Abstract
Knowledge about Sphagnum-associated microbial communities, their structure and their origin is important to understand and maintain climate-relevant Sphagnum-dominated bog ecosystems. We studied bacterial communities of two cosmopolitan Sphagnum species, which are well adapted to different abiotic parameters (Sphagnum magellanicum, which are strongly acidic and ombrotrophic, and Sphagnum fallax, which are weakly acidic and mesotrophic), in three Alpine bogs in Austria by a multifaceted approach. Great differences between bacterial fingerprints of both Sphagna were found independently from the site. This remarkable specificity was confirmed by a cloning and a deep sequencing approach. Besides the common Alphaproteobacteria, we found a discriminative spectrum of bacteria; although Gammaproteobacteria dominated S. magellanicum, S. fallax was mainly colonised by Verrucomicrobia and Planctomycetes. Using this information for fluorescent in situ hybridisation analyses, corresponding colonisation patterns for Alphaproteobacteria and Planctomycetes were detected. Bacterial colonies were found in high abundances inside the dead big hyalocytes, but they were always connected with the living chlorocytes. Using multivariate statistical analysis, the abiotic factors nutrient richness and pH were identified to modulate the composition of Sphagnum-specific bacterial communities. Interestingly, we found that the immense bacterial diversity was transferred via the sporophyte to the gametophyte, which can explain the high specificity of Sphagnum-associated bacteria over long distances. In contrast to higher plants, which acquire their bacteria mainly from the environment, mosses as the phylogenetically oldest land plants maintain their bacterial diversity within the whole lifecycle.
Keywords: abiotic factors, bog ecosystem, FISH–CLSM, deep-sequencing, microbe+plant communities, S. magellanicum/fallax
Introduction
Bog ecosystems belong to the oldest vegetation forms, with more or less constant conditions for thousands of years. It covers 4 million km2, approximately 3% of the earth's surface, and have a high value for biodiversity conservation, as reservoir of fresh water, for human welfare and our world climate due to its extraordinary role in carbon sequestration (Raghoebarsing et al., 2005). The latter resulted in a net cooling effect on the global radiation balance (Dise, 2009). On the other side, these long-existing ecosystems are extremely sensitive to changing abiotic factors connected with climate change (Belyea and Malmer, 2004; Dise, 2009). When peatlands degrade, the stored carbon will be released. For example, drainage of peat soils results in CO2 and N2O emissions of globally 2–3 Gt CO2-eq per year (Joosten and Couwenberg, 2009). For Alpine bog ecosystems, profound changes are expected due to global change (Theurillat and Guisan, 2001). The bryophyte genus Sphagnum, consisting of approximately 300 different species, is distributed world wide and forms the dominant component of bog vegetation (Daniels and Eddy, 1985). Therefore, Sphagnum moss has been used globally as an indicator of climate change (Whinam and Copson, 2006; Granath et al., 2009), and microbial communities living in Sphagnum were shown early indicators of ecosystem disturbances in a microcosm experiment (Jassey et al., 2011). Hence, knowledge concerning microbial ecology is important to protect, maintain and manage Sphagnum bog ecosystems.
Sphagnum mosses form a unique habitat for microorganisms such as high acidity and low temperature; water saturation, together with extremely low concentrations of mineral nutrients, are characteristic abiotic factors. Furthermore, Sphagnum leaves are highly specialised; they form a special tissue of living, chlorophyll-containing chlorocytes and dead cell content-free hyalocytes, which are responsible for their huge potential to store water. Sphagnum species also produce bioactive secondary metabolites influencing microbial colonisation (Opelt et al., 2007a). So far, mainly the microbial populations involved in CH4 cycling living on dead Sphagna (Dedysh et al., 1998; Dedysh et al., 2001; Horn et al., 2003; Pankratov et al., 2008; Rahman et al., 2010) have attracted research interest. Recently, we could show that living Sphagnum mosses are colonised in high abundances with specific microorganisms, which fulfil important functions like nutrient supply and pathogen defence for moss growth and health (Opelt et al., 2007a; Opelt et al., 2007b). New questions thus arose: (i) do new molecular and microscopic techniques allow deeper insights into Sphagnum-associated bacterial diversity?; (ii) what are the main drivers of this diversity?; and (iii) how is this specific bacterial diversity acquired? Regarding the latter, from higher plants, we know that bacterial communities have a certain degree of plant specificity, but the majority of bacteria is environment-acquired and only a few bacterial strains are transferred within the lifecycle (reviewed in Berg and Smalla, 2009). Although for higher plants, the sporophyte generation makes up almost their whole life cycle, bryophytes have a dominant photosynthetically active gametophyte stage. Bryophytes represent the phylogenetically oldest group of land plants, and due to the specific communities (Opelt et al., 2007c), our hypothesis was that this diversity is transferred directly from the sporophyte to the gametophyte and vice versa.
The objective of this work was to study the structure and origin of Sphagnum-associated bacteria, which were detected in three different Alpine bogs in Austria. To analyse differences between different Sphagnum species, two dominant and cosmopolitan species were selected: Sphagnum magellanicum and Sphagnum fallax. S. magellanicum (section Sphagnum) is typical for strong acidic, oligotrophic and ombrotrophic habitats, whereas S. fallax (section Cuspidata) grows in weakly acidic, more mesotrophic situations influenced by minerotrophic groundwater (Daniels and Eddy, 1985). A polyphasic approach was applied to study bacterial communities on gametophytes and sporophytes: (i) microbial fingerprints by PCR–SSCP (single-strand conformation polymorphism) of 16S rRNA genes, (ii) clone libraries and phylogenetic analysis of clones, (iii) deep-sequencing of Alphaproteobacteria and (iv) fluorescent in situ hybridisation with universal and group-specific probes, coupled with confocal laser scanning microscopy (FISH–CLSM) and image analysis. Results taken together showed that Sphagnum gametophytes, as well as sporophytes, have a similar intimate and highly specific interaction with their associated bacteria.
Materials and methods
Experimental design and sampling procedure
To analyse differences between two Sphagnum species, S. magellanicum B (section Sphagnum) and S. fallax H. K (section Cuspidata) were selected. Both bryophytes belong to the typical and cosmopolitan vegetation in peat bogs (Daniels and Eddy, 1985). Adult gametophytes of moss species were sampled in three different natural habitats in Alpine bogs in Austria in September 2009 (Table 1). Ecological characters of the habitats were described by composition of plant communities and average Ellenberg's indicator values for vascular plants and bryophytes (Ellenberg et al., 1991). From each of three investigated bogs, four single replicates per Sphagnum species consisting of 15–20 plantlets were collected and stored separately. The living green parts of the plantlets were placed into sterile plastic bags and transported to the laboratory. S. fallax plants forming sporophytes were solely detected in the Rotmoos bog. Sporophyte samples of S. fallax consisting of enclosed spore capsules were collected and processed separately. In general, sporophytes of S. magellanicum are uncommonly found.
