Abstract
Hydrogen (H2) release from photosynthetic microbial mats has contributed to the chemical evolution of Earth and could potentially be a source of renewable H2 in the future. However, the taxonomy of H2-producing microorganisms (hydrogenogens) in these mats has not been previously determined. With combined biogeochemical and molecular studies of microbial mats collected from Elkhorn Slough, Monterey Bay, California, we characterized the mechanisms of H2 production and identified a dominant hydrogenogen. Net production of H2 was observed within the upper photosynthetic layer (0–2 mm) of the mats under dark and anoxic conditions. Pyrosequencing of rRNA gene libraries generated from this layer demonstrated the presence of 64 phyla, with Bacteriodetes, Cyanobacteria and Proteobacteria dominating the sequences. Sequencing of rRNA transcripts obtained from this layer demonstrated that Cyanobacteria dominated rRNA transcript pyrotag libraries. An OTU affiliated to Microcoleus spp. was the most abundant OTU in both rRNA gene and transcript libraries. Depriving mats of sunlight resulted in an order of magnitude decrease in subsequent nighttime H2 production, suggesting that newly fixed carbon is critical to H2 production. Suppression of nitrogen (N2)-fixation in the mats did not suppress H2 production, which indicates that co-metabolic production of H2 during N2-fixation is not an important contributor to H2 production. Concomitant production of organic acids is consistent with fermentation of recently produced photosynthate as the dominant mode of H2 production. Analysis of rRNA % transcript:% gene ratios and H2-evolving bidirectional [NiFe] hydrogenase % transcript:% gene ratios indicated that Microcoelus spp. are dominant hydrogenogens in the Elkhorn Slough mats.
Keywords: microbial mats, fermentation, hydrogen, hydrogenase, Microcoleus spp., pyrotags
Introduction
Microbial mats are among the most phylogenetically and physiologically diverse ecosystems on Earth (Ley et al., 2006; Kunin et al., 2008; Bolhuis and Stal, 2011). These systems undergo daily cycling of redox potential, increasing as oxygenic phototrophs supersaturate the upper strata with oxygen during the day and then decreasing as the mats become completely anoxic at night. Although the role of mats in global carbon and oxygen cycling is acknowledged and well studied (Nisbet and Fowler, 1999; Des Marais, 2003), their H2 ecology has been underexplored.
H2 release from microbial mats has likely been important for the chemical evolution of Earth, where its emission over long periods of geological time might be an important contributing factor to the oxidation of our atmosphere (Hoehler et al., 2001). Today, H2 is an important industrial chemical, where 108 m3 (at 1 atm) are sold annually in the US alone (Lee et al., 2010). Biological systems capable of producing industrially significant amounts of H2 have been an important focus of alternative energy research. Mat ecosystems could thus have a potential role as a renewable source of H2 in the future (Bender and Phillips, 2004). Knowledge of the metabolic processes that lead to H2 generation and the phylogenetic identity of associated H2-producing microorganisms (hydrogenogens) within highly complex microbial mat communities will be an important initial step towards realizing the potential of these systems as a viable H2 source. This knowledge will be useful in optimizing system parameters, such as light, carbon and nutrient concentrations, that enhance hydrogenogen activity and may facilitate future use of microbial mats as a commercial source of H2.
The activity of phototrophs and diazotrophs is well established in photosynthetic microbial mats (Des Marais, 2003) and both groups are capable of producing H2, either fermentatively or as a co-metabolic product of N2-fixation (Bothe et al., 2010). Recent molecular ecology studies of microbial mats have focused on use of the H2-evolving bidirectional [FeFe] hydrogenase as a biomarker for investigating hydrogenogens in photosynthetic mats (Boyd et al., 2009, 2010). These studies infer that non-phototrophic, anaerobes are important hydrogenogens in mat ecosystems. However, the class of hydrogenases targeted and the assays used in these studies did not include H2-evolving bidirectional [NiFe] hydrogenases harbored by phototrophic microorganisms. To our knowledge, no studies have examined the diversity of H2-evolving bidirectional [NiFe] hydrogenases in microbial mats that may be active in phototrophic hydrogenogens that produce H2 by fermentative processes. Thus, our current understanding of the overall microbial ecology of H2 cycling in microbial mats remains an incomplete picture.
Previous work, which employed biogeochemical approaches has served to constrain several aspects of H2 cycling in microbial mats (Skyring et al., 1988, 1989; Hoehler et al., 2001). However, robust linkages between microorganisms and processes that generate H2 remain to be established and the taxonomic identity of principal hydrogenogens is unknown. In this study we determined the location of hydrogenogens in Elkhorn Slough mats, the relative importance of co-metabolic production of H2 during N2-fixation vs fermentation and phototrophic vs heterotrophic H2 production. In parallel we used pyrosequencing of SSU rRNA genes and transcripts and sequencing of H2-evolving bidirectional [NiFe] hydrogenase genes and transcripts to obtain evidence for the taxonomic identity of dominant hydrogenogens.
Methods
Field site sampling and short-term maintenance of mats in a greenhouse
Photosynthetic microbial mats were sampled from a tidal zone in the Elkhorn Slough estuary at Monterey Bay, California (Latitude 36.830 °N, Longitude −121.785 °W) on November 9, 2009. The samples measured 18 × 23 cm2 and were ∼2 cm in depth. The samples were transported to the NASA Ames Research Center greenhouse within 1–2 h of collection from the slough. Upon arrival, mats were placed in acrylic aquaria transparent to ultraviolet radiation and covered with 4 l of water (∼3 cm of overlying water column) from the field site. The salinity of the field site water was 35‰ and had a pH of 8.0. Mats were incubated under natural solar irradiance and a regulated temperature (∼19 °C, in situ average) for up to 5 days, during which diel cycle experiments were carried out.
Analysis of H2 and organic acids
Replicate vials were prepared identically for each control or manipulation experiment as follows: small sub-cores (11 mm diameter, 15 mm depth or vertically sectioned for depth profile analyses) were cut from whole sections of intact microbial mat and placed in serum vials with 4 ml of field site water. Serum vials were capped with butyl rubber stoppers. The 10.5 ml headspace of the serum vials was left as air for light/daytime incubations and was thoroughly flushed with N2 (gas and liquid phase degassed) for dark/nighttime incubations. For H2 analysis, at least six replicate vials were sampled at each time point for each control or manipulation experiment. Organic acid production was analyzed in each of three replicates for each time point for each control or manipulation experiment. Manipulations included sectioning of microbial mat to identify the location of H2 production and suppression of diazotroph or phototroph (oxygenic and/or anoxygenic) activity to assess the roles of these microbial groups in H2 production. See supplementary methods for further information regarding H2 and organic acid analyses and details on how manipulation experiments were performed.
Analysis of N2-fixation
N2-fixation activity was determined using the acetylene reduction (ethylene production) assay in triplicate incubations for each time point for each control or manipulation experiment using standard methods (Stewart et al., 1967; Bebout et al., 1993).
