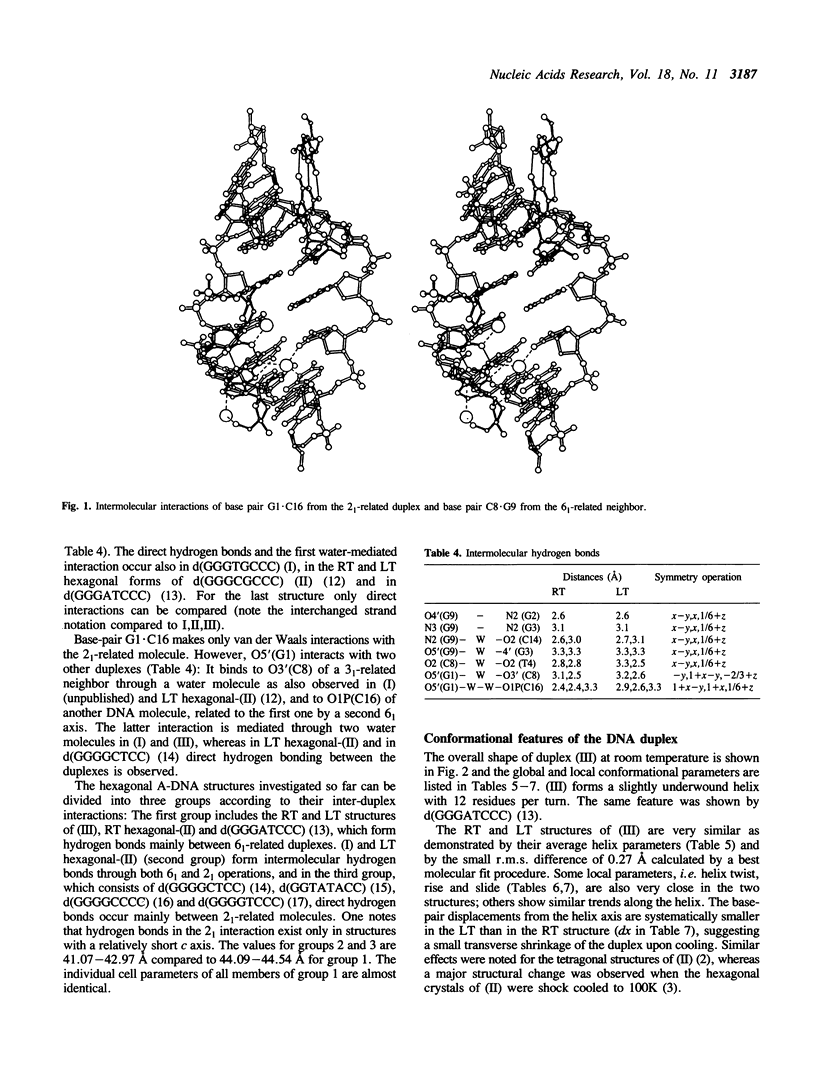
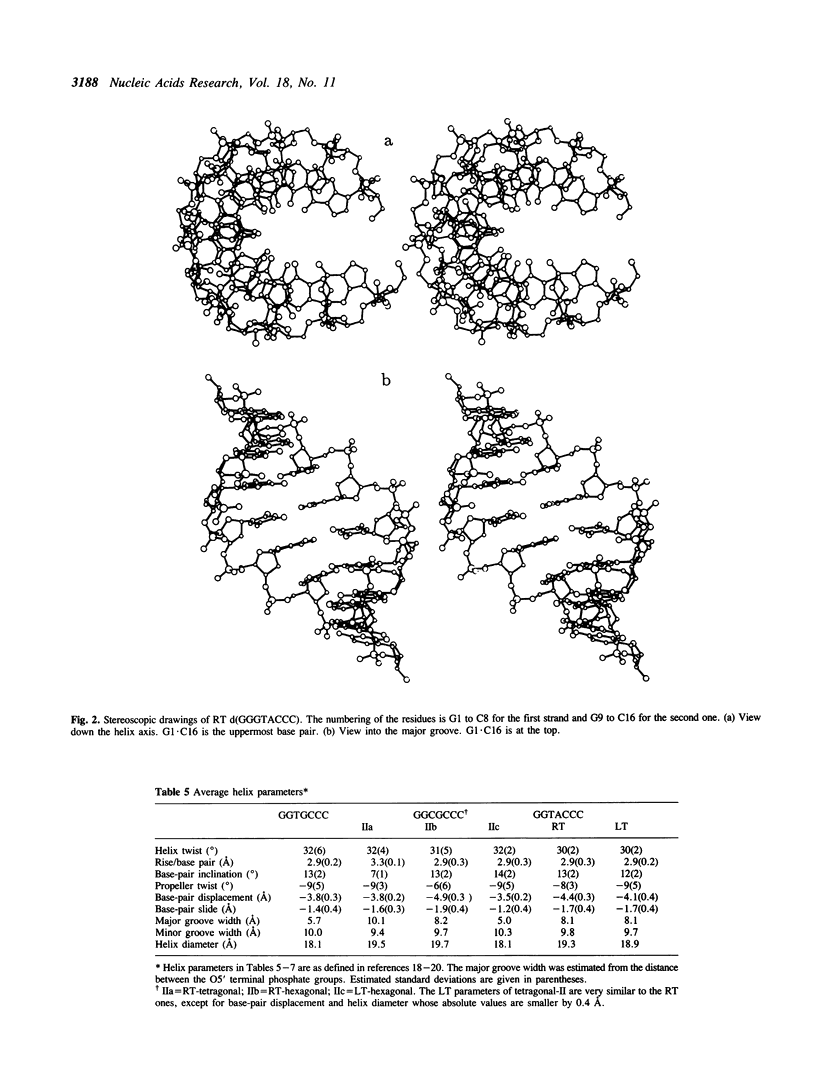
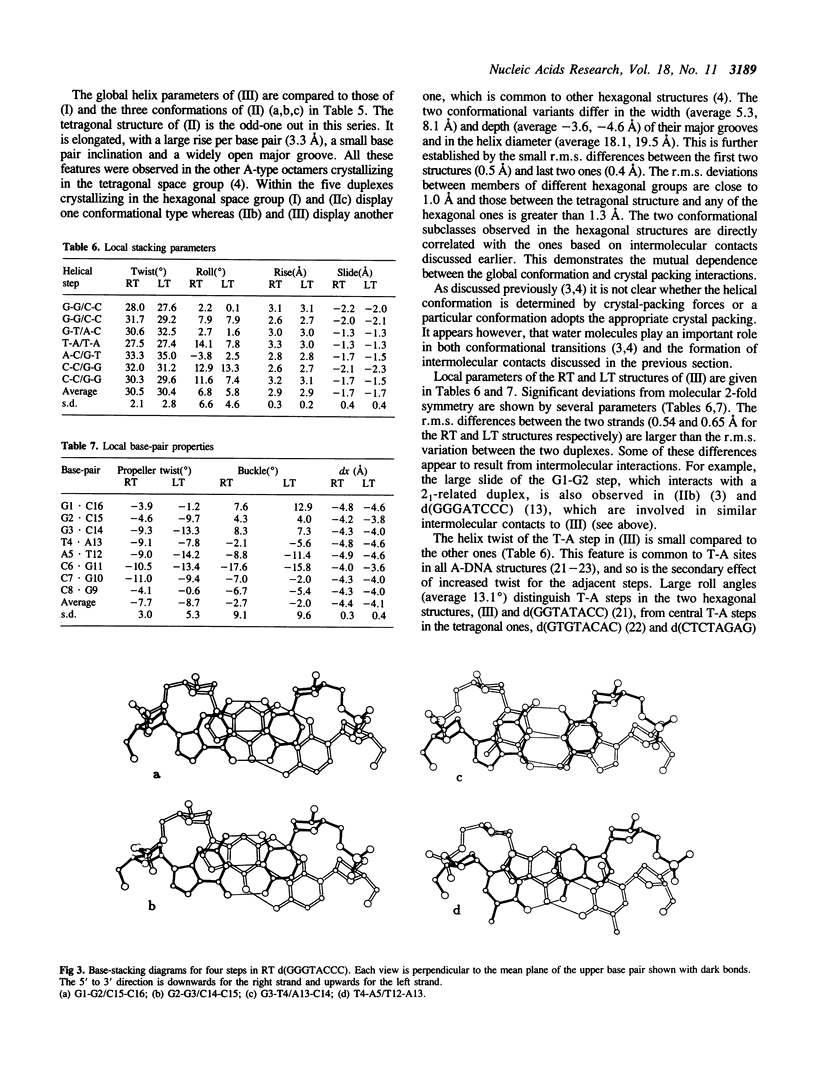
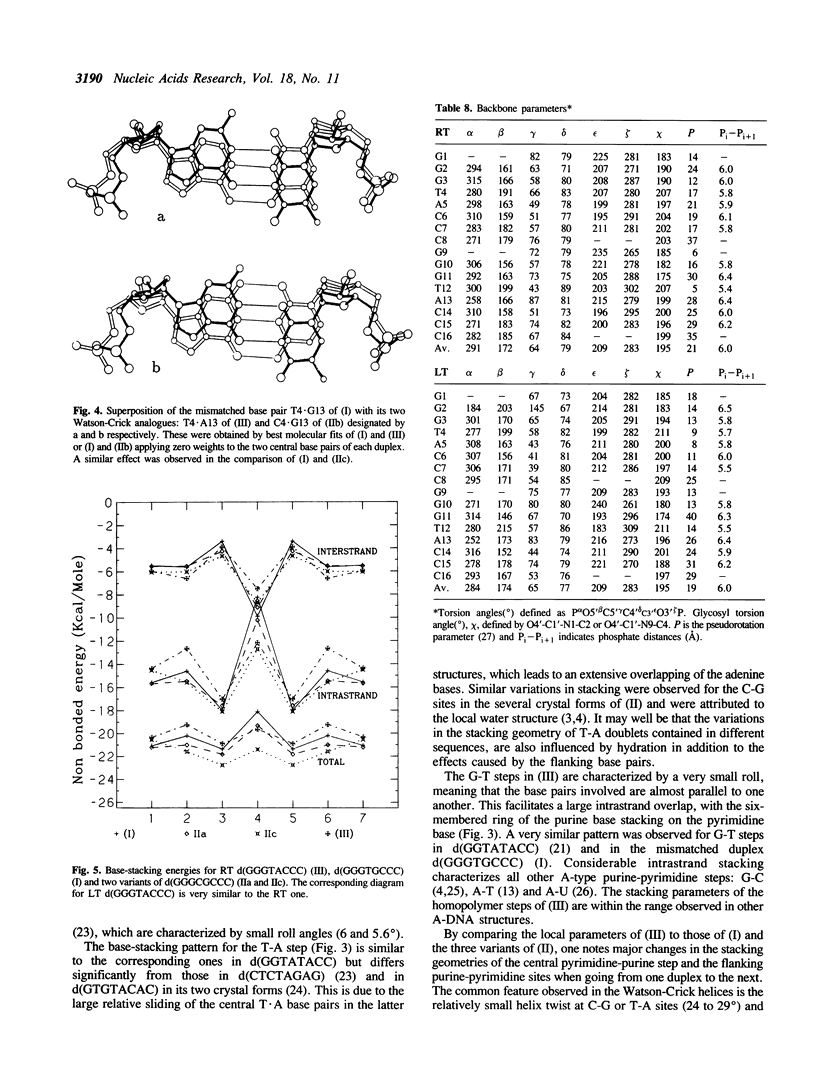
Abstract
The DNA fragment d(GGGTACCC) was crystallized as an A-DNA duplex in the hexagonal space group P6(1). The structure was analyzed at room temperature and low temperature (100K) at a resolution of 2.5 A. The helical conformations at the two temperatures are similar but the low-temperature structure is more economically hydrated than the room-temperature one. The structure of d(GGGTACCC) is compared to those of d(GGGTGCCC) and d(GGGCGCCC). This series of molecules, which consists of a mismatched duplex and its two Watson-Crick analogues, exhibits three conformational variants of the A-form of DNA, which are correlated with the specific intermolecular interactions observed in the various crystals. The largest differences in local conformation are displayed by the stacking geometries of the central pyrimidine-purine and the flanking purine-pyrimidine sites in each of the three duplexes. Stacking energy calculations performed on the crystal structures show that the mismatched duplex is destabilized with respect to each of the error-free duplexes, in accordance with helix-coil transition measurements.