Table 1. Sampling sites and ecological parameters of the habitats.
| Bog | Moss species | Sample numbera |
Geographical parameters |
Abiotic parametersc |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Coordinates | Altitude (m) | Soil reaction value | Nutrient value | Light value | Moisture value | Temperature value | |||
| Rotmoos (Styria) | S. magellanicum | RM1 | N47 41.030 E15 09.276 | 699 | 1.4 | 1.4 | 7.8 | 7.5 | 3.4 |
| RM2 | N47 41.021 E15 09.245 | 699 | — | — | — | — | — | ||
| RM3 | N47 40.971 E15 09.270 | 698 | — | — | — | — | — | ||
| RM4 | N47 41.017 E15 09.319 | 693 | — | — | — | — | — | ||
| S. fallax | RF1 | N47 40.908 E15 09.244 | 693 | 3.0 | 2.8 | 7.0 | 8.2 | 3.9 | |
| RF2 | N47 40.958 E15 09.175 | 691 | — | — | — | — | — | ||
| RF3, RFSb | N47 41.041 E15 09.232 | 690 | — | — | — | — | — | ||
| RF4 | N47 41.055 E15 09.264 | 689 | — | — | — | — | — | ||
| Wasenmoos (Salzburg) | S. magellanicum | WM1 | N47 18.373 E12 24.927 | 1216 | 1.5 | 1.4 | 7.6 | 7.2 | 3.2 |
| WM2 | N47 18.363 E12 24.944 | 1216 | — | — | — | — | — | ||
| WM3 | N47 18.337 E12 25.119 | 1214 | — | — | — | — | — | ||
| WM4 | N47 18.315 E12 25.126 | 1208 | — | — | — | — | — | ||
| S. fallax | WF1 | N47 18.387 E12 24.871 | 1211 | 2.6 | 2.7 | 6.9 | 7.8 | 3.6 | |
| WF2 | N47 18.391 E12 24.866 | 1215 | — | — | — | — | — | ||
| WF3 | N47 18.385 E12 24.882 | 1217 | — | — | — | — | — | ||
| WF4 | N47 18.347 E12 24.980 | 1213 | — | — | — | — | — | ||
| Pürgschachen Moor (Styria) | S. magellanicum | PM1 | N47 34.905 E14 20.402 | 637 | 1.6 | 1.4 | 7.7 | 7.8 | 3.6 |
| PM2 | N47 34.910 E14 20.454 | 639 | — | — | — | — | — | ||
| PM3 | N47 34.839 E14 20.497 | 640 | — | — | — | — | — | ||
| PM4 | N47 34.805 E14 20.493 | 639 | — | — | — | — | — | ||
| S. fallax | PF1 | N47 34.789 E14 20.398 | 638 | 2.5 | 2.3 | 6.8 | 7.3 | 3.8 | |
| PF2 | N47 34.814 E14 20.356 | 636 | — | — | — | — | — | ||
| PF3 | N47 34.824 E14 20.346 | 635 | — | — | — | — | — | ||
| PF4 | N47 34.848 E14 20.344 | 634 | — | — | — | — | — | ||
Abbreviations: F, Sphagnum fallax; M, Sphagnum magellanicum; P, Pürgschachen Moor; R, Rotmoos; RFS, sporophyte sample of S. fallax; S, sporophyte sample of S.fallax; W, Wasenmoos.
Letters indicate bogs and Sphagnum species: R, W, P, F, M and S. Arabic numerals specify replicates.
RFS was collected at the same sampling point as sample RF3.
Abiotic parameters were expressed by average Ellenberg's indicator values for vascular plant and bryophyte species. Numbers indicate properties of the habitat along ther ecological gradients (soil reaction: 1=extremely acidic, 9=calcareous; nutrient richness: 1=extremely nutrient poor, 9=extremely nutrient rich; light exposure: 1=deep shadowed, 9=full light exposed; moisture: 1=extremely dry, 9=extremely wet; temperature: 1=extremely cold, 9=extremely warm).
Total community DNA isolation
Before DNA isolation, the bacterial fraction associated with gametophytes was extracted according to the slightly modified protocol of Opelt and Berg, 2004. Briefly, 5 g of plant material were physically disrupted with sterile pestle and mortar, and resuspended in 10 ml of 0.85% NaCl. A volume of 2 ml of suspension were centrifuged at 13000 r.p.m. for 20 min at 4 °C, and the pellet was used for isolation of the total community DNA as described before (Martin-Laurent et al., 2001). For mechanical lysis, the cells were homogenised twice in a FastPrep FP120 Instrument (QBiogene, BIO101, Carlsbad, CA, USA) for 30 s at speed 5.0 m sec−1. Extraction of bacteria associated with sporophyte of S. fallax was carried out by grinding of 10 closed-spore capsules with the FastPrep FP120 cell disrupter (30 s, speed 5.0 m sec−1). Before grinding, the capsules were surface sterilised as described previously (Opelt et al., 2007a). Cell lysis and isolation of DNA was performed similarly to the cell pellets of gametophytes. The obtained DNA was purified using the FastDNA SPIN Kit for Soil (MP Biomedical, Solon, OH, USA) according to the manufacturer's protocol. Final aliquots of the total community DNA were further used for PCR-based approaches.
Microbial fingerprinting by PCR–SSCP of 16S rRNA genes
PCR-based SSCP analysis of the microbial 16S rRNA genes was carried out with universal bacterial primers Com1, Unibac-II-927rP (Schwieger and Tebbe, 1998). PCR and preparation of the single-stranded DNA were performed according to Schwieger and Tebbe (1998). The amplicons were separated using the TGGE Maxi system (Biometra, Göttingen, Germany) at 400 V and 26 °C for 26 h in 8% (wt/vol) acrylamid gel followed by silver staining. Gel profiles were digitalised by transmissive scanning (Epson perfection 4990 Photo, Long Beach, CA, USA) and further analysed with the GelCompare II version 5.1 software package (Applied Maths, Kortrijk, Belgium). A similarity matrix was constructed using Pearson's correlation coefficients (r) and cluster analysis was done by the unweighted pair group method with average linkages. The resulting clusters of samples were examined for statistical significance by applying permutation test with 10 000 random permutations of the single samples (Kropf et al., 2004).