Statistical analysis
To determine significant differences between H2 or organic acid production under control vs manipulated conditions, a one-way analysis of variance analysis was performed using the Tukey-HSD post hoc comparison within the JMP software package (v9; http://www.jmp.com/software/jmp9/). If a P-value of <0.05 was calculated between control and manipulated conditions, the difference was considered significant.
Nucleic acid isolation and cDNA synthesis
Microbial mat nucleic acids were isolated from at least three pooled mat cores, which were incubated in the same way as those analyzed for H2 and organic acid production. Mat cores were sampled periodically from serum vials and immediately transferred to 2 ml tubes, flash frozen in liquid nitrogen and stored at −80 °C. DNA and RNA were co-isolated from the upper photosynthetic layer (0–2 mm) of mat. DNA-free RNA was reverse transcribed to cDNA using random hexamers and a Superscript III first-strand synthesis kit (Invitrogen, Carlsbad, CA, USA). Further information regarding nucleic acid isolation, RNA purification and cDNA synthesis can be found in the supplementary methods.
Pyrotag sequencing of SSU rRNA genes and transcripts
The V8 hypervariable region of the SSU rRNA gene was amplified from DNA (rRNA gene) or cDNA (rRNA transcript) templates using the universal primer pair 926f/1392r, including the titanium adaptor sequences and a five-base barcode on the reverse primer according to the previously published protocols for pyrotag sequencing (Engelbrektson et al., 2010). Amplicons were quality checked, trimmed to a 200-bp sequence length and clustered (97% similarity) using PyroTagger (Kunin and Hugenholtz, 2010). Representative sequences from each cluster (OTU) were compared with the Greengenes NAST-aligned database (DeSantis et al., 2006) or the SILVA database (Pruesse et al., 2007) using PyroTagger (Kunin and Hugenholtz, 2010). Taxonomic affiliations of OTUs were further verified by querying the NCBI non-redundant nucleotide sequence databases (Altschul et al., 1990). If the best match was 100% identical over 200 bp, without multiple best matches to phylogenetically different taxa, then the OTU was conservatively assigned that taxonomic affiliation at the level of genus. If the OTU was <100%, but ⩾97% identical its taxonomic assignment was conservatively set to the level of order. OTUs with <97% identity matches were set to the level of phylum assigned by PyroTagger or if not assigned an identity by PyroTagger the OTU was unclassified (Kunin and Hugenholtz, 2010). Richness (Chao1 and ACE) and diversity (Simpson and Shannon) indices were calculated with the vegan package (v1.17–8) for R (v2.12; http://cran.r-project.org/web/packages/vegan/index.html). Hughes et al., 2001 and Hill et al., 2003 provide further information and critical review of these indices application in microbial ecology.
Design of degenerate bidirectional [NiFe] hydrogenase specific primers, PCR amplification, cloning and sequencing
A database of bidirectional [NiFe] hydrogenase gene sequences from a range of Bacteria and Archaea was compiled from the NCBI non-redundant nucleotide sequence database (Altschul et al., 1990) and the Integrated Microbial Genomes database (Markowitz et al., 2006). Gene sequences were converted to protein sequences to screen for the presence of bidirectional [NiFe] hydrogenase signature motifs L1 Ex[APV]xxxxRxCG[IL]Cxx[AS]Hx[IL][ACS][AGS][AGNSV][KR][ATV]xD and L2 DPC[IL]SC[AS][AST]H[ASTV]x[AG]xx[APV] (PROSITE format; Vignais and Billoud, 2007). Sequences with >2 mismatches to signature motifs were not included in the database. The screened bidirectional [NiFe] hydrogenase gene sequences were aligned using MUSCLE (Edgar, 2004) and the resulting alignments checked manually in Geneious (v5.3, http://www.geneious.com). The database of aligned bidirectional [NiFe] hydrogenase gene sequences was queried against candidate degenerate primers using PRIMROSE to determine the number of mismatches between primers and templates and direct selection of the optimal degenerate primer pair (Ashelford et al., 2002). The optimal degenerate primer pair that targeted the broadest diversity of Cyanobacteria and Chloroflexales corresponded to positions 13–19 (IEGHAKI) and positions 167–173 (WAVPGGV) in the M. chthonoplastes PCC7420 bidirectional [NiFe] hydrogenase protein sequence. Genomic DNA (gDNA) or cDNA was used as the template to amplify a ∼480-bp fragment of the bidirectional [NiFe] hydrogenase gene using the degenerate primers HoxH_F37 (5′-ATHGARGGHCAYGCBAARAT-3′) and HoxH_R518 (5′-ACNCCICCVGGNAYHGHCCA-3′) using the following PCR cycling conditions; 1 × 5 min, 95 °C (enzyme activation); 35 × 1 min, 95 °C (denature); 35 × 1 min, 56.5 °C (anneal); 35 × 1 min, 72 °C (extend) and 1 × 7 min, 72 °C (final extension). The 25 μl PCR reaction volume contained 12.5 μl GoTaq green master mix (contains 1.5 m MgCl2 and Taq enzyme; Promega, Madison, WI, USA), 8 μ of each primer (HoxH_F37/HoxH_R518), 20 μg bovine serum albumin and 1 μl template (10 ng genomic DNA or 1 μl of cDNA synthesis reaction). Positive and negative controls were also tested in PCR reactions (M. chthonoplastes PCC 7420 and Crocosphaera watsonii WH 8501, respectively) to evaluate the assay for efficiency and specificity. PCR products of the predicted size were excised from agarose gels after electrophoresis and purified using the Wizard SV gel and PCR clean-up system (Promega). Purified PCR product was ligated into the pCR2.1-topo vector (Invitrogen) and subsequently Sanger sequenced on a 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA).
Phylogenetic analysis of bidirectional [NiFe] hydrogenase clone sequences
Clone sequences were quality filtered, trimmed, translated to protein sequences, screened for L1 signature motif regions and aligned to a custom database of bidirectional [NiFe] hydrogenase protein sequences in Geneious (v5.3, http://www.geneious.com). Inferred hydrogenase amino-acid sequences were clustered at 97% similarity using CD-HIT (Huang et al., 2010) and representative sequences were queried against the NCBI non-redundant peptide sequence database (Altschul et al., 1990) to identify closest protein matches for phylogenetic analysis and tree building. Singletons were excluded from further analysis. Phylogenies were inferred using maximum-parsimony (PROTPARS; 1000 bootstraps) and maximum-likelihood (RAxML; PROTMIX rate distribution model; Dayhoff and WAG amino-acid substitution models; 1000 bootstraps) evolutionary models in ARB (Ludwig et al., 2004). Tree topologies from the different analyses were similar and the presented tree is based on a maximum-likelihood analysis, with bootstrap support of branch nodes indicated only when supported by all models. Representative clone sequences were deposited in GenBank (Accession numbers JF816258–JF816271).