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J Am Chem Soc. 1972 Nov 15;94(23):8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Definitions and nomenclature of nucleic acid structure parameters. EMBO J. 1989 Jan;8(1):1–4. doi: 10.1002/j.1460-2075.1989.tb03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Chevrier B., Podjarny A., Johnson J., de Bear J. S., Gough G. R., Gilham P. T., Moras D. Crystallographic structure of an RNA helix: [U(UA)6A]2. J Mol Biol. 1989 Oct 5;209(3):459–474. doi: 10.1016/0022-2836(89)90010-7. [DOI] [PubMed] [Google Scholar]
- Doster W., Cusack S., Petry W. Dynamical transition of myoglobin revealed by inelastic neutron scattering. Nature. 1989 Feb 23;337(6209):754–756. doi: 10.1038/337754a0. [DOI] [PubMed] [Google Scholar]
- Eisenstein M., Hope H., Haran T. E., Frolow F., Shakked Z., Rabinovich D. Low-temperature study of the A-DNA fragment d(GGGCGCCC). Acta Crystallogr B. 1988 Dec 1;44(Pt 6):625–628. doi: 10.1107/s0108768188004732. [DOI] [PubMed] [Google Scholar]
- Fratini A. V., Kopka M. L., Drew H. R., Dickerson R. E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982 Dec 25;257(24):14686–14707. [PubMed] [Google Scholar]
- Haran T. E., Berkovich-Yellin Z., Shakked Z. Base-stacking interactions in double-helical DNA structures: experiment versus theory. J Biomol Struct Dyn. 1984 Oct;2(2):397–412. doi: 10.1080/07391102.1984.10507575. [DOI] [PubMed] [Google Scholar]
- Hunter W. N., D'Estaintot B. L., Kennard O. Structural variation in d(CTCTAGAG). Implications for protein-DNA interactions. Biochemistry. 1989 Mar 21;28(6):2444–2451. doi: 10.1021/bi00432a015. [DOI] [PubMed] [Google Scholar]