SSCP is based on the differences in the conformation of single-stranded DNA fragments. The electrophoretic mobility of the single-stranded DNA fragments depends on their three-dimensional conformation. Each of the amplification products was identified by its electrophoretic distance on SSCP gel and the number of DNA fragments. According to the distance of the bands, the SSCP gels were theoretically divided into operational taxonomic units. The presence or absence of individual amplified product DNA bands in each group was scored. The obtained matrix was used to compare statistically (see statistics).
Construction and analysis of 16S rRNA gene clone libraries
Total community DNA samples of the mosses from the site of the highest diversity determined by fingerprints (S. magellanicum: Pürgschachen Moor and S. fallax: Wasenmoos) were pooled together and used as template. 16S rRNA gene fragments were amplified with 799f/1492r primers, which avoid plant-derived amplicons substantially (Lane, 1991; Chelius and Triplett, 2001). The clone libraries were constructed as previously described (Sun et al., 2008). PCR fragments for sequencing were generated with USP (5′-GTAAAACGACAACCAGT-3′) and RSP (5′-CAGGAAACAGCTATGACC-3′) vector-specific primers (Sigma-Aldrich, Taufkirchen, Germany). To exclude transformants with chloroplast-derived DNA, we applied restriction test with SphI-HF (New England Biolabs, Frankfurt am Main, Germany). Restriction profiles of chloroplast sequences were calculated in silico (data not shown). PCR products were sequenced with the Applied Biosystems 3130 Genetic Analyser (Foster City, CA, USA). The sequences were submitted to the EMBL Nucleotide Sequence Database under accession numbers FR832168–FR832348.
Taxonomic affiliation of the partial 16S rRNA gene sequences was defined by alignment with reference sequences from GenBank, using the BLASTn algorithm. A Bellerophon programme was applied to screen the clone libraries for the presence of chimeric sequences (Huber et al., 2004). Multi-alignments of the selected sequences were produced by ClustalX software version 2.0.12 (Larkin et al., 2007). Neighbour-joining phylogenetic trees were reconstructed using PHYLIP software package version 3.69 (Felsenstein, 1989). Confidence levels for the internal branches were assessed by bootstrap analysis with 100 resamplings.
FISH and CLSM
Single gametophytes of S. magellanicum and S. fallax were fixed with 4% paraformaldehyde/phosphate-buffered salt (3:1, v/v). Separated leaves and stems, sectioned with a razor blade, were stained by in-tube FISH (Grube et al., 2009). Sporophytes of S. fallax were fixed likewise gametophytes. For fixation, surface-sterilised capsules containing spores were disclosed by vortexing in phosphate buffer. Staining of spores was carried out on glass slides. The samples were hybridised with rRNA-targeting probes (genXpress, Wiener Neudorf, Austria) specific for Alphaproteobacteria and Planctomycetes, as dominant phylogenetic groups revealed by the clone libraries and with a set of universal bacterial probes. Hybridisation was carried out at 41 °C. The probes and corresponding stringency conditions are listed in Supplementary Table S1.
CLSM was performed with a Leica TCS SPE confocal microscope (Leica Microsystems, Mannheim, Germany). Fluorescent dyes Cy3 and Cy5 labelled to the FISH probes were sequentially excited with 532 and 635 nm laser beams, respectively; the emitted light was detected in the range of 556–607 and 657–709 nm, respectively. An additional channel (excitation at 488 nm; emission range 508–556 nm) was applied for acquiring the autofluorescence of the moss cells. Photomultiplier gain and offset were individually optimised for every channel and every field of view to improve the signal/noise ratio. Confocal stacks were acquired with a Leica ACS APO × 40 OIL CS objective (NA: 1.15) and a Leica ACS APO × 63 OIL CS objective (NA: 1.30) by applying a Z-step of 0.4–0.8 μm. Three-dimensional reconstructions were created with the software Imaris7.0 (Bitplane, Zurich, Switzerland).
Deep sequencing and bioinformatic analysis of Alphaproteobacteria
Pooled samples of the total-community DNA of S. fallax and S. magellanicum gametophytes were investigated by barcoded pyrosequencing approach (Binladen et al., 2007) with the Alphaproteobacteria-specific primers ADF 681f/1492r (Blackwood et al., 2005). Total-community DNA of S. fallax sporophyte was explored with universal bacterial primers 799f/1492r (Lane, 1991; Chelius and Triplett, 2001). Pyrosequencing libraries were generated by LGC Genomics, Berlin, Germany, using the Roche/454 GS FLX Titanium platform (454 Life Science Corporation, Brandford, CT, USA).
Raw-sequencing reads were quality filtered and trimmed by length (⩾150 bp). Rarefaction analysis was performed for phylotype clusters at 0.03, 0.05 and 0.1 genetic distance, corresponding to the levels of the species, genera and families, respectively (Schloss and Handelsman, 2006; Hur and Chun, 2004). Due to the different number of sequences among samples, the data were normalised considering the same number of sequences to all samples. Richness estimates and diversity indices were calculated for normalised data sets, using default settings in the open source software package QIIME (http://qiime.sourceforge.net/), which allows analysis of high-throughput community sequencing data (Caporaso et al., 2010).
Compositional analysis was performed using the BLAT pipeline integrated into the web interface SnoWMAn version 1.8 (https://epona.genome.tugraz.at/snowman/). Greengenes database was used as a reference database. Taxonomic assignment to family and genus levels was accomplished by the integrated Ribosomal Database Project classifier with 50% confidence threshold. For reads corresponding to S. fallax sporophyte, relative abundances of taxonomic groups within Alphaproteobacteria were recalculated to the total number of reads affiliated to the class.
Multivariate statistics: ecological analysis of bacterial and plant communities
Correspondence analysis was used to answer the question whether a correlation exists (1) between the independently sampled bacterial communities (defined as operational taxonomic units) of the different sampling points and (2) between bacterial communities and environmental data. For the latter, unweighted average indicator values by Ellenberg et al., 1991 for the vascular plants and bryophytes were applied as independent variables (Table 1). We used the canonical correspondence analysis for unimodal data of the software package Canoco 4.5 (Lepš and Smilauer, 2003). Significance of the environmental variables for the microbial communities was tested by Monte-Carlo test with 1000 permutations.