Results
H2 production
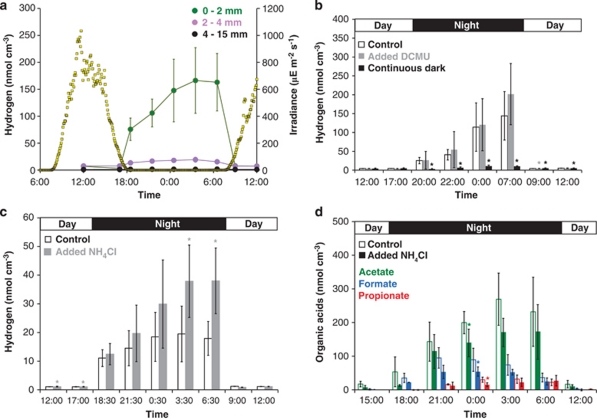
A time-course incubation of vertically sectioned mat was conducted to assess net H2 production in space and time. H2 production was observed predominantly at night under anoxic conditions within the upper photosynthetic layer (0–2 mm) of the mat and was an order of magnitude greater than that produced in lower layers (Figure 1a). H2 produced in intact mat cores (0–15 mm) was similar to H2 produced in the sectioned mat cores (0–2 mm; upper phototrophic layer) when calculated per surface area (exposed top side surface only; Supplementary Figure S1). This data further supported the observation that H2 production is principally occurring within the upper layer.
Figure 1.
Diel production of H2 or organic acids under manipulated conditions in Elkhorn Slough mats. Mat core incubations in vials commenced at dawn with H2 and organic acids allowed to accumulate over a diel cycle with N2 flushing of the vial at the end of the day and night periods. Vials in which organic acids were measured were reset at dawn by replacement with fresh field site water (amended as appropriate for manipulated conditions). Vertical error bars indicate the s.d. of six replicates (H2) or three replicates (organic acids). Asterisks denote significant differences between control and manipulated conditions (P values <0.05). (a) H2 production in different layers of microbial mat. Mat cores were sectioned at different depths, 0–2 mm, 2–4 mm and 4–15 mm. Yellow squares denote natural solar irradiance. μE, microeinsteins. (b) H2 production under control, DCMU (20 μ) added and continuous dark conditions. (c) H2 production under control and NH4Cl (8.8 m) added conditions. (d) Organic acid production under control and NH4Cl (8.8 m) added conditions.
Patterns of H2 and organic acid production observed in control and manipulation experiments reported here (Figure 1) were reproducible in mats sampled at other times during December–January 2008/2009 and August–October of 2009 (data not shown). However, the magnitude of H2 and organic acid production sometimes varied between diel cycle studies. This may have been due to community structure heterogeneity between different cores sampled from microbial mat pieces at different times or it may have been due to variable physiological states of hydrogenogens in the system.
H2 production under phototroph suppressed conditions
Manipulations of photosynthetic activity were performed to assess the role of different photosynthetic pathways and organisms in H2 production as explained below. In one set of experiments anoxygenic and oxygenic photosynthetic activity facilitated by photosystem (PSI) and photosytem II (PSII), respectively, was suppressed by depriving the mat of all light and thereby imposing continuously dark conditions over a complete diel cycle. In a second set of experiments performed under normal light conditions, 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), a well-known inhibitor of PSII was added to inhibit oxygenic photosynthesis specifically. Inhibition of PSII was confirmed using PAM fluorometry (Supplementary Table S1) and was further supported anecdotally by the absence of oxygen bubbles, which normally appear at the surface of the mat (Supplementary Figure S2). Light deprivation and the subsequent cessation of all photosynthetic activity during the day reduced nighttime H2 production on the following night by an order of magnitude. However, specific and complete inhibition of oxygenic phototrophy did not significantly affect H2 production under dark, anoxic conditions (Figure 1b). Together these results indicate that prior-day activity of photosynthetic microorganisms is the source of reducing equivalents for H2 production and further suggest that anoxygenic phototrophy, facilitated by PSI, is fully capable of serving this function in Elkhorn Slough mats. However, these data alone do not distinguish between H2 production occurring predominantly by (i) obligate oxygenic phototrophs that are replaced by anoxygenic phototrophs under the conditions of our experimental manipulation, (ii) oxygenic phototrophs capable of facultative anoxygenic phototrophy, (iii) anoxygenic phototrophs; or (iv) a combination of (i)–(iii).
Production of H2 and organic acids under N2-fixation suppressed conditions
To evaluate the relative importance of co-metabolic production of H2 during N2-fixation vs fermentative H2 production, both of which are consistent with the observed pattern of H2 production in space and time, we suppressed N2-fixation. This was performed by addition of a combined nitrogen source (NH4Cl). Nighttime N2-fixation was completely suppressed when mats were incubated in the presence of NH4Cl (Supplementary Figure S3), but H2 production was not suppressed (Figure 1c). During the same nighttime period acetate, formate and propionate were produced with H2 under dark and anoxic conditions (expected products of fermentation; Figure 1d). Lactate, butyrate and valerate were not detected (<200 n). Organic acid accumulation was not significantly different when N2-fixation was suppressed under the same conditions (Figure 1d), with the exception of one time point (0:00). Together these results indicate that N2-fixation is a relatively insignificant contributor to H2 formation and suggest that fermentation is the dominant mechanism of production.
SSU rRNA genes and transcripts
To profile the identity and activity of microorganisms in Elkhorn Slough microbial mats, we compared rRNA gene and transcript pyrosequence tags from four different time points, including day and night samples (Table 1). For each time point DNA and RNA templates were co-isolated from the upper photosynthetic layer (0–2 mm), as our biogeochemical data demonstrated that H2 was predominantly produced within this layer.
Table 1. Sampling times and statistics of rRNA gene and transcript pyrotag sequencing.
| Library | Time and date | Pool | Filtered reads (+) singletons | Filtered reads (−) singletons | OTUs (97%) (+) singletons | OTUs (97%) (−) singletons |
|---|---|---|---|---|---|---|
| No. 1 | Day, 1200, 10/11/2009 | DNA | 5796 | 5576 | 737 | 380 |
| No. 2 | Day, 1200, 10/11/2009 | RNA | 5756 | 5641 | 486 | 233 |
| No. 3 | Night, 0000, 11/11/2009 | DNA | 1191 | 1147 | 291 | 125 |
| No. 4 | Night, 0000, 11/11/2009 | RNA | 5310 | 5230 | 357 | 166 |
| No. 5 | Day, 1200, 12/11/2009 | DNA | 4487 | 4402 | 384 | 194 |
| No. 6 | Day, 1200, 12/11/2009 | RNA | 4825 | 4780 | 242 | 121 |
| No. 7 | Night, 0000, 13/11/2009 | DNA | 5573 | 5234 | 866 | 390 |
| No. 8 | Night, 0000, 13/11/2009 | RNA | 4091 | 4038 | 275 | 121 |
Richness estimates calculated using Chao1 and ACE indices suggested that we had not exhaustively sequenced the upper photosynthetic layer of the microbial mat community (Table 2). Microbial diversity was consistently estimated to be highest in the rRNA gene libraries as determined using either the Simpson (inverse) or Shannon indices, with each rRNA gene library value higher than the value for the corresponding rRNA transcript library (Table 2).
