Abstract
The availability of nutrients and energy is a main driver of biodiversity for plant and animal communities in terrestrial and marine ecosystems, but we are only beginning to understand whether and how energy–diversity relationships may be extended to complex natural bacterial communities. Here, we analyzed the link between phytodetritus input, diversity and activity of bacterial communities of the Siberian continental margin (37–3427 m water depth). Community structure and functions, such as enzymatic activity, oxygen consumption and carbon remineralization rates, were highly related to each other, and with energy availability. Bacterial richness substantially increased with increasing sediment pigment content, suggesting a positive energy–diversity relationship in oligotrophic regions. Richness leveled off, forming a plateau, when mesotrophic sites were included, suggesting that bacterial communities and other benthic fauna may be structured by similar mechanisms. Dominant bacterial taxa showed strong positive or negative relationships with phytodetritus input and allowed us to identify candidate bioindicator taxa. Contrasting responses of individual taxa to changes in phytodetritus input also suggest varying ecological strategies among bacterial groups along the energy gradient. Our results imply that environmental changes affecting primary productivity and particle export from the surface ocean will not only affect bacterial community structure but also bacterial functions in Arctic deep-sea sediment, and that sediment bacterial communities can record shifts in the whole ocean ecosystem functioning.
Keywords: arctic, bacteria, diversity, ecology, sediment
Introduction
The relationship between diversity and bioavailable energy, often measured as photosynthetic productivity, is best described by positive or hump-shaped functions in animal and plant communities (Waide et al., 1999; Mittelbach et al., 2001; Evans et al., 2005). Several explanations for this ecological pattern have been proposed, including effects of population size, biomass, competition, evolutionary, environmental or resource heterogeneity (Waide et al., 1999; Cardinale et al., 2009 and references therein). Although microbes dominate most ecosystems in terms of abundance, diversity and biomass (Whitman et al., 1998), we have only recently begun to understand to what degree these relationships may be extended to complex microbial communities (reviewed in Smith, 2007). Noticeably, the studies that have mainly focused on pelagic or simplified ecosystems came to different conclusions: no relationship of overall richness with productivity could be found in aquatic mesocosms (Horner-Devine et al., 2003), weak positive correlations were shown for global patterns of bacterioplankton diversity (Pommier et al., 2007; Fuhrman et al., 2008), and hump-shaped relationships were identified for genotypes of Pseudomonas fluorescens in microcosms (Kassen et al., 2000). As high-throughput fingerprinting methods have become available recently, the bacterial energy–diversity relationship may now begin to be addressed in complex aquatic or terrestrial communities (Fuhrman, 2009). Unraveling the relationships between environmental conditions, organism diversity and ecosystem functions remains a priority if we are to better understand effects of global change.
For benthic life, productivity–diversity relationships have been mostly studied along continental slopes, which constitute ideal natural laboratories because of relatively defined variations in energy availability with water depth. Benthic communities in the deep sea depend on the sedimentation of phytodetritus from the productive surface waters, but detritus flux to the seafloor decreases substantially with increasing water depth because of the grazing and remineralization of particles in the water column. As the main source of energy, phytodetritus flux to the deep sea has a strong impact on the abundance, biomass and biodiversity of various size classes of benthic organisms (Smith et al., 2008 and references therein). The input of phytodetritus to deep-sea sediments has also been shown to influence bacterial biomass and activity (Deming and Yager, 1992; Boetius and Lochte, 1994; Jorgensen and Boetius, 2007), but studies linking energy availability at the seafloor to bacterial diversity patterns are still rare (Polymenakou et al., 2005; Franco et al., 2007).
Here we tested for the first time bacterial energy–diversity relationships for complex natural communities in Arctic seafloor on a defined, regional scale, in order to minimize confounding factors from sampling across different ocean provinces. We chose depth transects across the Arctic continental slope, covering a range of phytodetritus fluxes, and representing mesotrophic to oligotrophic deep-sea settings. As a common proxy for phytodetritus input to sediments, we used the chlorophyll pigment content of surface sediments (Boetius and Damm, 1998; Soltwedel, 2000; Dell'Anno et al., 2002; Soltwedel et al., 2009). Along this natural energy gradient, we described the shape of the relationships between energy, bacterial activity and bacterial diversity at different taxonomic levels, and identified bacterial taxa that are most likely affected by changes in energy availability.
Materials and methods
Study site and contextual parameters
Sediment samples were collected on three transects down the Laptev Sea continental slope during RV Polarstern cruise ARK IX/4 in September 1993 (Boetius and Damm, 1998). The samples analyzed here included 17 stations from the outer Laptev Sea shelf into the deep Eurasian basin (Supplementary Figure S1). An opening of the ice cover occurred as a temporally and regionally restricted event between June and September for a period of 2–12 weeks at different stations (Fütterer, 1994), leaving the Eastern most transect largely ice free at the time of sampling. Sediment cores were horizontally sliced into 1-cm-thick layers and sediment samples from the same stations were used for measuring environmental parameters, potential enzyme activities and DNA extraction. Measurements of chlorophyll pigments, protein concentration and hydrolytic enzyme activities (esterase, lipase, peptidase, beta-glucosidase) have been previously published (Boetius and Damm, 1998). Corresponding phaeopigment concentrations of the same stations were retrieved through the Publishing Network for Geoscientific and Environmental Data (PANGAEA, doi:10.1594/PANGAEA).