Results
Insights into bacterial diversity by fingerprints and clone libraries
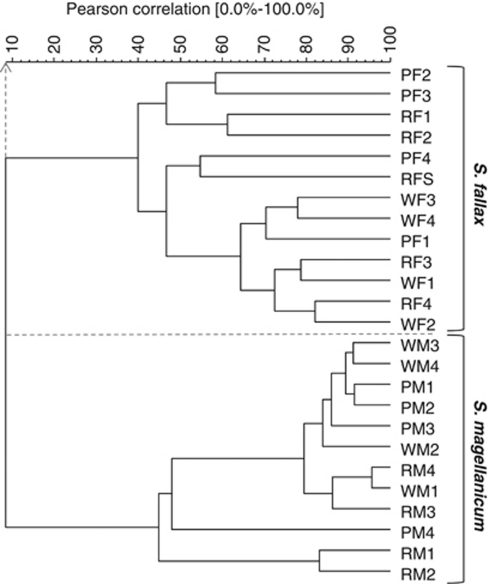
SSCP profiling of 16S rRNA genes amplified with universal primers showed highly diverse and specific bacterial communities on both Sphagnum species. Statistical analysis resulted in two distinct clusters of S. fallax- and S. magellanicum-originated profiles at similarity level of 9% (Figure 1). Within each species-specific cluster, only several of the samples from the different geographical sites were grouped together. Microbial fingerprints obtained from Sphagnum species of different bogs showed also a high similarity to each other, for example, S. magellanicum from Rotmoos and Wasenmoos (RM4, WM1).
Figure 1.
Dendrogram based on amplified 16S rRNA gene fragments of bacterial communities associated with S. magellanicum (M) and S. fallax (F; FS=sporophyte) from different sites in Austria (R, Rotmoos; W, Wasenmoos; P, Pürgschachen Moor) obtained by using eubacterial primers and separated by single-strand conformation polymorphism (SSCP). The patterns obtained were grouped by unweighted pair group method with average linkages.
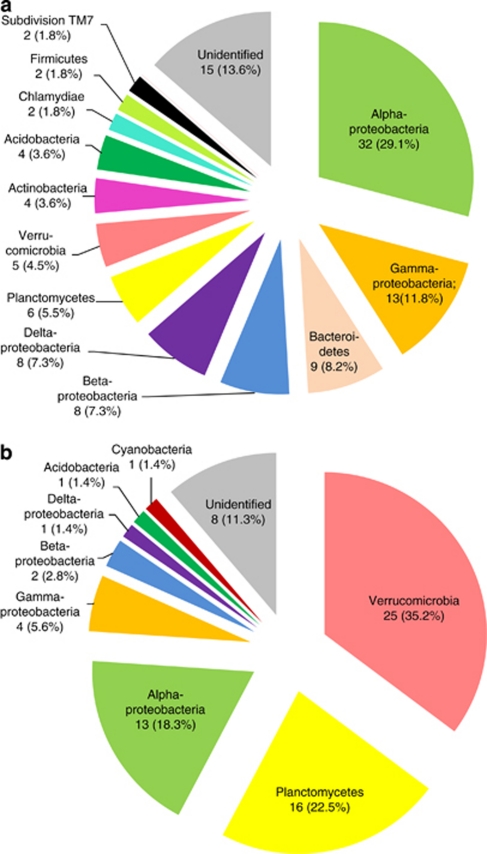
A first insight into the specific biodiversity was achieved by construction of two individual clone libraries: total community DNA isolated from S. magellanicum and S. fallax from one site was used as template. First, we solved the methodological problem to avoid the analysis of a high proportion of clones with plant-derived DNA. Introduction of a restriction assay with SphI before sequencing allowed to recognise and to eliminate plant-derived clones. Composition of microbial communities of S. fallax and S. magellanicum was clearly different (Figure 2). Altogether, S. magellanicum clone library consisted of 110 clones representing 12 taxonomic groups. The dominant bacterial fraction belonged to Alphaproteobacteria (29.1%), followed by sub-dominant Gammaproteobacteria (11.8%). S. fallax-associated community was recreated from 71 clones affiliated to eight taxa. Dominant groups belonged to Verrucomicrobia (35.2%), Planctomycetes (22.5%) and again Alphaproteobacteria (18.3%).
Figure 2.
Bacterial community composition revealed by 16S rRNA gene clone libraries. Absolute and relative abundances of the taxonomic groups are shown for the S. magellanicum- (a) and S. fallax-associated (b) bacterial communities.
BLAST analysis resulted in a high proportion of not-yet-cultivated bacteria. To perform the phylogenetic analysis of clones, we constructed a phylogenetic tree for each community. Sequences of Sphagnum-associated bacterial clones and reference strains clustered together within defined taxonomic groups with high bootstrap values (⩾50; Supplementary Figure S1). Only three clones were closely related (⩾97% similarity) to taxonomically described bacterial species. A certain number of clones possessed sequence similarities ⩾97% with clones from northern terrestrial habitats and acidic environments of different geographical regions. Interestingly, several clusters of Verrucomicrobia and Planctomycetes were solely formed by Sphagnum clones. BLASTn alignment of the sequences within these Sphagnum-specific clusters resulted in a sequence identity ⩽95% with database sequences. Thus, microbial diversity of Sphagnum comprised bacteria occurring also in other habitats, as well as Sphagnum-specific bacteria never found elsewhere, yet.
Deep insight into Alphaproteobacteria by pyrosequencing
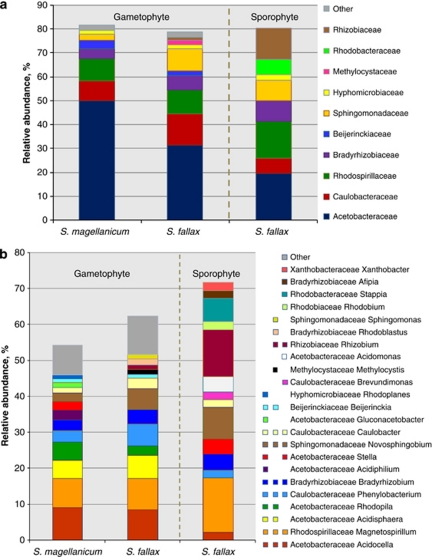
As a dominant component of both Sphagnum communities, Alphaproteobacteria group was selected for a deep sequencing study. The rarefaction analysis of the amplicon libraries is shown in Supplementary Figure S2. Comparison of the rarefaction analyses with the number of phylotype clusters estimated by Chao1 richness estimator revealed that pyrosequencing effort reached 57.7–68.9% of estimated richness at the taxonomic level of families (Supplementary Table S2). Richness estimates of the genera and species showed that 45.9–50.3% and 31.9–32.5% of estimated richness, respectively, was recovered. The deepest classification was obtained at the ranks of families and genera (Figure 3).
Figure 3.
Taxonomic classification of Alphaproteobacteria associated with S. fallax and S. magellanicum. Pyrosequencing reads are classified at family (a) and genus level (b) with a confidence threshold of 50%. Unclassified reads are not shown. Groups not reaching 1% of relative abundance are included in ‘Other'. Multi-coloured charts at the legend are shown for each genus and sample correspondingly.