- Hunter W. N., Kneale G., Brown T., Rabinovich D., Kennard O. Refined crystal structure of an octanucleotide duplex with G . T mismatched base-pairs. J Mol Biol. 1986 Aug 20;190(4):605–618. doi: 10.1016/0022-2836(86)90246-9. [DOI] [PubMed] [Google Scholar]
- Jain S., Sundaralingam M. Effect of crystal packing environment on conformation of the DNA duplex. Molecular structure of the A-DNA octamer d(G-T-G-T-A-C-A-C) in two crystal forms. J Biol Chem. 1989 Aug 5;264(22):12780–12784. [PubMed] [Google Scholar]
- Jain S., Zon G., Sundaralingam M. Base only binding of spermine in the deep groove of the A-DNA octamer d(GTGTACAC). Biochemistry. 1989 Mar 21;28(6):2360–2364. doi: 10.1021/bi00432a002. [DOI] [PubMed] [Google Scholar]
- Kneale G., Brown T., Kennard O., Rabinovich D. G . T base-pairs in a DNA helix: the crystal structure of d(G-G-G-G-T-C-C-C). J Mol Biol. 1985 Dec 20;186(4):805–814. doi: 10.1016/0022-2836(85)90398-5. [DOI] [PubMed] [Google Scholar]
- Lauble H., Frank R., Blöcker H., Heinemann U. Three-dimensional structure of d(GGGATCCC) in the crystalline state. Nucleic Acids Res. 1988 Aug 25;16(16):7799–7816. doi: 10.1093/nar/16.16.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall M., Brown T., Kennard O. The crystal structure of d(G-G-G-G-C-C-C-C). A model for poly(dG).poly(dC). J Mol Biol. 1985 Jun 5;183(3):385–396. doi: 10.1016/0022-2836(85)90009-9. [DOI] [PubMed] [Google Scholar]
