Abstract
Assembling individual genomes from complex community metagenomic data remains a challenging issue for environmental studies. We evaluated the quality of genome assemblies from community short read data (Illumina 100 bp pair-ended sequences) using datasets recovered from freshwater and soil microbial communities as well as in silico simulations. Our analyses revealed that the genome of a single genotype (or species) can be accurately assembled from a complex metagenome when it shows at least about 20 × coverage. At lower coverage, however, the derived assemblies contained a substantial fraction of non-target sequences (chimeras), which explains, at least in part, the higher number of hypothetical genes recovered in metagenomic relative to genomic projects. We also provide examples of how to detect intrapopulation structure in metagenomic datasets and estimate the type and frequency of errors in assembled genes and contigs from datasets of varied species complexity.
Keywords: metagenome, assembly, Illumina
Introduction
Next generation sequencing (NGS) technologies such as the Roche 454 and the Illumina/Solexa (Bennett, 2004; Margulies et al., 2005) are revolutionizing the study of natural microbial communities (DeLong et al., 2006; Konstantinidis et al., 2009; Qin et al., 2010). A major objective of metagenomic studies is to recover the genome sequence, complete or draft, of a genotype or species from a sample. Short-read (for example, 50–100 bp) NGS technologies are becoming increasingly popular due to their high-throughput, but it remains unclear whether these technologies can be used to robustly recover individual genomes from complex communities. Several recent studies have attempted to evaluate the sequencing errors and artifacts specific to each NGS platform (Gomez-Alvarez et al., 2009; Quince et al., 2009; Aird et al., 2011); however, most of these studies have not assessed assembly quality and/or have employed simple DNA samples (for example, single viral genomes) and thus, the relevance of their results for complex community samples remains to be evaluated. Moreover, the presence of closely related species in the sample may complicate the assembly of a single genotype.
Assembling genomes from metagenomes
To provide quantitative insights into the issues above, we generated a series of in silico metagenomes by spiking reference genome reads into a background metagenome (Lanier.Illumina) and compared the derived assembly against the assembly of the reference genome from the genome reads alone (Supplementary Figure S1). For this analysis, we used the Escherichia sp. strain TW10509, whose genome sequence we described previously (Luo et al., 2011) and which has no close relatives in the Lake Lanier sample (Supplementary Figure S2), as reference. The Lanier.Illumina dataset was described in detail elsewhere (Oh et al., 2011), originated from a freshwater planktonic community sample from Lake Lanier (Atlanta, GA, USA) and represents a total of 3640 Mbp sequence data (100 bp pair-ended reads; average G+C content ∼50%) obtained using the Illumina GA-II sequencer (Illumina, Inc., San Diego, CA, USA). The community complexity in the sequenced sample (in terms of species richness and evenness) was comparable to that of previously characterized open ocean communities.
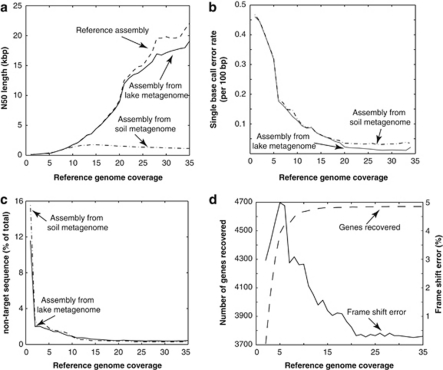
We varied the reference genome abundance, measured by the average coverage of the genome in the final in-silico generated metagenome, more than 30-fold (that is, 1 × to 35 × ). As expected, the fraction of the reference genome recovered increased exponentially in the low coverage range and reached a plateau at about 20 × coverage (Figure 1d). We also observed that greater than 20 × coverage did not improve the recovery of the target genome substantially; thus, obtaining greater coverage is not recommended (unless a different library insert size is used for closing purposes). Surprisingly, more than 10% of the total assembled contigs that belonged to the reference genome (that is, contained target sequences) were contaminated by non-target sequences at low coverage (1 × ) and this portion decreased to ∼0.2% when coverage exceeded 15 × (Figure 1c), which agrees with previous results from simulations (Mavromatis et al., 2007). Similar results were obtained when the reference genome represented an organism with close relatives in Lanier.Illumina (Supplementary Figures S3–S8), albeit the sequences of the relatives generally had a positive effect on the quality of the derived assemblies (Supplementary Figures S5–S6). We also quantitatively assessed the errors in the consensus sequences of the derived assemblies. About 1% of the total genes recovered in the Illumina assembly at 15 × coverage contained homopolymer-associated sequencing errors (that is, three or more consecutive identical DNA bases), resulting in truncated protein sequences or frameshifts. This number increased to about 3% when non-homopolymer-associated errors were also taken into account. Preliminary analyses revealed that the findings reported above were also applicable to a more complex soil metagenome, originating from a temperate (bulk) soil sample (Figures 1b and c), although the average length of the assembled contigs of the reference genome was consistently shorter in the soil spiked-in dataset (Figure 1a) owing to the higher complexity of the soil community (Supplementary Figure S9).
Figure 1.