Comparison of the classified reads revealed that the two investigated bryophyte species shared dominant bacterial groups and considerably differed in spectrum of sub-dominant and minor groups. Gametophytes of both moss species were dominated by members of the Acetobacteraceae family, followed by Caulobacteraceae and Rhodospirillaceae. Abundances of sub-dominant Bradyrhizobiaceae, Sphingomonadaceae, Methylocystaceae and Rhizobiaceae were clearly higher among S. fallax-associated Alphaproteobacteria than on S. magellanicum. A low number of Kordiimonadaceae and Phyllobacteriaceae were uniquely detected in S. magellanicum, whereas Rhodobacteraceae and Methylobacteraceae were detected only associated with S. fallax.
At genus level, more differences of the communities were revealed. In total, amplicon library of the gametophytes included sequences of 54 genera. S. magellanicum harboured 41 of them and S. fallax harboured 37. The Sphagnum species shared 24 bacterial genera. Both bryophytes were dominated by three alphaproteobacterial genera (Acidocella, Magnetospirillum and Acidisphaera), whereas sub-dominant genera (Rhodopila, Phenylobacterium, Bradyrhizobium, Novosphingobium and Caulobacter) occurred in different abundances.
Diversity of bacterial species was explored by Shannon diversity index (H‘) calculated at the genetic distance of 3%. Shannon values indicated higher diversity of Alphaproteobacteria for S. magellanicum (7.92) than for S. fallax (7.67).
Spatial structure of the bacterial communities
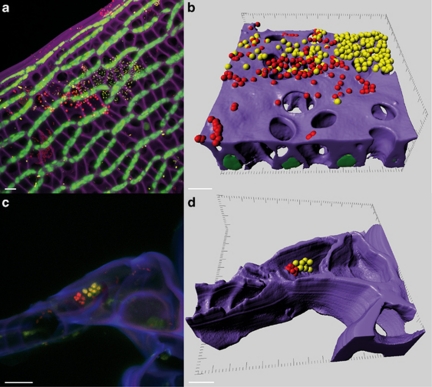
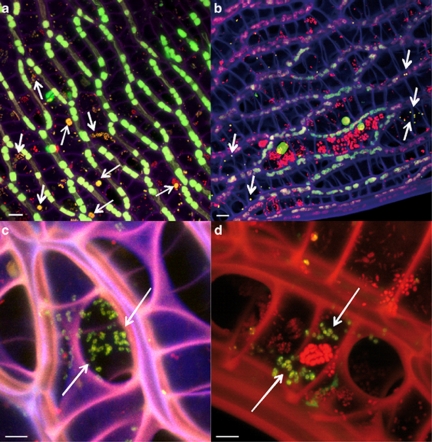
Due to the unique morphology of Sphagnum plantlets, the next step was to analyse the colonisation pattern of bacteria on/in Sphagnum. Especially the cell structure of leaves forms regular and peculiar microenvironments for the microbial communities; one-layer net of photosynthetically active cells (chlorocytes) is alternated with dead hyaline cells (hyalocytes). The latter possess large pores and are temporally filled with water. Combination of FISH, CLSM and computer-assisted three-dimensional reconstructions revealed colonisation patterns of Sphagnum gametophytes by two specific groups of microorganisms: Alphaproteobacteria and Planctomycetes. Bacterial micro-colonies were observed on the outer surface, as well as in the inner space of the gametophytes of both Sphagnum species (Figure 4). Three-dimensional reconstructions showed attachment of the bacteria to the cell wall of the Sphagnum cells. Internal spaces of the hyalocytes were densely colonised by micro-colonies closely associated to each other (Figure 5).
Figure 4.
Localisation of bacteria in moss gametophytes. Fluorescent in situ hybridisation (FISH) of S. fallax leaves showed colonisation of the outer surface (a and b) and hyaline cells (c and d). Violet: cell walls of Sphagnum cells; green: chlorophyll-containing Sphagnum chlorocytes; yellow: Alphaproteobacteria; red: other bacteria. Images acquired by confocal laser scanning microscopy (CLSM; panels a and c) and processed by 3D computer reconstruction using Imaris7.0 (b and d). Scale bar=10 μm.
Figure 5.
Localisation of bacteria in hyalocytes of Sphagnum. Internal space of hyalocytes of S. fallax (a and b) and S. magellanicum (c and d) hybridised with Alphaproteobacteria- and Planctomycetes-specific probes. Yellow: Alphaproteobacteria (a and c) or Planctomycetes (b and d) indicated by arrows; red: other bacteria; green: algae. Scale bar=10 μm (a and b) or 5 μm (c and d).
In general, using different probes (Supplementary Table S2), both Sphagnum species were characterised by similar colonisation patterns. Analyses using group-specific probes showed differences for the colonisation of Sphagnum leaves by Alphaproteobacteria and Planctomycetes. On both moss gametophytes, Alphaproteobacteria occurred in various morphological forms: coccoid, rod-shaped and vibroid cells, tetrads and sarcina-like aggregates (Figure 5a). Regarding the occurrence of Planctomycetes, we observed more colonies associated with S. fallax than S. magellanicum. Usually they formed less abundant colonies of coccoid cells (Figure 5d). Noteworthy, the outer cortex of the S. fallax stem tissues was occupied by Alphaproteobacteria in contrast to the non-colonised stems of S. magellanicum (Supplementary Figure S3).
Ecological factors driving the bacterial communities
Primary characterisation of the niches occupied by Sphagnum revealed differences in the composition of plant communities and abiotic conditions. S. magellanicum frequently grew together with S. fuscum, Eriophorum vaginatum, Vaccinium oxycoccos, Andromeda polifolia, Calluna vulgaris and Drosera rotundifolia, whereas S. fallax was mainly accompanied by S. angustifolium, S. palustre, Carex rostrata, Dryopteris carthusiana, Frangula alnus, Molinia caerulea and Vaccinium myrtillus. On the basis of Ellenberg's indicator values of bryophytes and vascular plants, both Sphagnum species showed distinctive preferences along ecological gradients (Table 1). In contrast to S. fallax, S. magellanicum habitats were characterised by lower values of soil reaction, moisture and temperature, lower amount of nutrients and more intensive exposure to the sun light.