Table 2. Richness and diversity estimates of OTUs (97% similarity).
| Library |
OTU richnessa |
OTU diversityb |
|||
|---|---|---|---|---|---|
| Observed | Chao1 | ACE | Simpsonc | Shannon | |
| No. 1 (Day 1d, DNA) | 737 | 1309 (±86) | 1334 (±22) | 52.84 | 5.08 |
| No. 2 (Day 1, RNA) | 486 | 880 (±70) | 952 (±19) | 5.30 | 3.15 |
| No. 3 (Night 1d, DNA) | 291 | 633 (±81) | 616 (±16) | 29.55 | 4.52 |
| No. 4 (Night 1, RNA) | 357 | 645 (±59) | 681 (±15) | 3.88 | 2.63 |
| No. 5 (Day 2d, DNA) | 384 | 723 (±72) | 715 (±16) | 14.10 | 3.77 |
| No. 6 (Day 2, RNA) | 242 | 469 (±62) | 433 (±13) | 3.38 | 2.18 |
| No. 7 (Night 2d, DNA) | 866 | 1679 (±107) | 1813 (±28) | 35.20 | 4.96 |
| No. 8 (Night 2, RNA) | 275 | 543 (±63) | 613 (±16) | 3.93 | 2.50 |
Chao1 and ACE (abundance-based coverage estimators) indices are non-parametric estimators of species (OTU) richness (total number of different OTUs in a given sample).
Shannon and Simpson diversity indices consider both species richness and evenness (relative abundance).
Inverse simpson values are reported.
Day 1 is 1200 hours, on 10 November, 2009; Night 1 is 0000 hours, on 11 November, 2009; Day 2 is 1200 hours, on 12 November, 2009; Night 2 is 0000 hours, on 13 November, 2009.
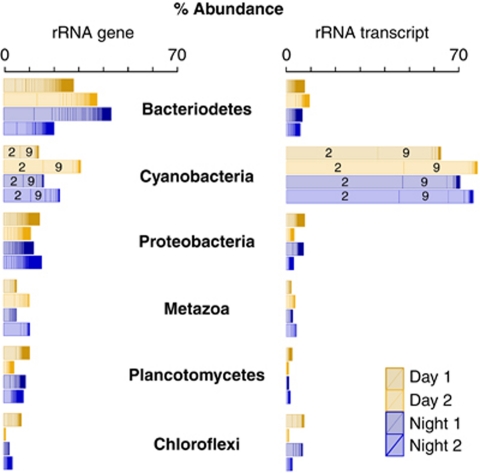
Analysis of all four rRNA gene libraries (n=17 047 sequences) revealed that 64 phyla/divisions across all three domains of life were present in the upper photosynthetic layer of Elkhorn Slough microbial mats (Supplementary Table S2). Abundant phyla/divisions included, Bacteriodetes (29%), Cyanobacteria (22%) and Proteobacteria (14%) (average of four libraries; Figure 2). A lower diversity of phyla/divisions were recovered from the upper photosynthetic layer of all four rRNA transcript libraries (n=19 982 sequences), with 49 phyla/divisions represented (Supplementary Table S3). Cyanobacteria dominated these libraries, representing 71% of all reads (average of four libraries; Figure 2). The rRNA transcript libraries were dominated by Microcoleus spp. affiliated OTU 2 (Library no. 2, Day 1, 37% Library no. 4, Night 1, 47% Library no. 6, Day 2, 48% Library no. 8, Night 2, 46% Figure 2) and Cyanobacteria affiliated OTU 9 (Library no. 2, Day 1, 22% Library no. 4, Night 1, 18% Library no. 6, Day 2, 27% Library no. 8, Night 2, 20% Figure 2). These OTUs are likely important members of the active microbial mat community during the day and also at night.
Figure 2.
Ranked barplot of the six most abundant phyla of the rRNA gene (left) and rRNA transcript (right) libraries. Day 1 is 12:00 on 10 November, 2009, Day 2 is 12:00 on 11 November, 2009, Night 1 is 0:00 on 11 November, 2009 and Night 2 is 0:00 on 13 November, 2009. OTUs within the same phyla are stacked in order of relative abundance and separated graphically into segments. The most abundant OTUs no. 2, Microcoleus spp. and no. 9, Cyanobacteria) are labeled according to their (arbitrary) OTU identification number assigned during PyroTagger clustering.
Relative activity of microbial populations was inferred at the transcript level from the ratio of the fraction of tags in the rRNA transcript library (active microorganisms) to those in the paired rRNA gene library (active+inactive microorganisms) for a given time point. OTUs were considered relatively active if the % rRNA transcripts recovered was greater than the % rRNA genes recovered that is, % rRNA transcripts:% rRNA genes ratio >1 (Rodriguez-Blanco et al., 2009; Jones and Lennon, 2010). These ratios may give additional insight into active microorganisms beyond those that we previously identified from our rRNA transcript libraries (the ‘abundant and active' OTUs 2 and 9) and identify those organisms that are relatively ‘rare and active' that are not obvious from examination of rRNA transcript libraries alone. We were specifically interested in detecting microorganisms correlated with H2 production and thus we report in Table 3 only those OTUs that were most relatively active at night and present in all our nighttime libraries. Microcoleus spp. (OTU 2) had the highest nighttime % transcript:% gene ratios of all OTUs analyzed (present in all libraries), 6.0 (Nov/11/2009) and 4.2 (Nov/13/2009). An OTU (225) with the second highest nighttime ratios was affiliated with the Desulfobacterales (Table 3). Bacteria within this order are exclusively sulfate-reducing bacteria (SRB) and these microorganisms may also be H2-consumers (hydrogenotrophs; Widell, 1987). The most relatively active nighttime OTUs present in all nighttime libraries were predominantly microbial groups within either the Cyanobacteria or Chloroflexi. These data correlate with our biogeochemical data and generally support the hypothesis that Cyanobacteria, including Microcoleus spp. (oxygenic phototrophs) and/or members of the Chloroflexi (anoxygenic phototrophs) are hydrogenogens in Elkhorn Slough mats.
