Abstract
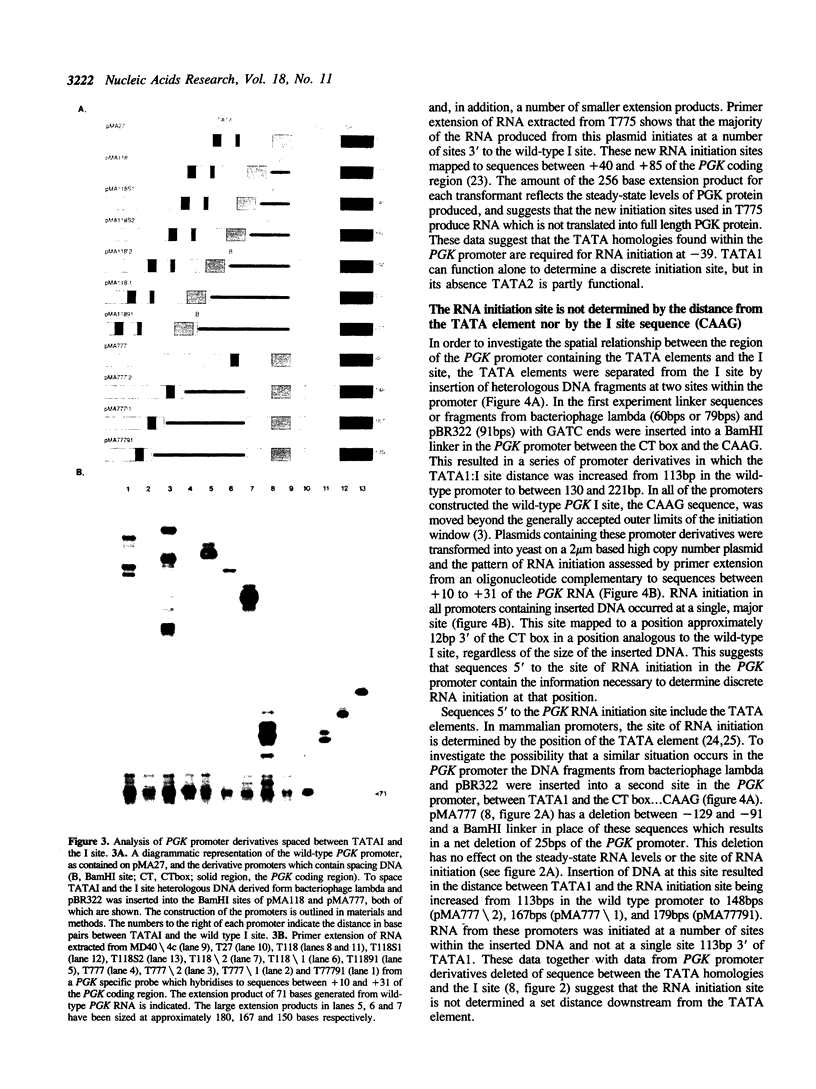
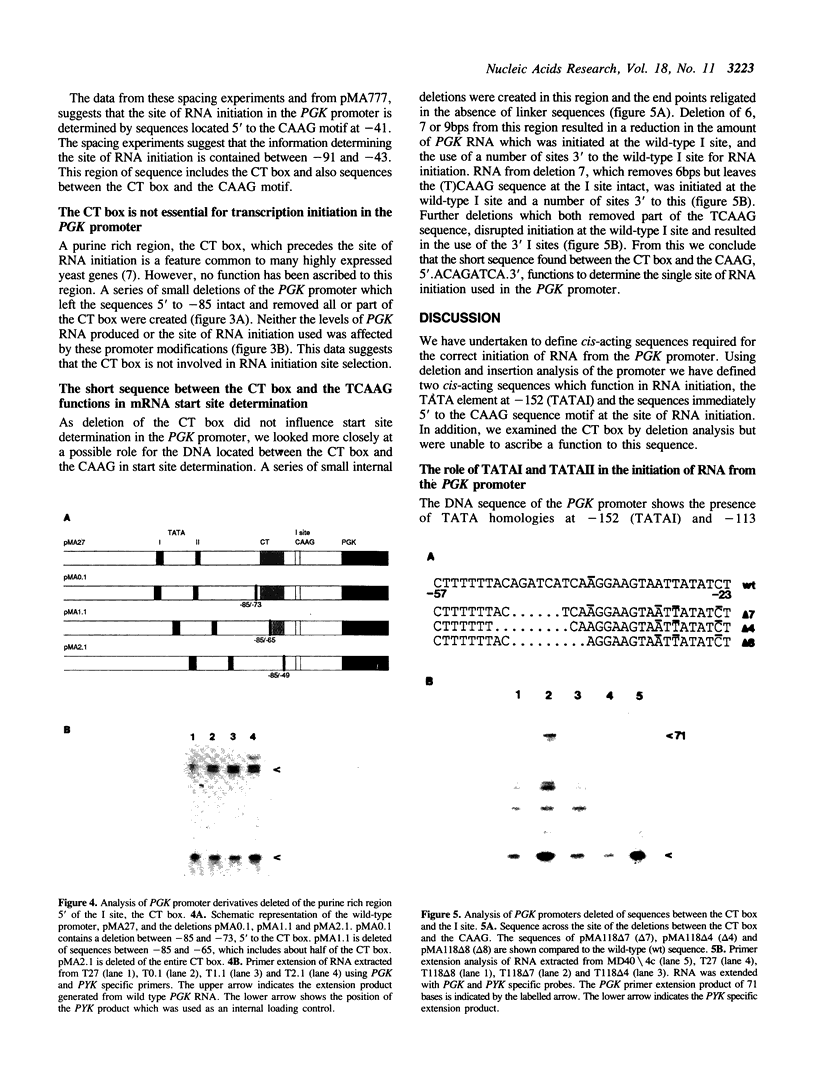
In the phosphoglycerate kinase (PGK) gene of yeast, as in other highly expressed yeast genes, the sequences surrounding the site of RNA initiation have a loosely conserved structure of a CT rich stretch followed by the tetranucleotide CAAG. Using internal deletions and insertions we have identified the elements in the PGK promoter which are required for correct RNA initiation at the CAAG sequence at -39. The results indicate that two different components of the PGK promoter contribute to correct RNA initiation, the TATA homologies, located at -152 and -113, and the sequences at the site of initiation. Both TATA elements can function in RNA initiation. Deletion of the upstream TATA element, TATAI, results in slightly heterogeneous RNA initiation, but the majority of the RNA initiates correctly. Deletion of both the PGK TATA elements results in the majority of the RNA initiating at sites downstream from the wild-type I site, within the structural gene between +40 to +80. The CT rich box is not essential for correct mRNA initiation as shown by deletion analysis. The site of RNA initiation in the PGK promoter appears to be determined by sequences located immediately 5' of the CAAG sequence motif. This short sequence, ACAGATC, when located the correct distance from the TATA elements may be sufficient to determine a discrete initiation site.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chen W., Struhl K. Yeast mRNA initiation sites are determined primarily by specific sequences, not by the distance from the TATA element. EMBO J. 1985 Dec 1;4(12):3273–3280. doi: 10.1002/j.1460-2075.1985.tb04077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson M. J., Mellor J., Fulton A. M., Roberts N. A., Bowen B. A., Kingsman S. M., Kingsman A. J. The identification and high level expression of a protein encoded by the yeast Ty element. EMBO J. 1984 May;3(5):1115–1119. doi: 10.1002/j.1460-2075.1984.tb01938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson M. J., Tuite M. F., Roberts N. A., Kingsman A. J., Kingsman S. M., Perkins R. E., Conroy S. C., Fothergill L. A. Conservation of high efficiency promoter sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Apr 24;10(8):2625–2637. doi: 10.1093/nar/10.8.