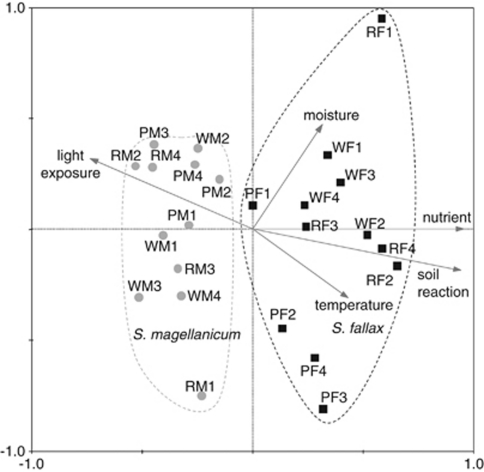
The influence of the environmental conditions on the microbial communities of bryophytes was examined using multivariate statistical analysis (Figure 6). By the Monte-Carlo permutation test, a statistical significance was proved for nutrient richness (P<0.001, correlation with first axis 0.8992) and for soil reaction (P<0.002, correlation with first axis 0.8876). Both have the character of co-variables.
Figure 6.
Canonical correspondence analysis biplot of operational taxonomic units identified by SSCP community fingerprints. Independent ecological gradients are given as unweighted average of indicator values by Ellenberg for vascular plants and bryophytes. Single fingerprints of S. fallax- and S. magellanicum-associated communities are depicted with black squares and gray circles, respectively. Names of the single SSCP patterns correspond to the names of the sample. Dashed ovals were drawn around samples of the same Sphagnum species. Species–environment correlations of the first and the second axes are 0.942 and 0.902, respectively. Sum of all Eigenvalues=0.563; significance for the first axis: P-value=0.002, F-ratio=2.795 tested by Monte-Carlo permutation test (1000 permutations).
Comparison of sporophyte- and gametophyte-associated communities
High similarity of microbial communities of S. fallax sporophyte and gametophyte was initially detected by molecular fingerprinting of 16S rRNA genes with universal bacterial primers (Figure 1). DNA pattern of the S. fallax sporophyte community shared up to 55% similarity with gametophyte samples. The microbial diversity of the sporophyte- and gametophyte-associated microbial communities was investigated in more detail by a deep sequencing approach for Alphaproteobacteria and by FISH–CLSM. For the amplicon library of Alphaproteobacteria associated with the sporophyte, the saturation at the family level reached 77.0% (Supplementary Figure S3). Sporophyte library remained unsaturated at the genera and species levels: 55.0% and 50.8% of the estimated richness was uncovered.
Alphaproteobacteria associated with sporophyte and gametophyte showed substantial similarities (Figure 3). Like in the gametophyte, dominant clusters in the sporophyte were affiliated with Acetobacteraceae, Rhodospirillaceae and Caulobacteraceae families. Bradyrhizobiaceae and Sphingomonadaceae were present at similar abundances in both gametophyte and sporophyte. Certain differences in contrast to gametophyte were found; Rhizobiaceae and Rhodobacteraceae presented a dominant fraction only in the sporophyte. Bejerinckiaceae, Methylocystaceae, Brucellaceae, Aurantimonadaceae and Methylobacteriaceae were detected only in the gametophyte. Alphaproteobacterial sequences from the sporophyte of S. fallax were classified into 14 genera. Sporophyte- and gametophyte-associated communities shared eight genera (Acidocella, Magnetospirillum, Phenylobacterium, Bradyrhizobium, Stella, Novosphingobacterium, Caulobacter, Rhizobium), but their abundances was different. Magnetospirillum, Rhizobium and Novosphingobium were detected in higher abundances in sporophyte than in the gametophyte. Six sub-dominant genera Stappia, Stella, Acidomonas, Rhodobium, Afipia and Xanthobacter were specific for the sporophyte. The genera Bejerinckia, Methylocystis, Rhodoblastus and Sphingomonas were exclusively found in the gametophyte.
Using FISH–CLSM, inside the sporophyte capsules, directly connected with spores, but embedded in a matrix, bacterial cells in high amount were observed (Supplementary Figure S4).
Discussion
The main aim of this study was to analyse both the structure and the origin of Sphagnum-associated bacterial communities. Remarkable differences in structural diversity between the S. fallax and S. magellanicum communities were found. Abiotic parameters, which also determine the occurrence of Sphagnum species inside the bog, were identified as drivers of the Sphagnum-specific community composition. To find out the origin of this specific bacterial diversity, we compared the gametophyte-associated communities with those of the sporophyte, and found a high similarity. This led to the conclusion that a high portion of bacterial populations is transferred during the whole life cycle from the gametophyte to the sporophyte and vice versa. In the following, we will answer our questions/hypotheses.
New molecular and microscopic techniques allowed deeper insights into the structure of Sphagnum-associated bacterial diversity. For example, using FISH, combined with CLSM and three-dimensional modelling. In comparison with Opelt and Berg (2004), we gained new insights into the spatial structure of Sphagnum-associated bacteria. Although the whole Sphagnum gametophytes were densely colonised by bacterial micro-colonies, especially the dead big hyalocytes of the branch, leaves were occupied by bacteria. Interestingly, all colonies and single cells were attached to cell walls connected with the living cells. This connection supports the symbiotic character of the moss–microbe interaction, which was also confirmed by first functional studies. For example, methanotrophic bacteria provide approximately 10–30% of Sphagnum carbon (Larmola et al., 2010). Moreover, Sphagnum harboured a high diversity of nitrogen-fixing bacteria and bacterial populations responsible for pathogen defense (Opelt et al., 2007a; Opelt et al., 2007b). In the CLSM images, differences in the occurrence of Alphaproteobacteria and Planctomycetes were found between both moss species. Although Alphaproteobacteria dominated colonisation of S. magellanicum, in microscopic pictures of S. fallax, more colonies of Planctomycetes were found. This was in accordance with our other results obtained by the analysis of microbial fingerprints and clone libraries. Besides of common Alphaproteobacteria, we found a discriminative spectrum of bacteria in clone libraries; although Gammaproteobacteria dominated S. magellanicum, S. fallax was mainly colonised by Verrucomicrobia and Planctomycetes. Unfortunately, representatives of Verrucomicrobia could not be detected by CLSM, although appropriate probes were applied. In contrast to the leaves, the stems were less colonised by bacteria. For stem tissues of S. fallax, similar colonisation patterns than for methanotrophs observed in S. cuspidatum were detected (Raghoebarsing et al., 2005). In contrast, the stem of S. magellanicum was not colonised by bacteria. A different morphology of the stem can explain this difference; the stem of S. magellanicum is more sclerotised than the stem of S. fallax. Furthermore, analysis of the bacterial communities by 16S rRNA gene clone libraries led to a highly discriminative community composition. Deep sequencing of the common Alphaproteobacteria showed more similarities especially for the dominant taxa, whereas differences for sub-dominant and minor taxa were calculated. Nevertheless, we reported the highest differences ever found between species in the plant kingdom (reviewed in Berg and Smalla, 2009). Altogether, our applied multifaceted approach led to matching results and to a comprehensive picture of specific Sphagnum-associated bacterial communities. Not to forget that results from the clone libraries, which yielded in a high amount of yet-not-described species, and the deep sequencing approach, where only up to half of the estimated richness was recovered, indicate a hidden, still unknown bacterial diversity, which has to be discovered.