Table 3. rRNA % transcript :% gene ratios of the most relatively active OTUs observed in all nighttime pyrotag libraries.
| OTUa | Most related sequencesb | Accession number | Identitiesc | E-valued | OTU taxonomic affiliatione | Ratiof night 1, 11/11/2009 | Ratiof night 2, 13/11/2009 |
|---|---|---|---|---|---|---|---|
| 2 | Microcoleus chthonoplastes | AM709630 | 200/200 | 6e-97 | Microcoleus spp. | 6.0 | 4.2 |
| Microcoleus chthonoplastes | GQ402023 | 200/200 | 6e-97 | ||||
| 225 | Desulfococcus sp. | EF442980 | 194/200 | 9e-89 | Desulfobacterales | 5.5 | 2.6 |
| Desulfococcus multivorans | AF418173 | 194/200 | 9e-89 | ||||
| 5 | Leptolyngbya fragilis | HQ832925 | 198/200 | 3e-94 | Oscillatoriales | 4.1 | 3.2 |
| Coral bacterium clone | FJ203604 | 193/200 | 4e-87 | ||||
| 9 | Mat bacterium clone | EU246323 | 200/200 | 7e-97 | Cyanobacteria | 3.3 | 3.3 |
| Phormidium cf. formosumb | EU196640 | 190/200 | 2e-83 | ||||
| 31 | Mat chloroflexales clone | AJ308496 | 200/200 | 7e-97 | Chloroflexales | 4.8 | 1.5 |
| Oscillochloris sp.b | AF149018 | 188/200 | 2e-78 | ||||
| 387 | Microbialite nematoda clone | GQ483712 | 200/200 | 7e-97 | Nematoda | 2.1 | 3.7 |
| Eumonhystera cf. simplexb | AY284692 | 177/200 | 6e-66 | ||||
| 379 | Wetland cytophagales clone | FJ516772 | 188/200 | 1e-80 | Bacteriodetes | 1.6 | 3.8 |
| Aquiflexum balticumb | NR025634 | 186/200 | 7e-78 | ||||
| 1176 | Mat chloroflexi clone | DQ329977 | 199/200 | 3e-95 | Chloroflexi | 1.9 | 2.9 |
| Oscillochloris sp.b | AF149018 | 178/200 | 5e-67 |
OTU identification number is arbitrary and wa designated by PyroTagger.
The NCBI non-redundant nucleotide sequence database was queried to identify closest matches to OTU sequences; closest pure culture match where a given OTU has its closest matches to only uncultured clone sequences.
BLASTN uses an identity matrix for nucleotide comparisons, see http://www.ncbi.nlm.nih.gov/BLAST/; total length of query was 200 bp.
E-value or expect value describes the significance of the match between two sequences, see http://www.ncbi.nlm.nih.gov/BLAST/.
For OTU taxonomic affiliation designation see methods (Pyrotag sequencing of SSU rRNA genes and transcripts).
rRNA % transcript:% gene ratios were calculated for sequences recovered from DNA or cDNA template-based libraries constructed from Night 1 (0000 hours) on 11 November, 2009 or Night 2 (0000 hours) on 13 November, 2009 samples by dividing the percentage abundance of pyrotags in the rRNA transcript library by the percentage abundance in the rRNA gene library.
H2-evolving bidirectional [NiFe] hydrogenase genes and transcripts
H2-evolving bidirectional [NiFe] hydrogenases are found in a diversity of Cyanobacteria, Chloroflexi and Proteobacteria (also other Bacteria and Archaea) and are the only known hydrogenases in these taxonomic groups capable of evolving H2 via fermentation (Tamagnini et al., 2007; Vignais and Billoud, 2007). Thus, genes encoding for these enzymes represent useful biomarkers for evaluating the diversity of hydrogenogens in photosynthetic microbial mats. [NiFe] hydrogenases are quite diverse (Vignais and Billoud, 2007) and therefore it is impossible to design degenerate primers that capture the entire diversity that may be present in highly complex ecosystems such as microbial mats. However, as our biogeochemical and pyrotag sequencing data indicated that the important hydrogenogens in this system are potentially members of the Cyanobacteria and Chloroflexales, we focused on amplifying bidirectional [NiFe] hydrogenases from these two groups, for which it was possible to design a single PCR-based assay.
Amplicons were generated that spanned the L1 signature motif region of the large subunit of the bidirectional [NiFe] hydrogenase. The primer pair HoxH_F37/HoxH_R518 designed in this study generated an amplicon of ∼480 bp, shorter than the bidirectional [NiFe] hydrogenase-specific primer pair HoxHf/HoxHr that produces an ∼1190-bp amplicon (Barz et al., 2010). However, HoxH_F37 and HoxH_R518 have substantially fewer primer-template mismatches to publicly available bidirectional [NiFe] hydrogenase genes than the bidirectional [NiFe] hydrogenase-specific primer pair HoxHf and HoxHr (Supplementary Table S4). Of the 39 publicly available Cyanobacteria or Chloroflexales bidirectional [NiFe] hydrogenases 38 had ⩽2 mismatches to HoxH_F37/HoxH_R518 compared with 9 of 39 for HoxHf/HoxHr, indicating that HoxH_F37/HoxH_R518 targets a larger portion of bidirectional [NiFe] hydrogenases from Cyanobacteria and Chloroflexales. Analysis of a selection of uptake [NiFe] hydrogenases showed that all had ⩾3 primer-template mismatches to HoxH_F37/HoxH_R518 (Supplementary Table S4). Amplicons of the expected size were generated from a selection of Cyanobacteria strains and one Chloroflexales strain that have a bidirectional [NiFe] hydrogenase, but these amplicons were not generated from Crocosphaera watsonii WH 8501, a cyanobacterium that does not have a bidirectional [NiFe] hydrogenase but has an uptake [NiFe] hydrogenase (Supplementary Table S5).
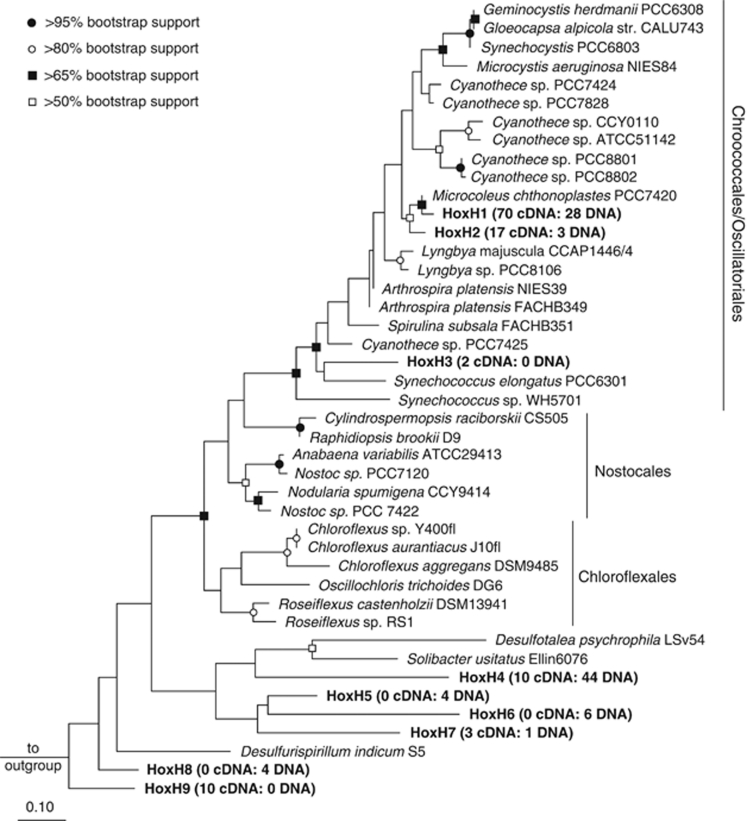
A total of 205 bidirectional [NiFe] hydrogenase gene and transcript fragments were amplified from DNA (91 clones) and RNA (cDNA; 114 clones) templates obtained from the upper photosynthetic layer (0–2 mm) of Elkhorn Slough mats under dark anoxic conditions at 0000 hours, on 11 November, 2009. The gene and transcript fragments formed nine distinct clusters (including five singleton clusters) at the 97% similarity threshold (Table 4; Figure 3). The majority of the sequences were affiliated with M. chthonoplastes PCC7420 (HoxH1 and HoxH2 clusters). The HoxH1 cluster represented 31% of gene clones and 61% of transcript clones (98% identity; >65% bootstrap support for branch point) and the HoxH2 cluster represented 3% of gene clones and 15% of transcript clones (96% identity; >50% bootstrap support for branch point). Another highly represented cluster, HoxH4 (48% of gene clones; 9% of transcript clones), was most closely related to Cyanothece sp. PCC8802 (70% identity; unresolved branch point). Clusters HoxH1 and HoxH2 were relatively active, with a % transcript:% gene ratio >1, whereas HoxH4 was relatively inactive with a % transcript:% gene ratio <1.