Community structure analysis
Total community DNA was extracted from 1 g of sediment using UltraClean Soil DNA Isolation Kits (MoBio Laboratories Inc., Carlsbad, CA, USA) and stored in a final volume of 100 μl Tris-EDTA buffer. DNA quantities were spectrophotometrically adjusted with a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA) for each step of the molecular protocol.
Automated ribosomal intergenic spacer analysis (ARISA)
A total of 42 samples composed of three sediment horizons (0–1, 1–2, 4–5 cm) were analyzed by ARISA. DNA quantities were standardized to 10 ng per reaction. PCR amplification, separation of fragments by capillary electrophoresis, evaluation of electrophoretic signals and subsequent binning into operational taxonomic units (OTUs) were done as reported elsewhere (Ramette, 2009). An OTU was considered present if it appeared in at least two of the three PCR replicates, and fingerprint profiles were standardized by dividing each individual peak area by the total area of peaks in a given profile.
454 massively parallel tag sequencing (MPTS)
A subset of 10 samples was selected for 454 massively parallel tag sequencing. Sequences were deposited in the GenBank Sequence Read Archive (http://www.ncbi.nlm.nih.gov) and their accession numbers are provided in the Supplementary material. To increase the representativeness of our analyses we combined samples from the upper two sediment layers (0–1 and 1–2 cm), after verifying that no significant differences were found in community structure between these two layers based on ARISA fingerprinting. Pigment concentrations for corresponding samples were averaged accordingly. Extracted DNA was amplified using primers targeting the V6 region of the bacterial 16S rRNA gene and including 454 Life Science's A or B sequencing adapters as published on http://vamps.mbl.edu. Fragments were sequenced by pyrosequencing on a Genome Sequencer FLX system (Roche, Basel, Switzerland) at the Marine Biological Laboratory (Woods Hole, MA, USA). Taxonomic assignments were performed with the Global Alignment for Sequence Taxonomy tool (Sogin et al., 2006; Huse et al., 2008). Sequence abundance was standardized by dividing by the total number of reads per sample.
In order to keep analyses over different taxonomic levels consistent, we used a subset of the 454 MPTS dataset for further analysis, in which only sequences with a complete assignment up to genus level were retained. A high Spearman's correlation between dissimilarity matrices of the reduced (20% of original) and the original dataset confirmed that ecological patterns were consistent in both datasets (Supplementary Table S1). When specifically investigating the response of different OTU categories to pigment concentration, we defined ‘less abundant' OTUs as those occurring with ⩽5 sequences in at least four samples and ‘common' OTUs as those occurring >5 sequences in at least four samples.
Statistical analyses
Chao1 richness estimates were calculated by re-sampling OTUs (defined at 3% sequence difference) based on the smallest dataset (n=7613 sequences), in order to obtain comparable estimates between samples. Despite the fact that diversity estimates based on 454 MPTS may be inflated by multiple factors (Kunin et al., 2009; Quince et al., 2009), we expect, however, that noise in the data is constant across samples, because the same experimental and sequencing protocols were used throughout. Although not allowing for absolute estimates of richness, the estimates still allow for relative comparisons between samples, where overall ecological relationships would not significantly change after denoising the datasets (Gobet et al., 2010; see Supplementary Information).
Bray–Curtis and Euclidean distances were used to calculate dissimilarity matrices for OTU tables and for environmental parameters, respectively. Mantel tests were used to compare Spearman's correlations of dissimilarity matrices between different datasets.
To avoid over-determination in modeling the community responses to environmental parameters, forward selection procedures were performed on groups of factors with redundancy analysis models. The best fitting models were selected using the Akaike Information Criterion. Space was modeled by using a polynomial of degree three of the spatial coordinates, from which the terms Y, XY, XY2, X3 and Y3 were finally retained after forward selection. Protein concentration, water depth, sediment depth and ice cover were kept as separate categories. The respective effects of various groups of variables on the variation in community composition were investigated by canonical variation partitioning (Legendre and Legendre, 1998; Ramette and Tiedje, 2007). Among different pigment measurements (Boetius and Damm, 1998), phaeopigments explained the highest amount of variation with 4% (P<0.001). Phaeopigments were therefore used to explore the response of the bacterial community to changes in phytodetritus input.
Directed dependencies between the response variables (ARISA diversity and enzymatic activity) and all groups of relevant contextual parameters were assessed in one causal model with path analysis (Legendre and Legendre, 1998). The RV coefficient (Robert and Escoufier, 1976) was used to derive a correlation matrix between groups of variables. Based on our previous statistical analyses, an initial model was tested and subsequently improved by comparing the fit of new models with the original matrix using chi-square tests. Other goodness-of-fit indices (for example, Bentler Comparative Fit Index, Bayesian Information Criterion) were used to further compare model performance.
All statistical analyses were performed in R (v.2.9.1; R Development Core Team 2009, http://www.R-project.org) using vegan, gmt, sem, and FactoMineR packages and custom R scripts.