Our research also addressed the question which factors drive the high specificity of Sphagnum-associated microbial communities? Using multivariate statistical analysis, abiotic factors, especially nutrient richness and pH, were identified to significantly influence the microbial communities. We used Ellenberg's indicator values, which are long-term indicator values established for plants to assess moisture, nitrogen and soil reaction (represents pH) in soil (Schaffers and Sỳkora, 2000). Interestingly, nutrient richness was identified as main influencing parameter in the nutrient-poor bog environment. The content of nutrients is important for both plant and microbial communities, although in the opposite way (Opelt et al., 2007c). pH expressed as soil reaction factor also significantly influenced Sphagnum-associated communities. This factor was often reported as the main driver, for example, in a global study of microbial communities in soil (Lauber et al., 2009). In microcosm experiments under controlled conditions, Jassey et al. (2011) identified similar abiotic drivers of the S. fallax-associated microbial community, such as pH, conductivity and temperature. Especially, due to the latter, they suggest microbial communities living with Sphagnum as early indicators for ecosystem disturbance, especially climate change. The key question is whether the abiotic factors modulate the composition of host-specific bacterial communities, or whether these factors are primary drivers of bacterial community composition. There are several facts supporting the first point. In Opelt et al. (2007a), the profile of secondary metabolites including antimicrobial substances was found to be different for S. fallax and S. magellanicum, which can be one reason for host specificity. A second hint is the specific colonisation of the sporophyte, which indicates a direct transfer of the host-specific bacteria. Furthermore, the high degree of specificity was not only shown at community level, but also at clone level. The same clones were found in Norwegian, Dutch, German and Sibirian bogs (NCBI database). On the other side, for methanotrophs, Larmola et al. (2010) found for transplanted Sphagnum species bacterial pattern and activity typical for the abiotic parameters of the destination site. However, this was an artificial experiment; the majority of approximately 300 Sphagnum species have very narrow ecological amplitudes (Daniels and Eddy, 1985). Moreover, Sphagnum mosses are not only able to adapt to their environment, but to change it; living Sphagna have extraordinarily high cation-exchange capacity and therefore acidify their environment by exchanging tissue-bound protons for basic cations in surrounding water (Soudzilovskaia et al., 2010). In conclusion, the highly specific core microbiome of Sphagnum, which is maintained during the whole life cycle, can be subsequently modified by abiotic factors.
To find out how this specific bacterial diversity is acquired, we decided to include also the sporophyte generation into our sampling design. The sporophyte develops from the zygote within the female sex organ or archegonium, and in its early development, is therefore nurtured by the gametophyte. Inside the sporophyte capsules, embedded within thousands of spores, we observed bacteria in high abundance and diversity. Although the proportion differed, we found a high degree of qualitative similarity between the microbial communities of the sporophyte and the gametophyte, including genera well known for their beneficial plant interaction, for example, Rhizobium, Bradyrhizobium and Caulobacter. The direct transfer of bacteria within the whole life cycle is an interesting observation. From microbial ecology studies of higher plants, we know that plants, which represent the diploid sporophyte, acquire their populations mainly from soil (Garbeva et al., 2004; Berg and Smalla, 2009), which was also found for endophytes (Hallmann et al., 1997; Berg et al., 2005). However, recently, it was shown that seeds also harbour microorganisms, which originate from the mother plants (Van Overbeek et al., 2011).
There are several reasons that suggest Sphagnum mosses as unique models to study plant-associated microbial diversity, as well as plant–microbe interaction: (i) they were the first land plants and had a long time of co-evolution, (ii) they harbour a highly specific microbial diversity, (iii) they are not influenced by soil due to their ombrotrophic lifestyle, (iv) they present a bridge between terrestrial and aquatic habitats, (v) they are easy to investigate microscopically, because they consist only of a one cell-layer network, (vi) they can be cultivated in vitro to study specific interactions, and (vii) they are relevant for our climate on earth. In our study, we found a remarkable bacterial diversity on Sphagnum mosses. This is, besides the impact on climate change, one more reason to protect this hidden beautiful biodiversity inside bogs.
Acknowledgments
We would like to thank Igor Tikhonovich (St Petersburg), Christin Zachow and Henry Müller (Graz) for inspiring discussions. This study was supported by the Austrian Science Foundation FWF with a grant to GB.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Belyea LR, Malmer N. Carbon sequestration in peatland: patterns and mechanisms of response to climate change. Glob Change Biol. 2004;10:1043–1052. [Google Scholar]
- Berg G, Krechel A, Ditz M, Sikora RA, Ulrich A, Hallman J. Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol Ecol. 2005;51:215–229. doi: 10.1016/j.femsec.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol. 2009;68:1–13. doi: 10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- Binladen J, Gilbert MTP, Bollbacj JP, Panitz F, Bendixen C, Nielsen R, et al. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE. 2007;2:1–9. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood CB, Adam O, Buyer JS. Phylum- and class-specific PCR primers for general microbial community analysis. Appl Environ Microbiol. 2005;71:6193–6198. doi: 10.1128/AEM.71.10.6193-6198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelius MK, Triplett EW. The diversity of archaea and bacteria in association with roots of Zea mays L. Microb Ecol. 2001;41:252–263. doi: 10.1007/s002480000087. [DOI] [PubMed] [Google Scholar]
- Daniels RE, Eddy A. Handbook of Europaen Sphagna. Natural Environment Research Council. Institute of Terrestrial Ecology; Cambrian News: Aberystwyth, UK; 1985. p. 262. [Google Scholar]
