Considering that 25% of Earth's terrestrial surface is underlain by permafrost (ground that has been continuously frozen for at least 2 years), our understanding of the diversity of microbial life in this extreme habitat is surprisingly limited. Taking into account the total mass of perennially frozen sediment (up to several hundred meters deep), permafrost contains a huge amount of buried, ancient organic carbon (Tarnocai et al., 2009). In addition, permafrost is warming rapidly in response to global climate change (Romanovsky et al., 2010), potentially leading to widespread thaw and a larger, seasonally thawed soil active layer. This concern has prompted the question: will permafrost thawing lead to the release of massive amounts of carbon dioxide (CO2) and methane (CH4) into the atmosphere? This question can only be answered by understanding how the microbes residing in permafrost will respond to thaw, through processes such as respiration, fermentation, methanogenesis and CH4 oxidation (Schuur et al., 2009).
Predicting future carbon fluxes is complicated by the diversity of permafrost environments, ranging from high mountains, southern boreal forests, frozen peatlands and Pleistocene ice complexes (yedoma) up to several hundred meters deep, which vary widely in soil composition, soil organic matter (SOM) quality, hydrology and thermal regimes (Figure 1). Permafrost degradation can occur in many forms: thaw can progress downward from seasonally-thawed ‘active layer' soils in warming climates or laterally because of changes in surface or groundwater flow paths (Grosse et al., 2011). Permafrost degradation can sometimes lead to dramatic changes in ecosystem structure and function, such as the formation of thermokarst bogs. Wildfires and other disturbances that remove vegetation and organic matter warm the ground, hastening permafrost degradation. The complexity of the Northern Arctic and Subarctic environments in terms of geology, vegetation, paleohistory and climate, suggests that understanding the microbial ecology in permafrost regions will require numerous studies throughout the Pan-Arctic.
Figure 1.
Several Alaskan permafrost features that affect carbon cycling in northern circumpolar soils. (a) Low-centered polygonal ground and thaw lakes from the North Slope; (b, c) polygonal tundra in Barrow; (d) thermokarst terrain in tussock tundra near Council; (e) subsurface core section containing organic matter in permafrost from Fairbanks.
Frozen conditions in permafrost efficiently preserve biological material from DNA to wooly mammoths. Low water potential, reduced protein flexibility and enzyme activity, limited membrane fluidity, and ice nucleation and melting are all potentially lethal, so it was long assumed that microbes were either dead or dormant when frozen. However, high ionic strength within pore water can depress the freezing point and preserve cell viability. Recent experiments demonstrated that permafrost microorganisms remain active at extremely low temperatures (Vishnivetskaya et al., 2006; Gilichinsky and Rivkina, 2011): indigenous bacteria incorporated 14C-labeled acetate into lipids down to −20 °C, many isolates showed psychroactive growth at −2.5 °C and acetotrophic methanogenesis continued between 5 °C and −17 °C. Thus, warming could induce SOM decomposition even before permafrost thaws completely. Microbial activity at low temperatures could transform complex organic compounds to soluble metabolites and gases, including the greenhouse gases (GHG): CO2, CH4 and N2O (Figure 2). Microbial transformations of carbon and nitrogen compounds in the soil active layer can in turn affect plant productivity and community composition, changing animal habitats and affecting human land use. Although these gas fluxes and landscape changes can be directly measured, we need more detailed, mechanistic models that integrate geochemical and structural parameters with microbial processes to explain and predict future changes in permafrost regions. In order to accurately predict the vulnerability of permafrost carbon to decomposition and the resulting fluxes of GHG products, we should understand how Arctic microbial communities respond to permafrost thawing. In addition to an increase in the rate of organic matter decomposition because of warming, there will most likely be shifts in the microbial community composition and in the relative consumption of different organic constituents.
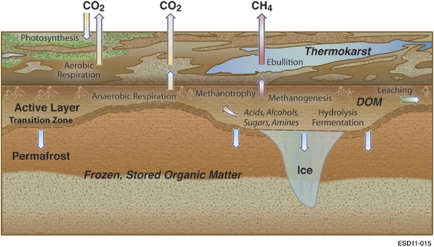
Figure 2.
Key biological processes in the carbon cycle of permafrost environments. Permafrost thawing at the transition zone introduces previously unavailable organic matter into the expanded active layer of soil. Enzymatic hydrolysis decomposes complex organic matter into soluble substrates for microbial fermentation, producing a mixture of organic acids, alcohols and microbial biomass. Methanogenic archaea convert acetate, methylated compounds or H2 and CO2 into CH4 that can be released to the atmosphere through ebullition, diffusion or aerenchyma. Methanotrophs oxidize some of this CH4, converting it to CO2.
Here we address how combining information about microbial identity (from phylotyping) and metabolic potential (from metagenomes) with information on what genes are expressed and functioning (from metatranscriptomes and metaproteomes) (VerBerkmoes et al., 2009; Jansson, 2011) has begun to open a new window of opportunity to map and predict biochemical processes in soils. They provide the tools to distinguish among different mechanisms for increased organic matter decomposition caused by increasing temperature (Conant et al., 2011). Two potential applications of this systems biology framework, discussed below, illustrate how these molecular techniques could be used to improve our ability to model SOM degradation rates and mechanisms, and to predict changes in CH4 and CO2 emissions from thawing permafrost.