Sequence errors and artifacts in assembled contigs of a target genotype from a complex metagenome. The assembly of a reference genome (Escherichia sp. TW10509) based solely on its own reads (reference assembly) was compared with the assembly of the genome from the in silico metagenome, which was composed of Lanier.Illumina spiked in with reads of the reference genome. (a) Comparison of N50, that is, the contig length that 50% of the entire assembly is contained in contigs no shorter than this length, between the latter and the reference assemblies over different reference genome coverage (abundance). (b) Single base call error rate decreased dramatically as reference genome abundance in the metagenome increased and reached a plateau at about 20 × coverage. (c) At low coverage, contigs from the metagenome assembly had a substantial portion of non-targeted (chimeric) sequences. (d) Frequency of frameshift errors as a function of the reference genome abundance. Results from similar analyses using a higher-complexity (Supplementary Figure S9) soil metagenome of similar size to the Lanier.Illumina metagenome are also shown for comparison.
Investigating intrapopulation genetic structure
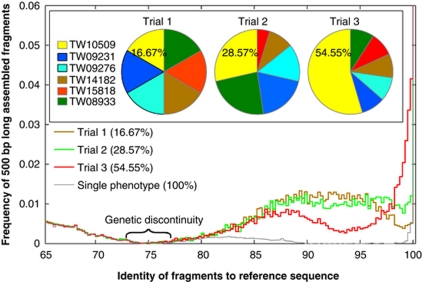
Natural populations are frequently composed of several closely related genotypes as opposed to a single genotype. It remains challenging to use metagenomics for the robust assessment of intrapopulation genetic structure, for example, to detect heterogeneous populations. To this end, we expanded the single genotype analysis to include five additional Escherichia sp. genomes, which show pairwise genetic relatedness ranging from 90% to 95% average nucleotide identity (ANI, (Konstantinidis and Tiedje, 2005); Supplementary Table S1 & Supplementary Figure S10). Regardless of the composition of the target population in the in silico generated metagenome, the six genomes were recovered as a discrete sequence cluster when all metagenomic reads were mapped on the reference Escherichia sp. strain TW10509 genome (Figure 2). The sequence-discrete clusters were obvious for other reference populations as long as no close relatives with higher than ∼85% ANI to the population were present in the metagenome. Furthermore, the shape of the coverage plot reliably reflected the target population genetic structure: when the population was homogeneous (that is, all genomes were spiked at similar abundances) the shape of the coverage plot approximated a normal distribution around the average ANI of the six genomes (∼92%); when the population structure was heterogeneous (for example, one genome more abundant than the others), the shape of the coverage plot deviated from the normal-like distribution and quantitatively reflected the variations in individual genome abundance (Figure 2). However, we were unable to recover robust assemblies of individual genotypes, even in trials where the target genotype consisted more than 50% of the population (Supplementary Figure S11) or when a high nucleotide identity cutoff in the assembly was used due to the fact that assemblers apply consensus strategy when encountering polymorphisms.
Figure 2.
Assessment of intrapopulation genetic structure based on sequence coverage plots. The total number of reads of the reference population spiked into the Lanier.Illumina metagenome was fixed at 35 × coverage, but the proportions of the different genotypes making up the population varied as represented by the pies (inset). The graph represents a coverage plot, constructed as previously described (Konstantinidis and DeLong, 2008), and shows the nucleotide identity (x axis) of all 500 bp long consecutive fragments of contigs assembled from the in silico generated metagenome (target and non-target) that map on the TW10509 genome sequence (y axis), which was used as reference. Note that a genetic discontinuity in the 75–80% nucleotide identity range was always observed, regardless of the genotype composition of the population. Also note that when the portion of TW10509 reads in the metagenome increased (from 16.67% in trial 1 to 54.55% in trial 3), the coverage plot reflected the shifts in the higher portion of reads in the 98–100% range (contributed by the TW10509 reads).
The results presented here reveal the errors and limitations as well as the strengths of metagenomics for population analysis, and provided practical standards and guidelines for experimental design and analysis (for example, Supplementary Table S2). Some of our results should be independent of the NGS platform used and therefore broadly applicable to short-read sequencing.
Acknowledgments
We thank Rachel Poretsky for useful discussions related to the manuscript. This work was supported by the US Department of Energy under Award No. DE-SC0004601 (to KTK) and contract No. DE-AC02-0SCH11231 to (NCK).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aird D, Ross MG, Chen WS, Danielsson M, Fennell T, Russ C, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, et al. Community genomics among stratified microbial assemblages in the ocean′s interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Braff J, Karl DM, DeLong EF. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific subtropical gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, DeLong EF. Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J. 2008;2:1052–1065. doi: 10.1038/ismej.2008.62. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci USA. 2011;108:7200–7205. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatis K, Ivanova N, Barry K, Shapiro H, Goltsman E, McHardy AC, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- Oh S, Caro-Quintero A, Tsementzi D, Deleon-Rodriguez N, Luo C, Poretsky R, et al. Metagenomic insights into the evolution, function and complexity of the planktonic microbial community of Lake Lanier, a temperate freshwater ecosystem. Appl Environ Microbiol. 2011;77:6000–6011. doi: 10.1128/AEM.00107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.