Results
Relationships of bacterial diversity and function with increasing energy availability
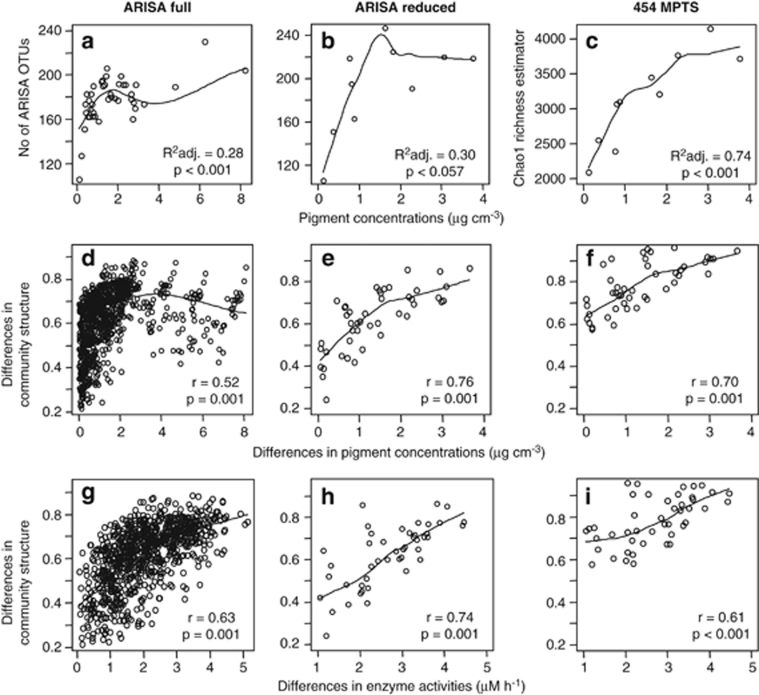
Changes in bacterial alpha-diversity (sample richness) and beta-diversity (changes in community structure between sites) were strongly related to changes in pigment concentrations (Figures 1a–f). OTU (as defined by ARISA, based on fingerprinting of the intergenic region of the 16S and 23S ribosomal genes) richness and pigment concentrations showed a strong positive, linear relationship until a pigment concentration of about 2 μg cm−3 sediment was reached. At higher pigment concentration levels, the relationship started to level off, and slightly increased again towards maximum pigment concentrations (Figure 1a). Patterns of bacterial community structure also showed high correlations with pigment concentrations (r=0.52, P=0.001; Figure 1d) and lower, yet significant correlations with spatial distance and water depth (r=0.24, P<0.001 and 0.14, P=0.011, respectively; Supplementary Figures S2a and b). When 454 MPTS (pyrosequencing of the variable V6 region of the 16S rRNA gene) was applied to a subset of samples to further explore the response of bacterial taxa to this range of energy availability, a similar linear relationship was found, appearing to level off at higher pigment concentrations (>3 μg cm−3; Figure 1c). The two molecular techniques, ARISA and 454 MPTS, revealed similar ecological patterns. Additional statistical tests further validated the technical concordance (Supplementary Table S1; Supplementary Information) and the usefulness of combining the two techniques to ecologically interpret the data.
Figure 1.
Changes in bacterial OTU richness, community structure and enzyme activity with pigment concentrations and correlation of changes in community structure with changes in enzyme activities for ARISA and 454 MPTS data. The plots in the left column of the figure (a, d, b) are based on the full ARISA dataset, the ones in the middle column (b, e, h) are based on a reduced ARISA dataset containing only samples used for 454 MPTS and plots in the right column (c, f, i) are based on 454 MPTS data. Linear regression R2 values and Spearman's correlations as tested by Mantel tests with 999 permutations are indicated in the plots.
Hydrolytic enzyme activities (esterase, lipase, peptidase, beta-glucosidase) showed a slightly lower, yet significant relationship with pigments (r=0.38, P=0.001; Supplementary Figure S3) and were also highly correlated with variations in bacterial community structure (r=0.63, P=0.001; Figures 1g–i). Changes in activity showed a low, but significant correlation with spatial distance (r=0.17, P=0.003), and no correlation with water depth (Supplementary Figures S2c and d). Similar results were obtained for oxygen consumption and carbon remineralization rates previously measured at 19 stations in the same area (Boetius and Damm, 1998). Oxygen consumption was significantly correlated with differences in pigment concentrations (r=0.59, P<0.001), spatial distance (r=0.26, P=0.01) and water depth (r=0.16, P=0.04), as were carbon remineralization rates with pigment concentrations (r=0.72, P<0.001), spatial distance (r=0.24, P=0.01) and water depth (r=0.31, P=0.004). Oxygen consumption and carbon remineralization were also correlated to changes in community structure (r=0.39, P=0.08 and r=0.66, P<0.001, respectively).