Currently, we cannot predict how microbes will use SOM released by permafrost thawing, or reliably estimate the temperature-dependent activities of the enzymes they produce to degrade this material. Current biogeochemical models segregate SOM into conceptual pools with different mean residence times (Smith et al., 1997). If most organic matter trapped in permafrost is difficult to degrade because of its chemical structure (for example, lignin) or its physical structure (for example, particulates or mineral complexes), then this humus comprises a recalcitrant pool that will slowly stimulate microbial growth and GHG production. Alternatively, if plant litter was rapidly frozen in permafrost, then microbes could quickly metabolize thawed polymers like cellulose or protein. Although chemical methods can be used to characterize this SOM, a complementary approach could use transcriptional and proteomic measurements of enzyme expression to analyze the microbial response to this SOM and infer its properties. Increased temperature may also cause changes in protein structure and conformation, protein adsorption, altered protein expression and shifts in microbial populations, which are not currently modeled (Waldrop et al., 2010; Wallenstein et al., 2011). We might expect soil warming to select for microbes producing enzymes that degrade SOM more efficiently at higher temperatures. If transcriptomic, proteomic and enzymatic analyses confirm this hypothesis through experiments replicating in situ conditions, then thermal acclimation of enzymes would need to be incorporated into models to accurately predict long-term C dynamics in response to permafrost thaw (Allison et al., 2010).
Predictions of soil GHG flux include increasingly sophisticated representations of processes in the subsurface carbon cycle (Figure 2), but these models are poorly parameterized for permafrost regions (Riley et al., 2011). 16S rRNA gene sequence data have identified both hydrogenotrophic and acetotrophic (methylotrophic) methanogen phylotypes in Arctic tundra samples, at substantial abundance (Wagner and Liebner, 2010). The two groups of methanogens differ in their substrates, syntrophic associations and isotopic fractionation of carbon: it is important to distinguish between the methanogenic pathways to predict the proportions of CH4 and CO2, as well as fluxes (Walter et al., 2008). Changes in methanogen abundance could also confuse estimates of the temperature and pH response factors. Comparative metagenomics can identify changes in the abundance of characteristic methanogen genes required for coenzyme biosynthesis and methyl-coenzyme M reductase production, as well as methanotrophic genes from CH4-oxidizing bacteria. Metaproteomics can measure peptides from highly abundant signature enzymes in soil (Chourey et al., 2010), such as methyl-coenzyme M reductase and CH4 monooxygenase. This high-resolution portrait will help determine how the CH4 flux changes in response to permafrost thawing, new SOM inputs and increased fermentation by other community members.
Until recently, the high microbial diversity of Arctic soils (Bartram et al., 2011) has hindered metagenomic studies. New technologies produce sequences on an unprecedented scale, at a fraction of the traditional cost. Accordingly, the first metagenomic analyses of permafrost are now becoming available. An analysis of one active layer soil and two-meter deep permafrost sample from the Canadian high Arctic identified signature genes for hydrogenotrophic methanogenesis, CH4 oxidation by type I methanotrophs, nitrification and carbohydrate degradation (Yergeau et al., 2010). Microbial abundances were 10–100 times lower in the permafrost than in the active layer sample, resulting in low DNA yields and complicating the downstream analysis. This report illustrates the feasibility and challenges of large-scale comparative analyses of metabolic potential in a complex permafrost microbiome. To improve the analysis of permafrost metagenomic data, more reference genome sequences should be produced through parallel techniques, such as genome sequencing of isolated or enriched microbes and single-cell genome sequencing.
Eventually, microbial activities will dictate whether permafrost environments will be a net source or sink of GHG in the coming decades and whether large-scale feedbacks to regional and global climate will develop because of increased CO2, N2O and CH4 emissions and vegetation changes in the Arctic. The new ‘omics' techniques of metagenomics, metatranscriptomics, metaproteomics and metabolomics are developing at an opportune time to provide process-level insights to microbial communities' responses to rapidly changing environments. We need this mechanistic understanding to extrapolate beyond observed events and consequently improve our ability to predict and quantify GHG flux from different permafrost ecosystems as a result of global warming. Biomolecular evidence of key processes in nitrogen and carbon cycling is essential for prioritizing and interpreting geochemical measurements and representing them in high-resolution ecosystem models. These detailed models could in turn help parameterize global climate models. As a scientific community, we advocate coordination and integration of our microbiological data with geochemical measurements to determine the magnitude and impacts of warming in high latitude, circumpolar permafrost systems.
Acknowledgments
We thank Diana Swantek for assistance preparing Figure 2 and Peter Thornton for helpful discussions. This work is funded in part by the US Department of Energy, Office of Science, Biological and Environmental Research (BER) Program. Oak Ridge National Laboratory is managed by UT Battelle, LLC under Contract No. DE-AC05-00OR22725 for the US Department of Energy. Additional support was provided by the US Department of Energy under Contract No. DE-AC02-05CH11231 with the Lawrence Berkeley National Laboratory.