Table 4. % transcript:% gene ratios of bidirectional [NiFe] hydrogenase sequence clusters.
| Sequence cluster | Most related sequencesa | Accession number | % ID | Transcriptb clones | Genec clones | Ratiod |
|---|---|---|---|---|---|---|
| HoxH1 | Microcoleus chthonoplastes PCC7420 | ZP_05027024 | 98 | 70 (61%) | 28 (31%) | 2.0 (61%:31%) |
| Arthrospira maxima CS328 | ZP_03273562 | 92 | ||||
| HoxH2 | Microcoleus chthonoplastes PCC7420 | ZP_05027024 | 96 | 17 (15%) | 3 (3%) | 4.5 (15%:31%) |
| Cyanothece sp. PCC7822 | YP_003888687 | 93 | ||||
| HoxH3 | Arthrospira platensis FACHB439 | AAQ63960 | 87 | 2 (2%) | 0 (0%) | —e |
| Arthrospira maxima CS328 | ZP_03273562 | 87 | ||||
| HoxH4 | Cyanothece sp. PCC8802 | YP_003135898 | 70 | 10 (9%) | 44 (48%) | 0.2 (9%:48%) |
| Cyanothece sp. PCC8801 | YP_002370357 | 70 | ||||
| HoxH5 | Roseiflexus castenholzii DSM13941 | YP_001431482 | 73 | 0 (0%) | 4 (4%) | — |
| Anaerolinea thermophila UNI1 | YP_004173889 | 73 | ||||
| HoxH6 | Anabaena variabilis ATCC29413 | YP_325153 | 68 | 0 (0%) | 6 (7%) | — |
| Synechococcus sp. PCC7335 | ZP_05039794 | 68 | ||||
| HoxH7 | Cyanothece sp. PCC8801 | YP_002370357 | 68 | 3 (3%) | 1 (1%) | 2.4 (3%:1%) |
| Solibacter usitatus Ellin6076 | YP_826256 | 68 | ||||
| HoxH8 | Roseiflexus sp. RS1 | YP_001277847 | 80 | 0 | 2 (2%) | — |
| Nostoc sp. PCC7120 | NP_484809 | 80 | ||||
| HoxH9 | Desulfurispirillum indicum S5 | YP_004113284 | 78 | 10 (9%) | 0 (0%) | — |
| Nostoc sp. PCC7120 | NP_484809 | 75 |
Amino-acid sequences inferred from nucleotide sequences.
Total transcript clones=114, including two singletons. Percentage abundance (rounded to nearest whole number) of a given clone recovered from the transcript library is denoted in parenthesis.
Total gene clones=91, including three singletons. Percentage abundance (rounded to nearest whole number) of a given clone recovered from the gene library is denoted in parenthesis.
% transcript:% gene ratios calculated by dividing percentage abundance of transcript clones by the percentage abundance of gene clones.
Ratio was not calculated as numerator or denominator was 0%.
Figure 3.
Maximum-likelihood phylogenetic tree of bidirectional [NiFe] hydrogenases. Branch points supported by all phylogenetic analyses (maximum likelihood using Dayhoff or WAG amino-acid correction and maximum parsimony evolutionary models) and their bootstrap support are denoted. The bar represents 0.1 changes per amino acid and the outgroup was Methanococcus voltae A3 HoxH.
Discussion
Analyses of rRNA and H2-evolving bidirectional [NiFe] hydrogenase genes and transcripts combined with biogeochemical measurements across a range of manipulations strongly suggest that fermentation of stored photosynthate by Microcoleus spp. is a major source of H2 in Elkhorn Slough mats.
H2 production occurred predominantly in the upper photosynthetic layer (0–2 mm) of Elkhorn Slough mats. Similar results have been reported in other photosynthetic mats from Guerrero Negro, Mexico (Hoehler et al., 2001), suggesting that quantitatively important hydrogenogens are active in the upper layer of geographically diverse mats. Elkhorn Slough mats also harbored SRB (Desulfobacterales; Table 3), a group known to be actively reducing sulfate over the full diel cycle in photosynthetic microbial mats (Canfield and Des Marais, 1991). These microorganisms include hydrogenotrophs (Widdel, 1987) and thus H2 production observed in our experiments is likely a measurement of net H2 production. Our observations cannot rule out that enhanced H2 consumption in deeper layers accounts for the lower net H2 production relative to the upper photosynthetic layer. Indeed, we anticipate that more stable physico-chemical conditions within the permanently dark, anoxic portion of the mat should allow for the development of closer associations between hydrogenogens and hydrogenotrophs, and thus a lower net efflux of H2 than in the highly dynamic upper phototrophic layer. However, we hypothesize that the observed rates of H2 and organic acid production are unlikely to be sustained in lower aphotic layers, which lack daily photosynthetic replenishment of the fixed carbon pool and therefore have a discrete reservoir of carbon that would be rapidly depleted. Further investigation of hydrogenotrophs is needed to better understand the complete H2 cycle and controls on net H2 production in microbial mats.
Previous research of microbial mats from Shark Bay, Australia had concluded that co-metabolic production of H2 during N2-fixation could be an important contributor to net H2 production as N2-fixation was observed to occur contemporaneously with H2 production (Skyring et al., 1989). Similar patterns of N2-fixation and H2 production were observed in mats from Elkhorn Slough. However, further examination of the role of N2-fixation in H2 production using N2-fixation suppression experiments indicated that H2-production is independent of N2-fixation and most likely fermentative due to the simultaneous production of organic acids (expected fermentation products) under dark and anoxic conditions.
