supporting Information). Magnetic detection with a miniaturized nuclear magnetic resonance (µNMR) system[17] showed the same trend, which is in agreement with the fluorescence measurement. Such higher signal levels obtained with the CD–ADA method can be attributed to the high valencies and small sizes of the CD–ADA binding partners, which promote the attachment of multiple NPs to a single biomarker. Conversely, in the direct labeling, at most one NP can be bound per marker; in the avidin–biotin method, the large footprint of the avidin molecule (molecular weight ca. 67 kDa) considerably lowers the avidin valency per nanoparticle, and because of its large size, the avidin sterically masks adjacent biotin sites, hindering multiple NP binding to a single biomarker.
Figure 3.
Performance in cell labeling experiments (SK-BR-3 cells) of the CD–ADA method (ADA–MFNP) relative to the direct labeling technique using nanoparticle–antibody conjugate (Ab-MFNP) and the avidin–biotin system (Avidin-MFNP). The analytical signal (fluorescence and magnetic) obtained with the different labeling methods was normalized against the signal from the direct labeling method and expressed as relative signal intensity. *P < 0.05 and **P < 0.01.
Amplifying analytical signal is a crucial task in detecting rare cells or scant biomarkers. Successive labeling with the CD–ADA system is a convenient strategy for signal amplification. Figure S9a in the Supporting Information shows an amplification method that is based on the alternating attachment of VT680-conjugated ADA–MFNPs and fluorescein isothiocyanate (FITC)-conjugated CD–MFNPs to cell surfaces. Confocal microscopy showed intense, co-localized fluorescence signals from both channels, thus confirming the amplification of the specific biomarkers (Figure S9b in the Supporting Information). Low nonspecific binding of CD–MFNPs and dose-dependent labeling of ADA–MFNPs with CD–MFNPs were observed from this system (Figure S10 in the Supporting Information). Further consecutive rounds of amplification indeed led to the increase of analytical signal, as measured by both flow cytometry and µNMR (Figure S11 in the Supporting Information).
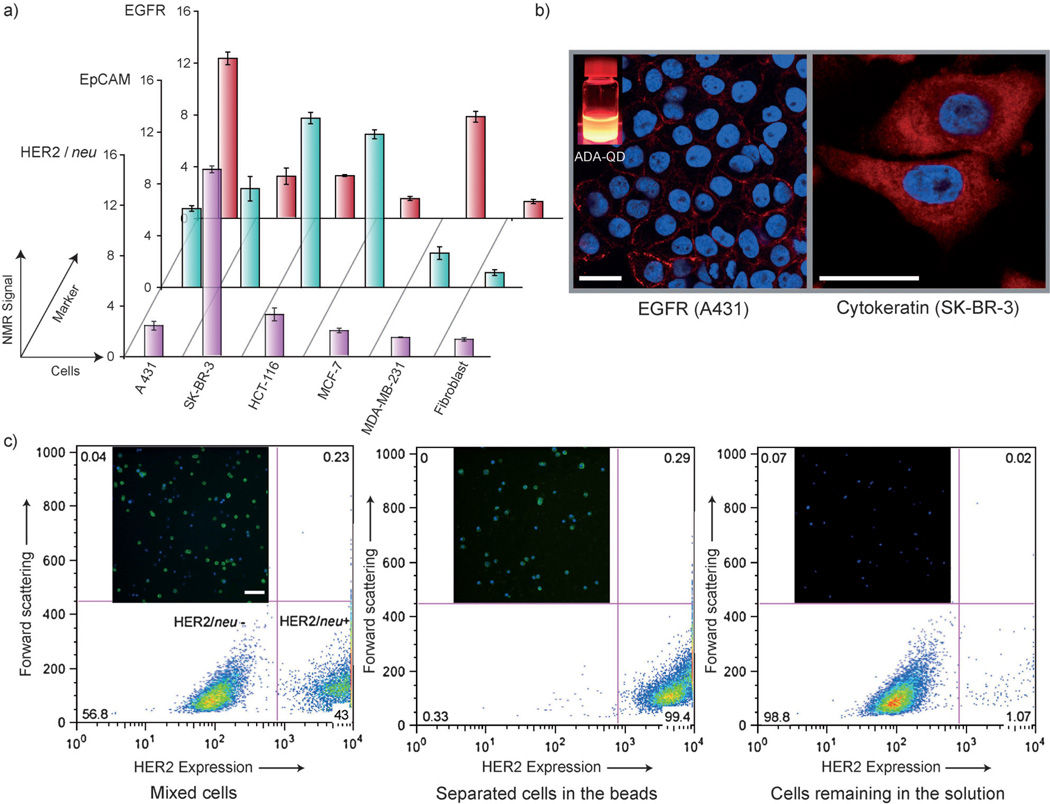
Cell-based assays require specific and efficient labeling of cellular biomarkers, which prompted us to apply this supramolecular method for various biodiagnostic configurations. We used the CD–ADA method for the rapid detection and profiling of cancer cells. A panel of human cancer cell lines (A431, SK-BR-3, HCT-116, MCF-7, and MDA-MB-231) and a control fibroblast cell line were labeled with CD–Abs against relevant cancer markers and subsequently tagged with ADA–MFNPs. Using the µNMR system, we measured the transverse relaxation rates (R2) of these samples to obtain the molecular expression profiles of these cells (Figure 4a). With the efficient magnetic labeling achieved with the CD–ADA system, reliable µNMR assays could be performed even with a small number of cells (ca. 200 cells). The results also showed direct correlation with those obtained by flow cytometry (Figure S12 in the Supporting Information), which required a much higher number of cells (ca. 10000 cells). To apply the CD–ADA labeling in more clinically relevant specimens, artificial clinical samples were prepared by spiking cancer cells (SK-BR-3) into whole blood. After lysing the red blood cells, samples were targeted with CD–HER2/neu and then labeled with ADA–MFNPs. Subsequent µNMR measurement detected a significantly higher R2 relaxation rate in blood samples containing cancer cells, thereby demonstrating the robustness of the supramolecular labeling technology (Figure S13 in the Supporting Information). Utility of the CD–ADA platform in multiplexed analysis was demonstrated by labeling EpCAM using CD–ADA interactions and by labeling HER2/neu receptors using avidin–biotin interactions on the same SK-BR-3 cells (Figure S14 in the Supporting Information).