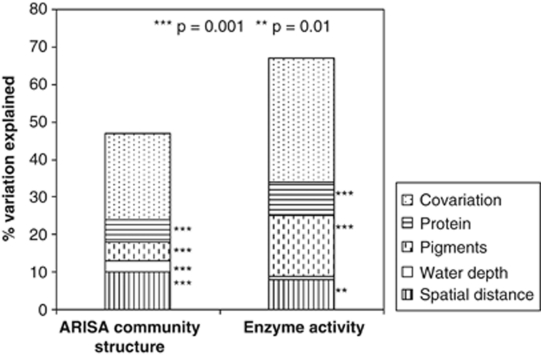
The effects of sedimentary pigments and protein (as proxies for phytodetritus input) on variation in bacterial community structure and function were further investigated by taking into account the confounding effects of spatial distance (geographic distance between samples), sediment depth and water depth (Figure 2). Variations in bacterial community structure and function were best explained by changes in pigment and protein concentrations. The full multivariate model explained 47% of the community variation, with pigment and protein concentrations significantly explaining 5% and 6% of the variation, respectively (P<0.001, based on 999 Monte Carlo permutation tests), and water depth and spatial distance explaining 3% and 10%, respectively. Sediment depth and ice cover could only explain a very small amount of variation in the biological data (1%). This was consistent with analysis of similarity results that showed no or only marginally significant differences in community structure between groups of samples originating from different sediment depths or ice covers (data not shown). Co-variation between explanatory variables, that is, variation that can be explained by the combined effects of several parameters, summed up to 23%. At the functional level, the variation in available energy as represented by pigment concentrations had the largest specific effects on the variation in enzyme activity (16% of the total variation, P<0.001; Figure 2), followed by protein concentration and spatial distance that explained 9% (P<0.001) and 8% (P<0.01) of the variation in enzyme activities, respectively. Water depth alone could not explain any significant part of the variation, whereas co-variation between variables overall accounted for 33%.
Figure 2.
Partitioning of the variation in bacterial community structure (ARISA) and enzyme activity (esterase, lipase, peptidase, beta-glucosidase). The specific effects of contextual parameters (protein concentration, pigment concentrations, water depth, spatial distance) and total co-variation between these parameters are represented. Statistical significance as determined by 999 Monte Carlo permutations under the full multivariate model is indicated by ***P<0.001 and **P<0.01.
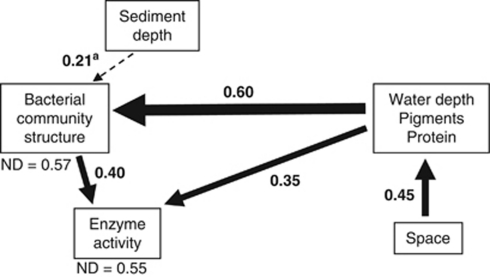
We further validated the causal relationship between bacterial community structure, activity and environmental parameters by path analysis. This method helps to determine the most plausible ecological models among a set of candidate models (Figure 3, Supplementary Figure S4). The strongest factor directly affecting changes in both bacterial community structure and their activity (enzymatic hydrolysis) was the energy gradient (combination of pigments and protein concentrations as indicators for the presence of labile organic matter, and depth for other flux-related processes; P<0.001). Noticeably, community structure and function were so tightly coupled that modeling a causal relationship in either direction resulted in very similar partial correlation values and overall path models (Supplementary Figure S4).
Figure 3.
Path analysis of the causal relationships between bacterial community structure, bacterial activity and contextual parameters. The Chi-square test is used to test whether the modeled relationships are significantly different from the original correlation matrix, with here a good agreement between the model and the data (P=0.76). The goodness-of-fit index (0.98) and Bentler Comparative Fit Index (1) indicate an optimal fit of the model. The Bayesian Information Criterion (−16.1) is another measure of the goodness of fit, and was the criterion that was iteratively minimized. The coefficient of non-determination (ND=1-R2) indicates the fraction of the variance in bacterial community structure and enzyme activity that is not explained by the model. aMarginally significant with P=0.078.
Response of individual taxa to changes in energy availability
The most abundant sequences in the complete 454 MPTS dataset were affiliated with the phylum Proteobacteria (51% of all sequences), followed by Actinobacteria (10%) and Acidobacteria (9%). On the class level, Gammaproteobacteria (26%), Deltaproteobacteria (14%), Actinobacteria (10%), Alphaproteobacteria (7%) and Acidobacteria (6%) contained the majority of all sequences. Taxa showing significant positive or negative relationships between their relative sequence abundance and pigment concentrations comprised the dominant fraction of the dataset, representing more than 50% of the sequences.
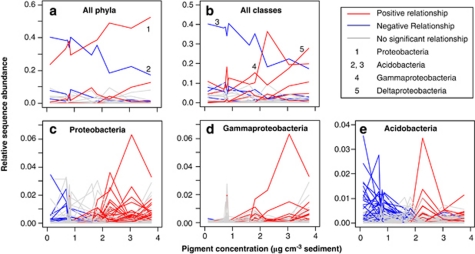
Already at coarse taxonomic resolution (that is, phylum and class levels), patterns of community structure with pigments were detected (Supplementary Table S2) and variable responses to changes in phytodetritus input were observed (Figures 4a,b). The major phylum Proteobacteria overall strongly responded positively to pigment concentration increase, whereas its corresponding classes showed positive, negative or no correlations (Figures 4a,b; Supplementary Table S3). Examples of classes showing positive linear relationships with pigments included Gammaproteobacteria and Flavobacteria (phylum Bacteroidetes), whereas no such relationship could be found for other bacterial classes, for example, Betaproteobacteria (Supplementary Table S3). Acidobacteria showed a negative, linear relationship with pigment concentrations. Taxa were also tested for quadratic relationships with pigment concentrations, but only very few significant correlations were found at high taxonomic resolution levels, for example, for the families Desulfuromonadaceae and Flavobacteriaceae.
Figure 4.