Inhibition of phototrophic activity generally or oxygenic phototrophy specifically, by respectively, depriving the microbial mats of solar irradiance or by inhibiting with DCMU suggested that phototrophs were important contributors to the H2 production observed during the subsequent nighttime period. However, biogeochemical data alone did not provide evidence to identify the type of phototroph or the taxonomic identity of microorganisms producing H2.
Our analysis of rRNA genes and transcripts demonstrated that Cyanobacteria, specifically Microcoleus spp. were important members of the active microbial community. We obtained results similar to an investigation of photosynthetic mats from Guerrero Negro, Mexico by Ley and colleagues (2006). In both studies, Cyanobacteria dominated the active microbial communities as determined by rRNA transcript analysis despite being substantially less significant within the total (active+inactive) community as determined by rRNA gene analysis. These observations underscore the need to investigate both the presence and activity of microorganisms in complex microbial ecosystems to gain a better understanding of their likely metabolic importance.
Some cyanobacterial strains isolated in pure culture from microbial mats are capable of fermentative H2 production from stored photosynthate, including glycogen (Stal and Moezelaar, 1997). No studies have been carried out on true Microcoleus sp. isolates to ascertain their capabilities of producing H2 (Garcia-Pichel et al., 1996; Moezelaar et al., 1996; Bolhuis et al., 2010). However, recent analysis of publicly available cyanobacterial genomes have shown that all Cyanobacteria (including M. chthonoplastes PCC7420) harboring a bidirectional [NiFe] hydrogenase also harbor a pyruvate:flavodoxin/ferredoxin oxidoreductase, leading to speculation that the bidirectional [NiFe] hydrogenase is used to shunt electrons (as H2) via a pyruvate:flavodoxin/ferredoxin oxidoreductase-like enzyme during fermentation (Barz et al., 2010). H2-evolving bidirectional [NiFe] hydrogenase clone libraries constructed from the upper photosynthetic layer of Elkhorn Slough mats using degenerate primers designed to equally target many Cyanobacteria and Chloroflexales were dominated by sequence clusters most closely affiliated with M. chthonoplastes PCC7420 (Figure 3). Overrepresentation of hydrogenases in the transcript library for clusters most closely affiliated with M. chthonoplastes PCC7420 (Table 4) provides further evidence that Microcoleus spp. are the main hydrogenogens in Elkhorn Slough mats.
Data obtained in this study indicate that anoxygenic phototrophy can drive H2 production in microbial mats. Facultative anoxygenic phototrophy in Cyanobacteria, including Microcoleus sp. has been reported (Garlick et al., 1977; Cohen et al., 1986; De Wit et al., 1988). However, misidentification of cyanobacterial strains as Microcoleus sp., including ‘strain 11' used by de Wit and colleagues (1988) has been documented (Garcia-Pichel et al., 1996; Bolhuis et al., 2010) and thus it is unclear if Microcoleus spp. are capable of anoxygenic phototrophy. The capability of Microcoleus spp. to switch between oxygenic and anoxygenic phototrophy might be important to their survival and proliferation in microbial mats where sulfurous compounds can be prevalent and inhibit PSII activity (Jorgensen et al., 1986). Alternatively, anoxygenic phototrophic Chloroflexi strains may have a role in H2 production. Analysis of nighttime rRNA gene and transcript libraries revealed that OTU 31 affiliated with Chloroflexales and OTU 1176 affiliated with Chloroflexi were overrepresented in the rRNA transcript libraries. These OTUs had % transcript:% gene ratios >1 in both nighttime samples (Table 3), indicating that they were consistently relatively active when H2 production was pronounced. Cloning of H2-evolving bidirectional [NiFe] hydrogenases did not indicate that Chloroflexales bidirectional [NiFe] hydrogenases were expressed. It should be noted that although degenerate primers were designed to amplify bidirectional [NiFe] hydrogenases from a diversity of Cyanobacteria and Chloroflexales, a relatively high number of mismatches (⩾2) between both forward and reverse primers to the bidirectional [NiFe] hydrogenase gene was determined for one of the Chloroflexales members, Oscillochloris trichoides DG6 (Supplementary Table S4). Thus, it is possible that Oscillochloris-like spp. or other as yet unknown Chloroflexales may contribute to the observed H2 produced in Elkhorn Slough mats without being represented in the hydrogenase gene and transcript libraries. In this regard it is noteworthy that the closest pure culture match to OTUs 31 and 1176, was Oscillochloris sp. BM (Table 3).
Although we do not rule out a role for Chloroflexales in H2 production, analysis of rRNA pyrotags and bidirectional [NiFe] hydrogenases indicates that Cyanobacteria, including Microcoleus spp., were relatively more active than Chloroflexales. Also, the dominance of Microcoleus spp. within the active fraction of the microbial community in the H2 producing layer suggests that the contribution of microorganisms other than Cyanobacteria to H2 production is likely minimal. Together, these biogeochemical data, manipulations and molecular data demonstrate that Microcoleus spp. are dominant hydrogenogens in Elkhorn Slough mats.