- Rabinovich D., Haran T., Eisenstein M., Shakked Z. Structures of the mismatched duplex d(GGGTGCCC) and one of its Watson-Crick analogues d(GGGCGCCC). J Mol Biol. 1988 Mar 5;200(1):151–161. doi: 10.1016/0022-2836(88)90340-3. [DOI] [PubMed] [Google Scholar]
- Saenger W., Hunter W. N., Kennard O. DNA conformation is determined by economics in the hydration of phosphate groups. 1986 Nov 27-Dec 3Nature. 324(6095):385–388. doi: 10.1038/324385a0. [DOI] [PubMed] [Google Scholar]
- Shakked Z., Guerstein-Guzikevich G., Eisenstein M., Frolow F., Rabinovich D. The conformation of the DNA double helix in the crystal is dependent on its environment. Nature. 1989 Nov 23;342(6248):456–460. doi: 10.1038/342456a0. [DOI] [PubMed] [Google Scholar]
- Shakked Z., Rabinovich D., Cruse W. B., Egert E., Kennard O., Sala G., Salisbury S. A., Viswamitra M. A. Crystalline A-dna: the X-ray analysis of the fragment d(G-G-T-A-T-A-C-C). Proc R Soc Lond B Biol Sci. 1981 Nov 24;213(1193):479–487. doi: 10.1098/rspb.1981.0076. [DOI] [PubMed] [Google Scholar]
- Shakked Z., Rabinovich D., Kennard O., Cruse W. B., Salisbury S. A., Viswamitra M. A. Sequence-dependent conformation of an A-DNA double helix. The crystal structure of the octamer d(G-G-T-A-T-A-C-C). J Mol Biol. 1983 May 15;166(2):183–201. doi: 10.1016/s0022-2836(83)80005-9. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]