Figure 4.
a) Rapid profiling of biomarkers using µNMR. The expression level of the target marker was represented as NMR signal ratio Δr2mAb/Δr2ø, where Δr2mAb and Δr2ø are cellular relaxivities for marker-specific and control MFNPs, respectively. b) ADA-linked QDs were used to fluorescently label cell surface marker EGFR in live cells (A431 cells) and intracellular marker cytokeratin in semipermeabilized cells (SK-BR-3 cells). Red fluorescence ADA–QD; blue fluorescence DAPI nuclear stain. Scale bars 25 µm. c) Cell sorting using ADA-functionalized magnetic beads. FITC-conjugated secondary antibody was used to distinguish the HER2-positive cells from the HER2-negative cells. Inset fluorescence micrographs show the clean magnetic separation of HER2-positive (green) and HER2-negative cells. Both cell types were stained with DAPI (blue). Scale bar 100 µm. The numbers in the four quadrants of the histogram correspond to the percentage of cells with the respective characteristics.
We next examined whether the CD–ADA method could be more generally extended to other substrate material. Two representative platforms, quantum dots (QDs) and magnetic beads, were chosen for cellular imaging and cell-sorting operations, respectively. For the QD-based immunostaining, we targeted an extracellular marker (EGFR) and an intracellular marker (cytokeratin) and subsequently applied ADA-decorated QDs. As for MFNPs, the supramolecular QD labeling was highly efficient with minimal nonspecific interactions; confocal microscopy showed bright staining on targeted cells (Figure 4 b), whereas control samples without primary CD–Abs targeting showed negligible fluorescence (Figure S15 in the Supporting Information). The magnetic sorting with the CD–ADA system was demonstrated by isolating HER2-positive SK-BR-3 cells from a heterogeneous cell mixture. The mixture was first incubated with CD–Abs, and subsequently the ADA-conjugated magnetic beads were added. As summarized in Figure 4c, the initial suspension contained two distinct cell populations as properly labeled by the secondary antibody conjugate. Using the magnetic sorting process we could effectively separate the HER2-positive cells from the HER2-negative cells.
In summary, we demonstrate the utility of a host–guest supramolecular system for modular and efficient NP targeting to cellular biomarkers. Because of the small size of the components and their steric advantages, as well as the amplifying and multiplexing capabilities of the method, we could achieve considerably better binding to cells than with commonly used methods. This coupling platform is versatile and can be easily adapted to various diagnostic techniques. We are currently exploring some of these applications involving cell-based therapies[18] and programming the assembly of cells to create microtissues.[19]
Supplementary Material
Footnotes
We thank M. Fernandez-Suarez, R. Mazitschek, T. Reiner, K. S. Yang, and K. Tran for helpful discussions; I. Bagayev and E. Tiglao for assistance in cell experiments; N. Sergeyev for providing the cross-linked iron oxide particles; Y. Fisher-Jeffes for reviewing the manuscript. This work was supported in part by National Institutes of Health Grants R01-EB0044626, R01-EB010011, T32 grant T32A79443, P50 grant P50A86355, CCNE contract U54A151884, and TPEN contract HHSN268201000044C.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201105670.