References
- Allison SD, Wallenstein MD, Bradford MA. Soil-carbon response to warming dependent on microbial physiology. Nature Geosci. 2010;3:336–340. [Google Scholar]
- Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl Environ Microbiol. 2011;77:3846–3852. doi: 10.1128/AEM.02772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey K, Jansson J, VerBerkmoes N, Shah M, Chavarria KL, Tom LM, et al. Direct cellular lysis/protein extraction protocol for soil metaproteomics. J Proteome Res. 2010;9:6615–6622. doi: 10.1021/pr100787q. [DOI] [PubMed] [Google Scholar]
- Conant RT, Ryan MG, Ågren GI, Birge HE, Davidson EA, Eliasson PE, et al. Temperature and soil organic matter decomposition rates – synthesis of current knowledge and a way forward. Glob Change Biol. 2011;17:3392–3404. [Google Scholar]
- Gilichinsky DA, Rivkina EM.2011Permafrost microbiologyIn: Reitner J, Thiel V (eds).Encyclopedia of Geobiology Springer: Verlag; 726–732. [Google Scholar]
- Grosse G, Harden J, Turetsky M, McGuire AD, Camill P, Tarnocai C, et al. Vulnerability of high-latitude soil organic carbon in North America to disturbance. J Geophys Res. 2011;116:G00K06. [Google Scholar]
- Jansson J. Towards ‘Tera-Terra': terabase sequencing of terrestrial metagenomes. Microbe. 2011;6:309–315. [Google Scholar]
- Riley WJ, Subin ZM, Lawrence DM, Swenson SC, Torn MS, Meng L, et al. Barriers to predicting changes in global terrestrial methane fluxes: analyses using CLM4Me, a methane biogeochemistry model integrated in CESM. Biogeosciences. 2011;8:1925–1953. [Google Scholar]
- Romanovsky VE, Smith SL, Christiansen HH. Permafrost thermal state in the polar Northern Hemisphere during the international polar year 2007–2009: a synthesis. Permafrost Periglac. 2010;21:106–116. [Google Scholar]
- Schuur EAG, Vogel JG, Crummer KG, Lee H, Sickman JO, Osterkamp TE. The effect of permafrost thaw on old carbon release and net carbon exchange from tundra. Nature. 2009;459:556–559. doi: 10.1038/nature08031. [DOI] [PubMed] [Google Scholar]
- Smith P, Smith JU, Powlson DS, McGill WB, Arah JRM, Chertov OG, et al. A comparison of the performance of nine soil organic matter models using datasets from seven long-term experiments. Geoderma. 1997;81:153–225. [Google Scholar]
- Tarnocai C, Canadell JG, Schuur EAG, Kuhry P, Mazhitova G, Zimov S. Soil organic carbon pools in the northern circumpolar permafrost region. Global Biogeochem Cycles. 2009;23:GB2023. [Google Scholar]
- VerBerkmoes NC, Denef VJ, Hettich RL, Banfield JF. Systems biology: functional analysis of natural microbial consortia using community proteomics. Nat Rev Microbiol. 2009;7:196–205. doi: 10.1038/nrmicro2080. [DOI] [PubMed] [Google Scholar]
- Vishnivetskaya TA, Petrova MA, Urbance J, Ponder M, Moyer CL, Gilichinsky DA, et al. Bacterial community in ancient Siberian permafrost as characterized by culture and culture-independent methods. Astrobiology. 2006;6:400–414. doi: 10.1089/ast.2006.6.400. [DOI] [PubMed] [Google Scholar]
- Wagner D, Liebner S.2010Methanogenesis in Arctic permafrost habitatsIn: Timmis KN (ed).Handbook of Hydrocarbon and Lipid Microbiology Springer: Berlin Heidelberg; 655–663. [Google Scholar]
- Waldrop MP, Wickland KP, White R, III, Berhe AA, Harden JW, Romanovsky VE. Molecular investigations into a globally important carbon pool: permafrost-protected carbon in Alaskan soils. Glob Change Biol. 2010;16:2543–2554. [Google Scholar]
- Wallenstein M, Allison SD, Ernakovich J, Steinweg JM, Sinsabaugh R.2011Controls on the temperature sensitivity of soil enzymes: a key driver of In situ enzyme activity ratesIn: Shukla G, Varma A (eds).Soil Enzymology Springer: Berlin Heidelberg; 245–258. [Google Scholar]
- Walter KM, Chanton JP, Chapin FS, III, Schuur EAG, Zimov SA. Methane production and bubble emissions from Arctic lakes: isotopic implications for source pathways and ages. J Geophys Res. 2008;113:G00A08. [Google Scholar]
- Yergeau E, Hogues H, Whyte LG, Greer CW. The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR and microarray analyses. ISME J. 2010;4:1206–1214. doi: 10.1038/ismej.2010.41. [DOI] [PubMed] [Google Scholar]