Examples of typical behaviors of taxa (a, b) and individual OTUs (c–e) with pigment concentrations. Significant positive and negative Spearman's rank correlation values are displayed in red and blue, respectively, whereas no significant relationships are in gray. The proportions of significantly correlated taxa and OTUs are for phyla (21% positive, 16% negative), classes (40%, 20%), Proteobacteria (4.6%, 0.6%), Gammaproteobacteria (3.8%, 0.1%), Acidobacteria (4.8%, 1.9%).
The significant relationships were, in some cases, consistent at various taxonomic levels, for example, the class Gammaproteobacteria, the family Psychromonadaceae and the genus Psychromonas were significantly positively related to phytodetritus input (Supplementary Table S3). Yet, a more complex picture emerged when considering the highest resolution level, that is of individual sequences (Figures 4c–e). Proteobacteria and the class Gammaproteobacteria showed strong positive relationships with pigment concentrations (R2=0.81, P<0.001 and R2=0.56, P=0.008, respectively), but OTUs consisting of individual sequences assigned to these taxa varied in their response from positive to negative, the same being true for OTUs assigned to Acidobacteria which overall showed negative relationships with pigments (R2=0.80, P<0.001). When the responses of less abundant and more common types to changes in pigment concentration were compared, 26% of the less abundant types showed significant relationships with phytodetritus input, whereas 46% of the more common types showed significant relationships, both being dominated by positive, linear correlations (data not shown).
Discussion
Change in richness with increasing energy availability
Our results suggest an overall positive response of bacterial OTU richness to energy availability in the form of phytodetritus, which was strongest at oligotrophic conditions defined by low levels of pigment concentrations (<2–3 μg cm−3 sediment) (Figures 1a–c). Positive relationships between diversity and food availability have also been described for benthic meio- and megafaunal organisms (Vanaverbeke et al., 1997; Soltwedel et al., 2009), and suggest that bacteria and animals of different size classes may be structured by similar ecological mechanisms. Increasing phytodetritus input sustains increase in bacterial abundance and biomass (Wei et al., 2010), potentially enabling more species to coexist. This would be in line with the ‘more individuals' hypothesis (Srivastava and Lawton, 1998) of the species–energy theory (Wright, 1983). A leveling-off of richness emerging from the inclusion of sites with higher phytodetritus supply (mesotrophic sites at the upper slope and close to the ice edge) may result from other resources, for example oxygen, becoming limited (Levin et al., 2001 and references therein). From a technical point of view, a weaker relationship of ARISA OTU richness with energy availability at high pigment concentrations may also be because of a saturation of the molecular method to further detect changes, as observed with fingerprinting techniques (for example, Cho and Tiedje, 2000). Other types of community dynamics may also cause this type of relationship, like competition or predation, which may put a limit to the number of coexisting species (Rex, 1976; Levin et al., 2001). Further studies of natural and experimental systems are needed to test and decipher the mechanisms responsible for the establishment and maintenance of energy–diversity relationship in bacterial communities, and if these can be extended to the global scale.
Changes in community structure and function with increasing energy availability
Not only bacterial richness but also community structure and function were affected by energy availability, suggesting a tight coupling between community structure and functions in organic matter remineralization, such as hydrolytic enzyme activity and oxygen consumption. A close association between community structure and functional patterns has also recently been reported for other Arctic regions (Teske et al., 2011). This may imply that changes in bacterial community structure could directly translate into functional changes that may even affect overall ecosystem functioning, for example carbon retention and nutrient remineralization. Additional analyses will be needed to investigate not only the role of quantity but also quality of phytodetrital material for the specific functional response of benthic bacterial communities, which may alter organic matter recycling at the seafloor.
Community variation was mainly explained by changes in energy availability, but with respect to the effects of sediment depth, results differed between variation partitioning (Figure 2) and path analysis (Figure 3). Variation partitioning and path analysis consist of two distinct ecological modeling methods. With the former, the response and the explanatory variables are defined a priori, while with the latter, different ecological scenarios are evaluated within a causal modeling framework using goodness-of-fit statistics (more details about the techniques are given in the Supplementary Information). It is therefore not surprising that different, yet complementary ecological insights may be obtained when the two approaches are combined. Effects of sediment depth detected by path analysis could be because of changes in other environmental parameters across sediment depth, such as oxygen availability (Boetius and Damm, 1998). Because some of the community variation was explained by pure effects of the categories ‘spatial distance' and ‘water depth', this may suggest isolation-by-distance processes, but also the effects of other, unmeasured biogeochemical parameters that would also be spatially structured (Legendre and Legendre, 1998). Furthermore, the relation between phytodetritus input and bacterial community structure could also be indirectly produced by top-down effects, such as nanoflagellate grazing (Danovaro et al., 1998; Lebaron et al., 1999; Lindström, 2000), viral infection (Danovaro and Serresi, 2000) or food-dependent differences in benthic fauna composition that would affect grazing, defecation and bioturbation. Yet, we identified reproducible ecological patterns of diversity on all taxonomic levels investigated, demonstrating that bacterial diversity is not just randomly distributed along a well-defined energy gradient but compares well with response patterns of other benthic organisms.
