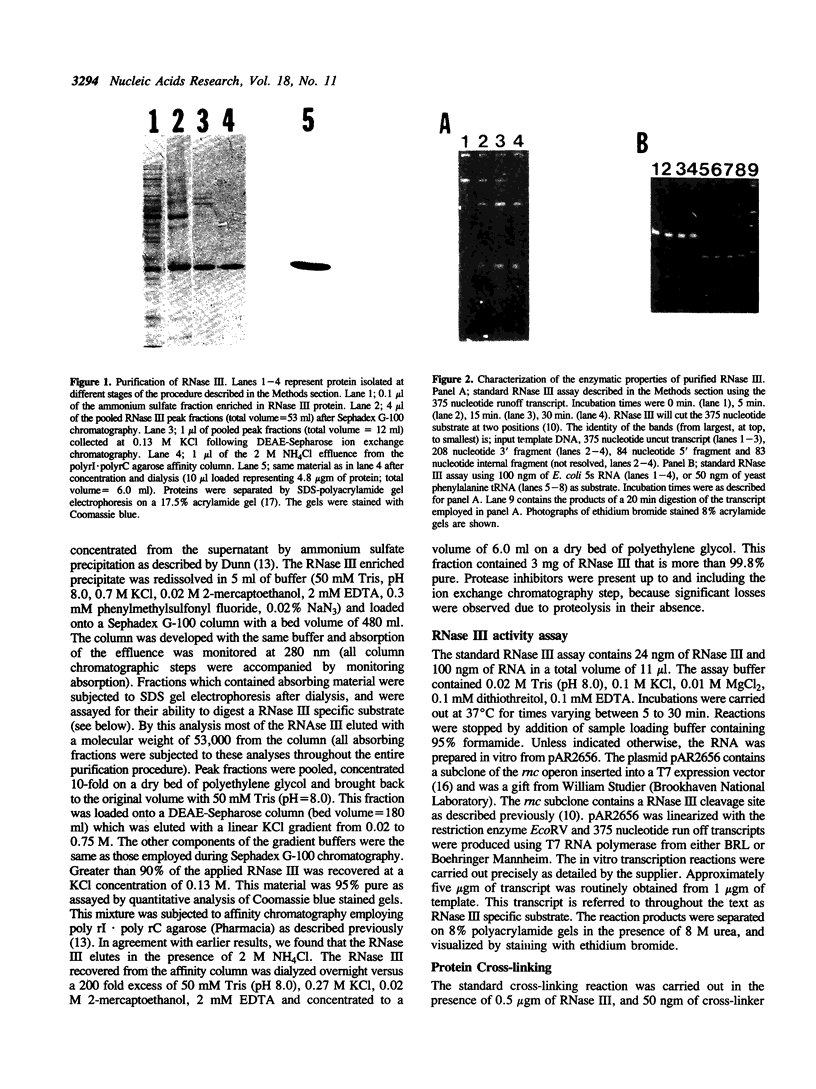
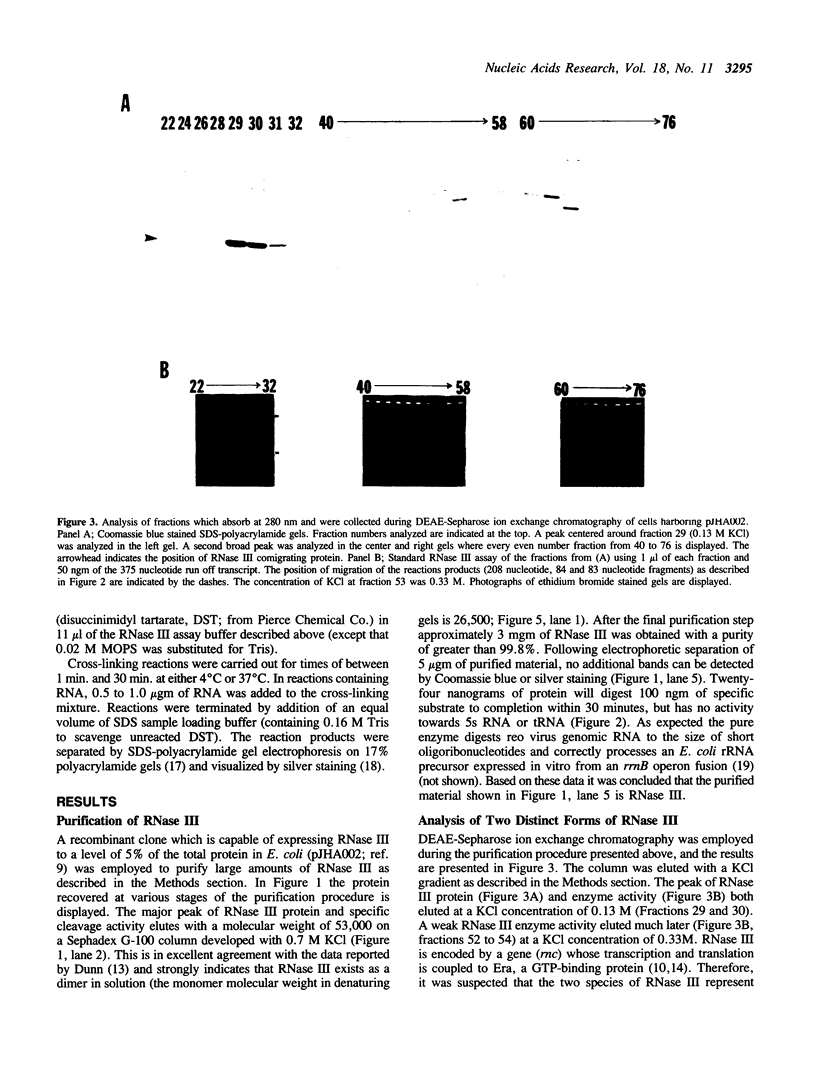
Abstract
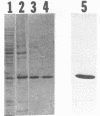
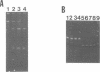
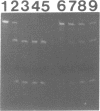
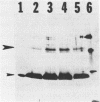
An Escherichia coli double strand specific endoribonuclease, RNase III, was cloned, expressed in large amounts, and purified to homogeneity. Enzyme activity was monitored by assaying fractions for the ability to correctly process exogenous RNA containing specific RNase III cleavage sites. DEAE-Sepharose ion exchange chromatography in the presence of a linear KCl gradient (from 0.02 M to 0.75 M) demonstrated that RNase III exists as two distinct forms. One form elutes at a KCl concentration of 0.13 M and the other elutes at 0.33 M. The presence of stoichiometric amounts of the GTP-binding protein Era during purification results in the conversion of the low salt form into the high salt form. Size exclusion chromatography demonstrated that both forms exist as a dimer in solution. In order to investigate the nature of the dimer, protein cross-linking was performed and cross-linked products were detected by silver staining. The protein-protein dimer can be visualized at protein:cross-linker molar ratios as low as 1:15 within 1 minute of exposure to cross-linker in 0.1 M KCl. Upon addition of substrate RNA to the cross-linking reaction a second form of the protein-protein dimer (with a slightly smaller apparent molecular weight) becomes prominent. Induction of the new form is absolutely dependent upon the addition of substrate mRNA to the reaction mixture. We postulate that the RNase III dimer undergoes a dramatic conformational change upon recognition of RNA which we are able to trap by cross-linking.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahnn J., March P. E., Takiff H. E., Inouye M. A GTP-binding protein of Escherichia coli has homology to yeast RAS proteins. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8849–8853. doi: 10.1073/pnas.83.23.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. W., Baum P. R., Gesteland R. F. Processing of adenovirus 2-induced proteins. J Virol. 