Acknowledgments
We thank Michael Kubo, Adrienne Frisbee, Angela Detweiler and Erich Fleming for technical support. We thank Tijana Glavina del Rio, Susannah Tringe and Stephanie Malfatti of the Joint Genome Institute for assistance obtaining and analyzing amplicon pyrosequencing. Funding was provided by the U.S. Department of Energy (DOE) Genomic Sciences Program under contract SCW1039. Work at LLNL was performed under the auspices of the U.S. Department of Energy at Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344. Work at LBNL was performed under the auspices of the U.S. Department of Energy at Lawrence Berkeley National Laboratory under Contract DE-AC02-05CH11231. DW was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ashelford KE, Weightman AJ, Fry JC. PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Res. 2002;30:3481–3489. doi: 10.1093/nar/gkf450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barz M, Beimgraben C, Staller T, Germer F, Opitz F, Marquardt C, et al. Distribution analysis of hydrogenases in surface waters of marine and freshwater environments. PLoS ONE. 2010;5:e13846. doi: 10.1371/journal.pone.0013846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebout BM, Fitzpatrick MW, Paerl HW. Identification of the sources of energy for nitrogen fixation and physiological characterization of nitrogen-fixing members of a marine microbial mat community. Appl Environ Microbiol. 1993;59:1495–1503. doi: 10.1128/aem.59.5.1495-1503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J, Phillips P. Microbial mats for multiple applications in aquaculture and bioremediation. Bioresour Technol. 2004;94:229–238. doi: 10.1016/j.biortech.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Bolhuis H, Severin I, Confurius-Guns V, Wollenzien UI, Stal LJ. Horizontal transfer of the nitrogen fixation gene cluster in the cyanobacterium Microcoleus chthonoplastes. ISME J. 2010;4:121–130. doi: 10.1038/ismej.2009.99. [DOI] [PubMed] [Google Scholar]
- Bolhuis H, Stal LJ.2011Analysis of bacterial and archaeal diversity in coastal microbial mats using massive parallel 16S rRNA gene tag sequencing ISME Je-pub ahead of print 5 May 2011; doi: 10.1038/ismej.2011.52 [DOI] [PMC free article] [PubMed]
- Bothe H, Schmitz O, Yates MG, Newton WE. Nitrogen fixation and hydrogen metabolism in cyanobacteria. Microbiol Mol Biol R. 2010;74:529. doi: 10.1128/MMBR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd ES, Hamilton TL, Spear JR, Lavin M, Peters JW. [FeFe]-hydrogenase in yellowstone national park: evidence for dispersal limitation and phylogenetic niche conservatism. ISME J. 2010;4:1485–1495. doi: 10.1038/ismej.2010.76. [DOI] [PubMed] [Google Scholar]
- Boyd ES, Spear JR, Peters JW. [FeFe] hydrogenase genetic diversity provides insight into molecular adaptation in a saline microbial mat community. Appl Environ Microbiol. 2009;75:4620–4623. doi: 10.1128/AEM.00582-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield DE, Des Marais DJ. Aerobic sulfate reduction in microbial mats. Science. 1991;251:1471–1473. doi: 10.1126/science.11538266. [DOI] [PubMed] [Google Scholar]
- Cohen Y, Jorgensen BB, Revsbech NP, Poplawski R. Adaptation to hydrogen sulfide of oxygenic and anoxygenic photosynthesis among cyanobacteria. Appl Environ Microbiol. 1986;51:398–407. doi: 10.1128/aem.51.2.398-407.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit R, van Boekel WHM, van Gemerden H. Growth of the cyanobacterium Microcoleuschthonoplastes on sulfide. FEMS Microbiol Ecol. 1988;53:203–209. [Google Scholar]
- Des Marais DJ. Biogeochemistry of hypersaline microbial mats illustrates the dynamics of modern microbial ecosystems and the early evolution of the biosphere. Biol Bull. 2003;204:160–167. doi: 10.2307/1543552. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, et al. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 2010;4:642–647. doi: 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- Garcia-Pichel F, Prufert-Bebout L, Muyzer G. Phenotypic and phylogenetic analyses show Microcoleus chthonoplastes to be a cosmopolitan cyanobacterium. Appl Environ Microbiol. 1996;62:3284–3291. doi: 10.1128/aem.62.9.3284-3291.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlick S, Oren A, Padan E. Occurence of facultative anoxygenic photosynthesis among filamentous and unicellular cyanobacteria. J Bacteriol. 1977;129:623–629. doi: 10.1128/jb.129.2.623-629.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TC, Walsh KA, Harris JA, Moffett BF. Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol. 2003;43:1–11. doi: 10.1111/j.1574-6941.2003.tb01040.x. [DOI] [PubMed] [Google Scholar]
- Hoehler TM, Bebout BM, Des Marais DJ. The role of microbial mats in the production of reduced gases on the early Earth. Nature. 2001;412:324–327. doi: 10.1038/35085554. [DOI] [PubMed] [Google Scholar]
- Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA. 2010;107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen BB, Cohen Y, Revsbech NP. Transition from anoxygenic to oxygenic photosynthesis in a Microcoleus chthonoplastes cyanobacterial mat. Appl Environ Microbiol. 1986;51:408–417. doi: 10.1128/aem.51.2.408-417.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Hugenholtz P. PyroTagger: A fast, accurate pipeline for analysis of rRNA amplicon pyrosequence data. Open J. 2010;1:e1. [Google Scholar]
- Kunin V, Raes J, Harris J, Spear J, Walker J, Ivanova N, et al. Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol Syst Biol. 2008;4:e198. doi: 10.1038/msb.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Vermaas WFJ, Rittmann BE. Biological hydrogen production: prospects and challenges. Trends Biotechnol. 2010;28:262–271. doi: 10.1016/j.tibtech.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Ley RE, Harris JK, Wilcox J, Spear JR, Miller SR, Bebout BM, et al. Unexpected diversity and complexity of the guerrero negro hypersaline microbial mat. Appl Environ Microbiol. 2006;72:3685–3695. doi: 10.1128/AEM.72.5.3685-3695.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, Padki A, et al. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 2006;34:D344–D348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moezelaar R, Bijvank SM, Stal LJ. Fermentation and sulfur reduction in the mat-building cyanobacterium Microcoleus chthonoplastes. Appl Environ Microbiol. 1996;62:1752–1758. doi: 10.1128/aem.62.5.1752-1758.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisbet EG, Fowler CMR. Archaean metabolic evolution of microbial mats. Proc R Soc Lond B Biol Sci. 1999;266:2375–2382. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Blanco A, Ghiglione JF, Catala P, Casamayor EO, Lebaron P. Spatial comparison of total vs active bacterial populations by coupling genetic fingerprinting and clone library analyses in the NW Mediterranean Sea. FEMS Microbiol Ecol. 2009;67:30–42. doi: 10.1111/j.1574-6941.2008.00591.x. [DOI] [PubMed] [Google Scholar]
- Skyring GW, Lynch RM, Smith GD. Acetylene reduction and hydrogen metabolism by a cyanobacterial/sulfate-reducing bacterial mat ecosystem. Geomicrobiol J. 1988;6:25–31. [Google Scholar]
- Skyring GW, Lynch RM, Smith GD.1989Quantitative relationship between carbon, hydrogen and sulfur metabolism in cyanobacterial matsIn Cohen Y, Rosenberg E (eds),Microbial mats: Physiological ecology of benthic microbial communities American Society for Microbiology: Washington, DC; 170–179. [Google Scholar]
- Stal LJ, Moezelaar R. Fermentation in cyanobacteria. FEMS Microbiol Rev. 1997;21:179–211. [Google Scholar]
- Stewart WD, Fitzgerald GP, Burris RH. In situ studies on N2-fixation using the acetylene reduction technique. Proc Natl Acad Sci USA. 1967;58:2071–2078. doi: 10.1073/pnas.58.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnini P, Leitao E, Oliveira P, Ferreira D, Pinto F, Harris DJ, et al. Cyanobacterial hydrogenases: diversity, regulation and applications. FEMS Microbiol Rev. 2007;31:692–720. doi: 10.1111/j.1574-6976.2007.00085.x. [DOI] [PubMed] [Google Scholar]
- Vignais PM, Billoud B. Occurrence, classification, and biological function of hydrogenases: an overview. Chem Rev. 2007;107:4206–4272. doi: 10.1021/cr050196r. [DOI] [PubMed] [Google Scholar]
- Widdel F. New types of acetate-oxidizing, sulfate-reducing Desulfobacter species, D hydrogenophilus sp. nov, D latus sp. Nov and D curvatus sp. nov. Arch Microbiol. 1987;148:286–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.