Specific taxa–energy relationships
Bacterial OTU richness generally increased with increasing phytodetritus input, with indications of a plateau at higher phytodetritus input levels, whereas the response varied from positive to negative for the relative sequence abundance of individual taxonomic groups of bacteria. Taxa showing significant relationships with phytodetritus input were usually sequence abundant. Common taxa, such as the Gammaproteobacteria and Acidobacteria with strong relationships to energy availability may serve as indicator taxa for certain environmental conditions, for example high vs low phytodetritus availability, and could be helpful for future monitoring studies of benthic ecosystems in the Arctic Ocean. Although most of the sediment in the deep sea may be formed by sinking particles, pelagic and benthic communities have very contrasted community compositions (Zinger et al., 2011), suggesting that the ecological response we observed specifically originates from sediment bacterial communities and not from pelagic communities. The strong positive correlations with energy availability in the Gammaproteobacteria, a globally ubiquitous group in marine sediments, imply that these organisms may include many opportunistic, fast growing bacteria. This is not surprising, considering the fact that members of the Gammaproteobacteria have been described as copiotrophs (Glöckner et al., 1999). Similarly, Flavobacteria exhibited a strong positive correlation with energy availability, consistent with the association of copiotrophic attributes to the phylum Bacteroidetes (Fierer et al., 2007), and a strong responsiveness of Flavobacteria to phytoplankton blooms (Pinhassi et al., 2004). In contrast, Acidobacteria may be especially adapted to oligotrophic conditions (Fierer et al., 2007), indicated by their significant negative relationship with pigments. Our results are consistent with other studies that showed differing responses of bacterial taxa to changes in productivity (Horner-Devine et al., 2003; Pommier et al., 2007). This suggests that varying ecological strategies and broad-scale patterns of co-existence or avoidance between bacterial groups may exist. Also at the level of individual OTUs, different relationships were observed, indicating niche differentiation even in closely related bacterial types. Future experimental studies on microbial energy–diversity relationships using quantitative methods such as fluorescence in situ hybridization or quantitative PCR, which generally target specific microbial populations, would need to consider this biological variability in their assays. Interestingly, common types were more likely to show significant relationships with food input than less abundant types, suggesting that the more abundant types may be actively growing and mediating most ecosystem functions (Pedros-Alio, 2006). Although a large number of sequences within a given OTU may increase the chance to detect significant relationships with environmental parameters, the fact that 26% of less abundant types exhibited relationships with food availability indicates that both common and less abundant types may respond to changes in energy availability. This is consistent with observations made in other ecosystems in which the ‘rare' biosphere has been shown to display biogeographic patterns (Galand et al., 2009) or temporal patterns with changing environmental conditions (Brazelton et al., 2010).
Further implications
Beyond the description of energy availability–diversity relationships for complex bacterial communities, this study strongly suggests that any environmental changes affecting primary productivity and particle export will cause shifts in bacterial community structure and function in the Arctic, which in turn could affect key processes such as carbon cycling (Deming and Baross, 1993; Klages et al., 2003). Already now, structural shifts of Arctic ecosystems in response to changing environmental conditions have been observed (Grebmeier et al., 2006) and may further affect benthic–pelagic coupling (Aagaard et al., 1999). Our samples were collected at a time when the Laptev Sea was largely ice-covered throughout the year, explaining why short-term changes in ice cover occurring during our study were mostly not reflected in changes in bacterial community structure and function. Since then, a rapid decline in sea ice cover has occurred, leaving most of this area ice free during the Arctic summer (Serreze et al., 2007). Such long-term changes in ice cover are predicted to result in changing primary productivity and particle flux (Arrigo et al., 2008; Lalande et al., 2009; Wassmann et al., 2010). Our study thus offers a unique ecological baseline against which ecosystem shifts can be assessed in the future, especially by incorporating bacterial community dynamics in a region increasingly influenced by global change.
Acknowledgments
We thank Mitchell Sogin, Linda Amaral-Zettler, Hilary Morrison and Katerina Andreishcheva at the MBL (Woods Hole) for their assistance with 454 massively parallel tag sequencing. Sue Huse and Lucie Zinger are acknowledged for their help with the calculation of OTU richness estimates. We would also like to thank the scientific party and crew of RV Polarstern for sampling during the expedition ARK IX-4. We thank three anonymous reviewers for their helpful comments and suggestions. This study has been funded by the Leibniz programme of the DFG to AB. The work was also financially supported by the Helmholtz Association and the Max Planck Society.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aagaard K, Darby D, Falkner K, Flato G, Grebmeier JM, Measures C, et al. 1999Marine Science in the Arctic: A Strategy (ARCUS) ARCotUS: Fairbanks; p84. [Google Scholar]