Contributor Information
Sarit S. Agasti, Center for Systems Biology, Massachusetts General Hospital/Harvard Medical School, 185 Cambridge St., Boston, MA 02114 (USA).
Monty Liong, Center for Systems Biology, Massachusetts General Hospital/Harvard Medical School, 185 Cambridge St., Boston, MA 02114 (USA).
Carlos Tassa, Center for Systems Biology, Massachusetts General Hospital/Harvard Medical School, 185 Cambridge St., Boston, MA 02114 (USA).
Hyun Jung Chung, Center for Systems Biology, Massachusetts General Hospital/Harvard Medical School, 185 Cambridge St., Boston, MA 02114 (USA).
Stanley Y. Shaw, Center for Systems Biology, Massachusetts General Hospital/Harvard Medical School, 185 Cambridge St., Boston, MA 02114 (USA)
Hakho Lee, Email: hlee@mgh.harvard.edu, Center for Systems Biology, Massachusetts General Hospital/Harvard Medical School, 185 Cambridge St., Boston, MA 02114 (USA).
Ralph Weissleder, Email: rweissleder@mgh.harvard.edu, Center for Systems Biology, Massachusetts General Hospital/Harvard Medical School, 185 Cambridge St., Boston, MA 02114 (USA); Department of Systems Biology, Harvard Medical School, 200 Longwood Ave., Alpert 536, Boston, MA 02115 (USA).
References
- 1.Sha MY, Xu HX, Natan MJ, Cromer R. J. Am. Chem. Soc. 2008;130:17214–17215. doi: 10.1021/ja804494m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu XY, Liu HJ, Liu JQ, Haley KN, Treadway JA, Larson JP, Ge NF, Peale F, Bruchez MP. Nat. Biotechnol. 2003;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 3.Galanzha EI, Shashkov EV, Kelly T, Kim JW, Yang LL, Zharov VP. Nat. Nanotechnol. 2009;4:855–860. doi: 10.1038/nnano.2009.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Gaster RS, Hall DA, Nielsen CH, Osterfeld SJ, Yu H, Mach KE, Wilson RJ, Murmann B, Liao JC, Gambhir SS. Nat. Med. 2009;15:1327–1332. doi: 10.1038/nm.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lee H, Yoon TJ, Figueiredo JL, Swirski FK, Weissleder R. Proc. Natl. Acad. Sci.USA. 2009;106:12459–12464. doi: 10.1073/pnas.0902365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Ferrari M. Nat. Rev. Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]; b) Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 6.Veiseh O, Gunn JW, Zhang M. Adv. Drug Delivery Rev. 2010;62:284–304. doi: 10.1016/j.addr.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Sletten EM, Bertozzi CR. Angew. Chem. 2009;121:7108–7133. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wu C, Jin Y, Schneider T, Burnham DR, Smith PB, Chiu DT. Angew. Chem. 2010;122:9626–9630. doi: 10.1002/anie.201004260. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2010;49:9436–9440. doi: 10.1002/anie.201004260. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Rossin R, Renart Verkerk P, van den Bosch SM, Vulders R, Verel I, Lub J, Robillard MS. Angew. Chem. 2010;122:3447–3450. doi: 10.1002/anie.200906294. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2010;49:3375–3378. doi: 10.1002/anie.200906294. [DOI] [PubMed] [Google Scholar]; d) Haun JB, Devaraj NK, Hilderbrand SA, Lee H, Weissleder R. Nat. Nanotechnol. 2010;5:660–665. doi: 10.1038/nnano.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Wang S, Su H, Chen K-J, Armijo AL, Lin W-Y, Wang Y, Sun J, Kamei K-I, Czernin J, Radu CG, Tseng H-R. Angew. Chem. 2009;121:4408–4412. [Google Scholar]; Angew. Chem. Int. Ed. 2009;48:4344–4348. doi: 10.1002/anie.200900063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Uhlenheuer DA, Petkau K, Brunsveld L. Chem. Soc. Rev. 2010;39:2817–2826. doi: 10.1039/b820283b. [DOI] [PubMed] [Google Scholar]; b) Lagona J, Mukhopadhyay P, Chakrabarti S, Isaacs L. Angew. Chem. 2005;117:4922–4949. doi: 10.1002/anie.200460675. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2005;44:4844–4870. doi: 10.1002/anie.200460675. [DOI] [PubMed] [Google Scholar]