1973 Aug;12(2):241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell J. C., Régnier P., Chen S. M., Nakamura Y., Grunberg-Manago M., Court D. L. Autoregulation of RNase III operon by mRNA processing. EMBO J. 1989 Nov;8(11):3401–3407. doi: 10.1002/j.1460-2075.1989.tb08504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Dunn J. J. RNase III cleavage of single-stranded RNA. Effect of ionic strength on the fideltiy of cleavage. J Biol Chem. 1976 Jun 25;251(12):3807–3814. [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Fishel R., Kolodner R. Gene conversion in Escherichia coli: the recF pathway for resolution of heteroduplex DNA. J Bacteriol. 1989 Jun;171(6):3046–3052. doi: 10.1128/jb.171.6.3046-3052.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada T., Kawakami K., Chen S. M., Takiff H. E., Court D. L., Nakamura Y. Temperature-sensitive lethal mutant of era, a G protein in Escherichia coli. J Bacteriol. 1989 Sep;171(9):5017–5024. doi: 10.1128/jb.171.9.5017-5024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler P., Keil T. U., Hofschneider P. H. Isolation and characterization of a ribonuclease 3 deficient mutant of Escherichia coli. Mol Gen Genet. 1973 Oct 16;126(1):53–59. doi: 10.1007/BF00333481. [DOI] [PubMed] [Google Scholar]
- Kole R., Baer M. F., Stark B. C., Altman S. E. coli RNAase P has a required RNA component. Cell. 1980 Apr;19(4):881–887. doi: 10.1016/0092-8674(80)90079-3. [DOI] [PubMed] [Google Scholar]
- Lozeron H. A., Anevski P. J., Apirion D. Antitermination and absence of processing of the leftward transcript of coliphage lambda in the RNAase III-deficient host. J Mol Biol. 1977 Jan 15;109(2):359–365. doi: 10.1016/s0022-2836(77)80039-9. [DOI] [PubMed] [Google Scholar]
- March P. E., Ahnn J., Inouye M. The DNA sequence of the gene (rnc) encoding ribonuclease III of Escherichia coli. Nucleic Acids Res. 1985 Jul 11;13(13):4677–4685. doi: 10.1093/nar/13.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March P. E., Lerner C. G., Ahnn J., Cui X., Inouye M. The Escherichia coli Ras-like protein (Era) has GTPase activity and is essential for cell growth. Oncogene. 1988 Jun;2(6):539–544. [PubMed] [Google Scholar]