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye G., Leung D. W., Tatchell K., Hall B. D., Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P., Frisque R. J., Gluzman Y. Identification of a promoter component involved in positioning the 5' termini of simian virus 40 early mRNAs. Proc Natl Acad Sci U S A. 1981 Jan;78(1):100–104. doi: 10.1073/pnas.78.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy A. M., Helser T. L., Zitomer R. S. Sequences required for transcriptional initiation of the Saccharomyces cerevisiae CYC7 genes. Mol Cell Biol. 1987 Oct;7(10):3785–3791. doi: 10.1128/mcb.7.10.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C., Meeks-Wagner D. W., Fangman W. L., Botstein D. A rapid, efficient method for isolating DNA from yeast. Gene. 1986;42(2):169–173. doi: 10.1016/0378-1119(86)90293-3. [DOI] [PubMed] [Google Scholar]
- Nagawa F., Fink G. R. The relationship between the "TATA" sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
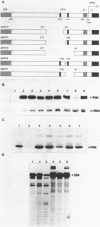
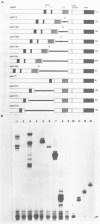
- Ogden J. E., Stanway C., Kim S., Mellor J., Kingsman A. J., Kingsman S. M. Efficient expression of the Saccharomyces cerevisiae PGK gene depends on an upstream activation sequence but does not require TATA sequences. Mol Cell Biol. 1986 Dec;6(12):4335–4343. doi: 10.1128/mcb.6.12.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., Hereford L. M., Skryabin K. G. Characterization of two types of yeast ribosomal DNA genes. J Bacteriol. 1978 Apr;134(1):295–305. doi: 10.1128/jb.134.1.295-305.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rudolph H., Hinnen A. The yeast PHO5 promoter: phosphate-control elements and sequences mediating mRNA start-site selection. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1340–1344. doi: 10.1073/pnas.84.5.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
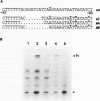
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano P. G., Tatchell K. Transcription and regulatory signals at the mating type locus in yeast. Cell. 1984 Jul;37(3):969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Deletion mapping a eukaryotic promoter. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4461–4465. doi: 10.1073/pnas.78.7.4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]