- Arrigo KR, van Dijken G, Pabi S. Impact of a shrinking arctic ice cover on marine primary production. Geophys Res Lett. 2008;35:L19603. [Google Scholar]
- Boetius A, Damm E. Benthic oxygen uptake, hydrolytic potentials and microbial biomass at the Arctic continental slope. Deep-Sea Res Part I-Oceanogr Res Pap. 1998;45:239–275. [Google Scholar]
- Boetius A, Lochte K. Regulation of microbial enzymatic degradation of organic matter in deep-sea sediments. Mar Ecol Prog Ser. 1994;104:299–307. [Google Scholar]
- Brazelton WJ, Ludwig KA, Sogin ML, Andreishcheva EN, Kelley DS, Shen C-C, et al. Archaea and bacteria with surprising microdiversity show shifts in dominance over 1000-year time scales in hydrothermal chimneys. Proc Natl Acad Sci USA. 2010;107:1612–1617. doi: 10.1073/pnas.0905369107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale BJ, Hillebrand H, Harpole WS, Gross K, Ptacnik R. Separating the influence of resource ‘availability' from resource ‘imbalance' on productivity–diversity relationships. Ecol Lett. 2009;12:475–487. doi: 10.1111/j.1461-0248.2009.01317.x. [DOI] [PubMed] [Google Scholar]
- Cho JC, Tiedje JM. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl Environ Microbiol. 2000;66:5448–5456. doi: 10.1128/aem.66.12.5448-5456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danovaro R, Marrale D, Della Croce N, Dell'Anno A, Fabiano M. Heterotrophic nanoflagellates, bacteria, and labile organic compounds in continental shelf and deep-sea sediments of the Eastern Mediterranean. Microb Ecol. 1998;35:244–255. doi: 10.1007/s002489900080. [DOI] [PubMed] [Google Scholar]
- Danovaro R, Serresi M. Viral density and virus-to-bacterium ratio in deep-sea sediments of the Eastern Mediterranean. Appl Environ Microbiol. 2000;66:1857–1861. doi: 10.1128/aem.66.5.1857-1861.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Anno A, Mei ML, Pusceddu A, Danovaro R. Assessing the trophic state and eutrophication of coastal marine systems: a new approach based on the biochemical composition of sediment organic matter. Mar Pollut Bull. 2002;44:611–622. doi: 10.1016/s0025-326x(01)00302-2. [DOI] [PubMed] [Google Scholar]
- Deming JW, Baross JA.1993The early diagenesis of organic matter: bacterial activityIn: Engel M, Macko S (eds).Organic Geochemistry Plenum Press: New York; 119–144. [Google Scholar]
- Deming JW, Yager PL.1992Natural bacterial assemblages in deep-sea sediments - towards a global viewIn: Rowe GT, Pariente V (eds)Deep-Sea Food Chains and the Global Carbon Cycle Kluwer Academic Publishers: Dordrecht, The Netherlands; 11–27. [Google Scholar]
- Evans KL, Warren PH, Gaston KJ. Species–energy relationships at the macroecological scale: a review of the mechanisms. Biol Rev Cambridge Philos Soc. 2005;80:1–25. doi: 10.1017/s1464793104006517. [DOI] [PubMed] [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- Franco MA, De Mesel I, Diallo MD, Van der Gucht K, Van Gansbeke D, Van Rijswijk P, et al. Effect of phytoplankton bloom deposition on benthic bacterial communities in two contrasting sediments in the southern North Sea. Aquat Microb Ecol. 2007;48:241–254. [Google Scholar]
- Fuhrman JA. Microbial community structure and its functional implications. Nature. 2009;459:193–199. doi: 10.1038/nature08058. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL, et al. A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA. 2008;105:7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer DK.1994The expedition ARCTIC ‘93 Leg ARK-IX/4 of RV “Polarstern” 1993 Rep Polar Res 149p244. [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Natl Acad Sci USA. 2009;106:22427–22432. doi: 10.1073/pnas.0908284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner FO, Fuchs BM, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobet A, Quince C, Ramette A. Multivariate Cutoff Level Analysis (MultiCoLA) of large community data sets. Nucleic Acids Res. 2010;38:e155. doi: 10.1093/nar/gkq545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebmeier JM, Overland JE, Moore SE, Farley EV, Carmack EC, Cooper LW, et al. A major ecosystem shift in the northern Bering Sea. Science. 2006;311:1461–1464. doi: 10.1126/science.1121365. [DOI] [PubMed] [Google Scholar]
- Horner-Devine MC, Leibold MA, Smith VH, Bohannan BJM. Bacterial diversity patterns along a gradient of primary productivity. Ecol Lett. 2003;6:613–622. [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PloS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen BB, Boetius A. Feast and famine—microbial life in the deep-sea bed. Nat Rev Microbiol. 2007;5:770–781. doi: 10.1038/nrmicro1745. [DOI] [PubMed] [Google Scholar]
- Kassen R, Buckling A, Bell G, Rainey PB. Diversity peaks at intermediate productivity in a laboratory microcosm. Nature. 2000;406:508–512. doi: 10.1038/35020060. [DOI] [PubMed] [Google Scholar]
- Klages M, Boetius A, Christensen JP, Deubel H, Piepenburg D, Schewe I, et al. 2003The benthos of Arctic Seas and its role for the carbon cycle at the seafloorIn: Stein R, Macdonald RW (eds).The Arctic Organic Carbon Cycle Springer: Heidelberg; 139–167. [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2009;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Lalande C, Belanger S, Fortier L. Impact of a decreasing sea ice cover on the vertical export of particulate organic carbon in the northern Laptev Sea, Siberian Arctic Ocean. Geophys Res Lett. 2009;36:L21604. [Google Scholar]