- Mayer J. E., Schweiger M. RNase III is positively regulated by T7 protein kinase. J Biol Chem. 1983 May 10;258(9):5340–5343. [PubMed] [Google Scholar]
- Oakley B. R., Kirsch D. R., Morris N. R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980 Jul 1;105(2):361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Whiteley H. R. Bacillus subtilis RNAase III cleavage sites in phage SP82 early mRNA. Cell. 1983 Jul;33(3):907–913. doi: 10.1016/0092-8674(83)90033-8. [DOI] [PubMed] [Google Scholar]
- Pines O., Yoon H. J., Inouye M. Expression of double-stranded-RNA-specific RNase III of Escherichia coli is lethal to Saccharomyces cerevisiae. J Bacteriol. 1988 Jul;170(7):2989–2993. doi: 10.1128/jb.170.7.2989-2993.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier C., Dondon L., Grunberg-Manago M., Régnier P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5' end. EMBO J. 1987 Jul;6(7):2165–2170. doi: 10.1002/j.1460-2075.1987.tb02484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. D., Webster R. E., Zinder N. D. Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem. 1968 Jan 10;243(1):82–91. [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56(1):125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Saito H., Richardson C. C. Processing of mRNA by ribonuclease III regulates expression of gene 1.2 of bacteriophage T7. Cell. 1981 Dec;27(3 Pt 2):533–542. doi: 10.1016/0092-8674(81)90395-0. [DOI] [PubMed] [Google Scholar]
- Steen R., Dahlberg A. E., Lade B. N., Studier F. W., Dunn J. J. T7 RNA polymerase directed expression of the Escherichia coli rrnB operon. EMBO J. 1986 May;5(5):1099–1103. doi: 10.1002/j.1460-2075.1986.tb04328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiff H. E., Chen S. M., Court D. L. Genetic analysis of the rnc operon of Escherichia coli. J Bacteriol. 1989 May;171(5):2581–2590. doi: 10.1128/jb.171.5.2581-2590.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson N., Apirion D. Molecular cloning of the gene for the RNA-processing enzyme RNase III of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Feb;82(3):849–853. doi: 10.1073/pnas.82.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]