- 10.a) Gomez-Casado A, Dam H, Yilmaz MD, Florea D, Jonkheijm P, Huskens J. J. Am. Chem. Soc. 2011;133:10849–10857. doi: 10.1021/ja2016125. [DOI] [PubMed] [Google Scholar]; b) Mulder A, Auletta T, Sartori A, Del Ciotto S, Casnati A, Ungaro R, Huskens J, Reinhoudt DN. J. Am. Chem. Soc. 2004;126:6627–6636. doi: 10.1021/ja0317168. [DOI] [PubMed] [Google Scholar]; c) Mulder A, Huskens J, Reinhoudt DN. Org. Biomol. Chem. 2004;2:3409–3424. doi: 10.1039/b413971b. [DOI] [PubMed] [Google Scholar]
- 11.Hennig A, Bakirci H, Nau WM. Nat. Methods. 2007;4:629–632. doi: 10.1038/nmeth1064. [DOI] [PubMed] [Google Scholar]
- 12.a) Escalante M, Zhao Y, Ludden MJ, Vermeij R, Olsen JD, Berenschot E, Hunter CN, Huskens J, Subramaniam V, Otto C. J. Am. Chem. Soc. 2008;130:8892–8893. doi: 10.1021/ja802843m. [DOI] [PubMed] [Google Scholar]; b) Hwang I, Baek K, Jung M, Kim Y, Park KM, Lee DW, Selvapalam N, Kim K. J. Am. Chem. Soc. 2007;129:4170–4171. doi: 10.1021/ja071130b. [DOI] [PubMed] [Google Scholar]
- 13.Lee DW, et al. Nat. Chem. 2011;3:154–159. doi: 10.1038/nchem.928. see the Supporting Information. [DOI] [PubMed] [Google Scholar]
- 14.a) Kim C, Agasti SS, Zhu ZJ, Isaacs L, Rotello VM. Nat. Chem. 2010;2:962–966. doi: 10.1038/nchem.858. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Meng HA, Xue M, Xia TA, Zhao YL, Tamanoi F, Stoddart JF, Zink JI, Nel AE. J. Am. Chem. Soc. 2010;132:12690–12697. doi: 10.1021/ja104501a. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhao YL, Li Z, Kabehie S, Botros YY, Stoddart JF, Zink JI. J. Am. Chem. Soc. 2010;132:13016–13025. doi: 10.1021/ja105371u. [DOI] [PubMed] [Google Scholar]; d) Patel K, Angelos S, Dichtel WR, Coskun A, Yang YW, Zink JI, Stoddart JF. J. Am. Chem. Soc. 2008;130:2382–2383. doi: 10.1021/ja0772086. [DOI] [PubMed] [Google Scholar]; e) Park C, Kim H, Kim S, Kim C. J. Am. Chem. Soc. 2009;131:16614–16615. doi: 10.1021/ja9061085. [DOI] [PubMed] [Google Scholar]
- 15.Pittet MJ, Swirski FK, Reynolds F, Josephson L, Weissleder R. Nat. Protoc. 2006;1:73–79. doi: 10.1038/nprot.2006.11. [DOI] [PubMed] [Google Scholar]
- 16.a) Tassa C, Duffner JL, Lewis TA, Weissleder R, Schreiber SL, Koehler AN, Shaw SY. Bioconjugate Chem. 2010;21:14–19. doi: 10.1021/bc900438a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hayashida O, Sato D. Polym. Adv. Technol. 2010;21:96–99. [Google Scholar]; c) Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JRJ, Banaszak Holl MM. Chem. Biol. 2007;14:107–115. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]; d) Munoz EM, Correa J, Fernandez-Megia E, Riguera R. J. Am. Chem. Soc. 2009;131:17765–17767. doi: 10.1021/ja9074826. [DOI] [PubMed] [Google Scholar]
- 17.a) Lee H, Sun E, Ham D, Weissleder R. Nat. Med. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Haun JB, Castro CM, Wang R, Peterson VM, Marinelli BS, Lee H, Weissleder R. Sci. Transl. Med. 2011;3 doi: 10.1126/scitranslmed.3002048. 71ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao W, Teo GSL, Kumar N, Karp JM. Mater. Today. 2010;13:14–21. doi: 10.1016/S1369-7021(10)70056-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Dutta D, Pulsipher A, Luo W, Yousaf MN. J. Am. Chem. Soc. 2011;133:8704–8713. doi: 10.1021/ja2022569. [DOI] [PubMed] [Google Scholar]; b) Gartner ZJ, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2009;106:4606–4610. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.