- Lebaron P, Servais P, Troussellier M, Courties C, Vives-Rego J, Muyzer G, et al. Changes in bacterial community structure in seawater mesocosms differing in their nutrient status. Aquat Microb Ecol. 1999;19:255–267. [Google Scholar]
- Legendre L, Legendre P. Numerical Ecology. Elsevier Science: Amsterdam; 1998. [Google Scholar]
- Levin LA, Etter RJ, Rex MA, Gooday AJ, Smith CR, Pineda J, et al. Environmental influences on regional deep-sea species diversity. Annu Rev Ecol Syst. 2001;32:51–93. [Google Scholar]
- Lindström ES. Bacterioplankton community composition in five lakes differing in trophic status and humic content. Microb Ecol. 2000;40:104–113. doi: 10.1007/s002480000036. [DOI] [PubMed] [Google Scholar]
- Mittelbach GG, Steiner CF, Scheiner SM, Gross KL, Reynolds HL, Waide RB, et al. What is the observed relationship between species richness and productivity. Ecology. 2001;82:2381–2396. [Google Scholar]
- Pedros-Alio C. Marine microbial diversity: can it be determined. Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Pinhassi J, Sala MM, Havskum H, Peters F, Guadayol O, Malits A, et al. Changes in bacterioplankton composition under different phytoplankton regimens. Appl Environ Microbiol. 2004;70:6753–6766. doi: 10.1128/AEM.70.11.6753-6766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenakou PN, Bertilsson S, Tselepides A, Stephanou EG. Links between geographic location, environmental factors, and microbial community composition in sediments of the Eastern Mediterranean Sea. Microb Ecol. 2005;49:367–378. doi: 10.1007/s00248-004-0274-5. [DOI] [PubMed] [Google Scholar]
- Pommier T, Canback B, Riemann L, Bostrom KH, Simu K, Lundberg P, et al. Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol. 2007;16:867–880. doi: 10.1111/j.1365-294X.2006.03189.x. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Meth. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Ramette A. Quantitative community fingerprinting methods for estimating the abundance of operational taxonomic units in natural microbial communities. Appl Environ Microbiol. 2009;75:2495–2505. doi: 10.1128/AEM.02409-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette A, Tiedje JM. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc Natl Acad Sci USA. 2007;104:2761–2766. doi: 10.1073/pnas.0610671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex MA. Biological accommodation in the deep-sea benthos: comparative evidence on the importance of predation and productivity. Deep-Sea Res Part I-Oceanogr Res Pap. 1976;23:975–987. [Google Scholar]
- Robert P, Escoufier Y. A unifying tool for linear multivariate statistical methods: the RV-coefficient. J Roy Stat Soc C Appl Stat. 1976;25:257–265. [Google Scholar]
- Serreze MC, Holland MM, Stroeve J. Perspectives on the Arctic's shrinking sea-ice cover. Science. 2007;315:1533–1536. doi: 10.1126/science.1139426. [DOI] [PubMed] [Google Scholar]
- Smith CR, De Leo FC, Bernardino AF, Sweetman AK, Arbizu PM. Abyssal food limitation, ecosystem structure and climate change. Trends Ecol Evol. 2008;23:518–528. doi: 10.1016/j.tree.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Smith VH. Microbial diversity–productivity relationships in aquatic ecosystems. FEMS Microbiol Ecol. 2007;62:181–186. doi: 10.1111/j.1574-6941.2007.00381.x. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltwedel T. Metazoan meiobenthos along continental margins: a review. Prog Oceanogr. 2000;46:59–84. [Google Scholar]
- Soltwedel T, Jaeckisch N, Ritter N, Hasemann C, Bergmann M, Klages M. Bathymetric patterns of megafaunal assemblages from the arctic deep-sea observatory HAUSGARTEN. Deep-Sea Res Part I-Oceanogr Res Pap. 2009;56:1856–1872. [Google Scholar]
- Srivastava DS, Lawton JH. Why more productive sites have more species: an experimental test of theory using tree-hole communities. Am Nat. 1998;152:510–529. doi: 10.1086/286187. [DOI] [PubMed] [Google Scholar]
- Teske A, Durbin A, Ziervogel K, Cox C, Arnosti C. Microbial community composition and function in permanently cold seawater and sediments from an Arctic Fjord of Svalbard. Appl Environ Microbiol. 2011;77:2008–2018. doi: 10.1128/AEM.01507-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaverbeke J, Arbizu PM, Dahms HU, Schminke HK. The Metazoan meiobenthos along a depth gradient in the Arctic Laptev Sea with special attention to nematode communities. Polar Biol. 1997;18:391–401. [Google Scholar]
- Waide RB, Willig MR, Steiner CF, Mittelbach G, Gough L, Dodson SI, et al. The relationship between productivity and species richness. Annu Rev Ecol Syst. 1999;30:257–300. [Google Scholar]
- Wassmann P, Slagstad D, Ellingsen I. Primary production and climatic variability in the European sector of the Arctic Ocean prior to 2007: preliminary results. Polar Biol. 2010;33:1641–1650. [Google Scholar]
- Wei CL, Rowe GT, Escobar-Briones E, Boetius A, Soltwedel T, Caley MJ, et al. Global patterns and predictions of seafloor biomass using random forests. PLoS One. 2010;5:e15323. doi: 10.1371/journal.pone.0015323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DH. Species-energy theory: an extension of species-area theory. Oikos. 1983;41:496–506. [Google Scholar]
- Zinger L, Amaral-Zetter LA, Fuhrman JA, Horner-Devine MC, Huse SM, Mark Welch D, et al. A community ecology approach reveals contrasted patterns between seawater and seafloor bacteria. PLoS One. 2011;6:e24570. doi: 10.1371/journal.pone.0024570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.