- Dedysh SN, Panikov NS, Tiedje JM. Acidophilic methanotrophic communities from Sphagnum peat bogs. Appl Environ Microbiol. 1998;64:922–929. doi: 10.1128/aem.64.3.922-929.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedysh SN, Derakshani M, Liesack W. Detection and enumeration of methanotrophs in acidic Sphagnum peat by 16S rRNA fluorescence in situ hybridization, including the use of newly developed oligonucleotide probes for Methylocella palustris. Appl Environ Microbiol. 2001;67:4850–4857. doi: 10.1128/AEM.67.10.4850-4857.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dise NB. Environmental science. Peatland response to global change. Science. 2009;326:810–811. doi: 10.1126/science.1174268. [DOI] [PubMed] [Google Scholar]
- Ellenberg H, Weber HE, Dull R, Wirth V, Werner W.1991Zeigerwerte von Pflanzen in Mitteleuropa Verlag Erich Goltze KG: Goettingen; p 248. [Google Scholar]
- Felsenstein J. PHYLIP – phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Garbeva P, van Jeen JA, van Elsas JD. Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol. 2004;42:243–270. doi: 10.1146/annurev.phyto.42.012604.135455. [DOI] [PubMed] [Google Scholar]
- Granath G, Wiedermann MM, Strengbom J. Physiological responses to nitrogen and sulphur addition and raised temperature in Sphagnum balticum. Oecologia. 2009;161:481–490. doi: 10.1007/s00442-009-1406-x. [DOI] [PubMed] [Google Scholar]
- Grube M, Cardinale M, Vieira de Castro Junior J, Müller H, Berg G. Species-specific structural and functional diversity of bacterial communities in lichen symbiosis. ISME J. 2009;3:1105–1115. doi: 10.1038/ismej.2009.63. [DOI] [PubMed] [Google Scholar]
- Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW. Endophytic bacteria in agricultural crops. Can J Microbiol. 1997;43:5–914. [Google Scholar]
- Horn MA, Matthies C, Küsel K, Schramm A, Drake HL. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl Environ Microbiol. 2003;69:74–83. doi: 10.1128/AEM.69.1.74-83.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences I multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- Hur I, Chun J. A method for comparing multiple bacterial community structures from 16S rDNA clone library sequences. J Microbiol. 2004;42:9–13. [PubMed] [Google Scholar]
- Jassey VEJ, Gilbert D, Binet P, Toussaint M, Chiapusioa G. Effect of a temperature gradient on Sphagnum fallax and its associated living microbial communities: a study under controlled conditions. Can J Microbiol. 2011;57:226–235. doi: 10.1139/W10-116. [DOI] [PubMed] [Google Scholar]
- Joosten H, Couwenberg J.2009Are emissions reductions from peatlands MRV- able?Report in: Wetlands International, Ede. p14
- Kropf S, Läuter J, Eszlinger M, Krohn K, Paschke R. Nonparametric multiple test procedures with data-driven order of hypotheses and with weighted hypotheses. J Stat Plan Infer. 2004;125:31–47. [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E, Goodfellow M (eds)Nucleic acid techniques in bacterial systematic Wiley: New York; 115–175. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Larmola T, Tuittila ES, Tiirola M, Nykänen H, Martikainen PJ, Yrjälä K, et al. The role of Sphagnum mosses in the methane cycling of a boreal mire. Ecology. 2010;91:2356–2365. doi: 10.1890/09-1343.1. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepš J, Smilauer P.2003Multivariate analysis of ecological data using Canoco Cambridge University Press: Cambridge, UK; p282. [Google Scholar]
- Martin-Laurent F, Philippot L, Hallet S, Chaussod R, Germon JC, Soulas G, et al. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl Environ Microbiol. 2001;67:2354–2359. doi: 10.1128/AEM.67.5.2354-2359.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opelt K, Berg C, Berg G. The bryophyte genus Sphagnum is a reservoir for powerful and extraordinary antagonists and potentially facultative human pathogens. FEMS Microbiol Ecol. 2007b;61:38–53. doi: 10.1111/j.1574-6941.2007.00323.x. [DOI] [PubMed] [Google Scholar]
- Opelt K, Berg C, Schönmann S, Eberl L, Berg G. High specificity but contrasting biodiversity of Sphagnum-associated bacterial and plant communities in bog ecosystems independent of the geographical region. ISME J. 2007c;1:502–516. doi: 10.1038/ismej.2007.58. [DOI] [PubMed] [Google Scholar]
- Opelt K, Berg G. Diversity and antagonistic potential of bacteria associated with bryophytes from nutrient-poor habitats of the Baltic Sea Coast. Appl Environ Microbiol. 2004;70:6569–6579. doi: 10.1128/AEM.70.11.6569-6579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opelt K, Chobot V, Hadacek F, Schönmann S, Eberl L, Berg G. Investigations of the structure and function of bacterial communities associated with Sphagnum mosses. Environ Microbiol. 2007a;91:2795–2809. doi: 10.1111/j.1462-2920.2007.01391.x. [DOI] [PubMed] [Google Scholar]
- Pankratov TA, Serkebaeva YM, Kulichevskaya IS, Liesack W, Dedysh SN. Substrate-induced growth and isolation of Acidobacteria from acidic Sphagnum peat. ISME J. 2008;2:551–560. doi: 10.1038/ismej.2008.7. [DOI] [PubMed] [Google Scholar]
- Raghoebarsing AA, Smolders AJP, Schmid MC, Rijpstra WIC, Wolters-Arts M, Derksen JM, et al. Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature. 2005;436:1153–1156. doi: 10.1038/nature03802. [DOI] [PubMed] [Google Scholar]
- Rahman T, Crombie A, Chen Y, Stralis-Pavese N, Bodrossy L, Meir P, et al. Environmental distribution and abundance of the facultative methanotroph Methylocella. ISME J. 2010;5:1061–1066. doi: 10.1038/ismej.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffers AP, Sỳkora KV. Reliability of Ellenberg indicator values for moisture, nitrogen and soil reaction: a comparison with field measurements. J Veg Sci. 2000;11:225–244. [Google Scholar]
- Schloss PD, Handelsman J. Toward a census of bacteria in soil. PLoS Comput Biol. 2006;2:786–793. doi: 10.1371/journal.pcbi.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieger F, Tebbe CC. A new approach to utilize PCR–single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl Environ Microbiol. 1998;64:4870–4876. doi: 10.1128/aem.64.12.4870-4876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudzilovskaia NA, Cornelissen JHC, During HJ, van Logtestijn RSP, Lang SI, Aerts R. Similar cation exchange capacities among bryophyte species refute a presumed mechanism of peatland acidification. Ecology. 2010;91:2716–2726. doi: 10.1890/09-2095.1. [DOI] [PubMed] [Google Scholar]
- Sun L, Qiu F, Zhang X, Dai X, Dong X, Song W. Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microb Ecol. 2008;55:415–424. doi: 10.1007/s00248-007-9287-1. [DOI] [PubMed] [Google Scholar]
- Theurillat JP, Guisan A. Potential impact of climate change on vegetation in the European Alps: A review. Climatic Change. 2001;50:77–109. [Google Scholar]
- Whinam J, Copson G. Sphagnum moss: an indicator of climate change in the sub-Antarctic. Polar Record. 2006;42:43–49. [Google Scholar]
- Van Overbeek L, Franke AC, Nijhuis EHM, Groeneveld RMW, Nunes da Rocha U, Lotz LAP. Bacterial communities associated with Chenopodium album and Stellaria media seeds from arable soils. Microb Ecology. 2011;62:257–264. doi: 10.1007/s00248-011-9845-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.