Abstract
Antigen receptor loci are regulated to promote allelic exclusion, but the mechanisms are not well understood. Assembly of a functional T cell receptor (TCR) β chain gene triggers feedback inhibition of Vβ-to-DJβ recombination in double positive (DP) thymocytes which correlates with reduced Vβ chromatin accessibility and a locus conformational change that separates Vβ from DJβ gene segments. We previously generated a Tcrb allele that maintained Vβ accessibility but was still subject to feedback inhibiton in DP thymocytes. We have now further analyzed the contributions of chromatin accessibility and locus conformation to feedback inhibition using two novel TCR alleles. We show that reduced Vβ accessibility and increased distance between Vβ and DJβ gene segments both enforce feedback inhibition in DP thymocytes.
Introduction
A defining characteristic of T- and B-lymphocytes is their ability to create and express unique antigen receptors that can recognize a vast array of foreign pathogens. To achieve receptor diversity, antigen receptor variable domains are encoded by multiple variable (V), diversity (D), and joining (J) gene segments that are joined by a process known as V(D)J recombination (1). Recombination Activating Genes 1 and 2 (RAG1/2) mediate V(D)J recombination by: (i) binding to recombination signal sequences (RSSs)3 that flank antigen receptor gene segments, (ii) bringing two RSSs (one with a 12 and one with a 23 bp spacer) into a synaptic complex, and (iii) generating DNA double strand breaks between the coding sequences and RSSs. Hairpin-sealed coding ends are subsequently opened by the Artemis endonuclease and ligated by non-homologous end joining proteins to form antigen receptor coding joints. Because RAG1/2-generated double strand breaks are potentially toxic, V(D)J recombination is highly regulated.
B cell receptor and T cell receptor (TCR) genes undergo stepwise recombination in developing B and T lymphocytes, respectively (2–4). Igh rearranges in pro-B cells and Igk and Igl rearrange in pre-B cells; Tcrb, Tcrg and Tcrd rearrange in CD4−CD8− double negative (DN) thymocytes and Tcra rearranges in CD4+CD8+ double positive (DP) thymocytes. Moreover, Igh and Tcrb rearrangements are ordered such that D-to-J recombination precedes V-to-DJ recombination. This regulation is achieved, in part, by cis-elements such as enhancers and promoters that alter the chromatin landscape to make RSSs accessible to RAG1/2 (5). Accessible chromatin is characterized by active transcription, by histone H3 and H4 acetylation, by histone H3 lysine 4 trimethylation (H3K4me3), by displacement and removal of nucleosomes, and by hypomethylation of CpG dinucleotides (2, 4). H3K4me3-modified nucleosomes also stimulate V(D)J recombination by docking RAG2 (6, 7) and enhancing the catalytic activity of the RAG1/2 complex (8).
Antigen receptor loci also undergo changes in their conformation during lymphocyte development (9). A contracted locus conformation is thought to promote V(D)J recombination by facilitating the interaction between RSSs separated by great distances (e.g. Vβ and Dβ RSSs, VH and DH RSSs). Detailed analysis of contracted Igh loci revealed that VH segments spanning 2.5 megabases are all situated proximal to DH RSSs, presumably affording them all an opportunity for recombination (10). This interpretation is supported by the behavior of Pax5 deficient pro B-cells, in which Igh contraction and distal VH recombination are both impaired (11).
Antigen receptor loci are also regulated to enforce allelic exclusion (12–14). For Igh and Tcrb, allelic exclusion is manifest at the V-to-DJ step and is thought to occur in two phases: 1) an initiation phase, in which V-to-DJ recombination is regulated so that it is not attempted simultaneously on the two alleles, and 2) a maintenance phase, in which V-to-DJ recombination is terminated by a feedback mechanism once an in-frame rearrangement is produced. Feedback inhibition of Igh recombination in pre B-cells and of Tcrb recombination in DP thymocytes is associated with epigenetic and locus conformational changes. Thus, whereas Igh and Tcrb alleles are by multiple criteria accessible in pro-B cells and DN thymocytes, respectively, their V gene segments display reduced accessibility in pre-B cells and DP thymocytes (2, 13, 14). In addition, unrearranged Igh and Tcrb alleles, while contracted in pro-B and DN thymocytes, respectively, become decontracted in pre B-cells and DP thymocytes (15, 16). These changes could inhibit recombination by limiting RAG1/2 binding to V segment RSSs and the likelihood of RSS synapsis.
Several genetically modified Igh and Tcrb alleles have been created to assess the significance of these changes for feedback inhibition. Two Tcrb alleles with large deletions (βLD and Vβ1 NT) (17, 18) moved the otherwise distant Vβ10 gene segment into proximity of DJβ gene segments and increased its accessibility in DP thymocytes. Disruption of allelic exclusion was detected on Vβ1 NT alleles only, but no data evaluated whether altered Vβ10 recombination reflected a loss of feedback inhibition in DP thymocytes as opposed to dysregulated rearrangement in DN thymocytes. Another study simply inserted a Vβ gene segment just upstream of DJβ gene segments (19). While allelic exclusion was perturbed at the level of Vβ recombination, whether this reflected a loss of feedback inhibition in DP thymocytes was not evaluated in this study either. Bates et al. generated a modified Igh allele in which a VH gene segment was introduced just upstream of DH gene segments (20). This allele clearly displayed a disruption of feedback inhibition in pre-B cells. However, as the genetic manipulation moved the VH into an accessible chromatin domain and also modulated distance, the individual effects of accessibility and distance could not be distinguished.
Jackson et al. previously generated a Tcrb allele in which Vβ accessibility was maintained in DP thymocytes by introducing the Tcra enhancer (Eα) into the middle of the Vβ array (EαKI allele) (21). Despite accessible Vβ chromatin, feedback inhibition of Vβ-to-DJβ recombination was maintained in DP thymocytes, indicating that parameters other than chromatin accessibility must be essential to enforce feedback inhibition in DP thymocytes. We have now further analyzed contributions of gene segment accessibility and proximity to feedback inhibition through the generation of two novel TCR alleles. Our results establish that reduced RSS accessibility and increased distance between RSSs both contribute to feedback inhibition of Vβ-to-DJβ recombination in DP thymocytes.
Materials and Methods
Mice and gene targeting
Wild-type 129, Rag2−/− (22) and Rag2−/− mice containing a rearranged Tcrb tg (23) were purchased from Taconic. EαKI, EαKI Rag2−/−, and EαKI Rag2−/− Tcrb tg mice were previously described (21). All mice were used in accordance with protocols approved by the Duke University and Washington University Animal Care and Use Committees.
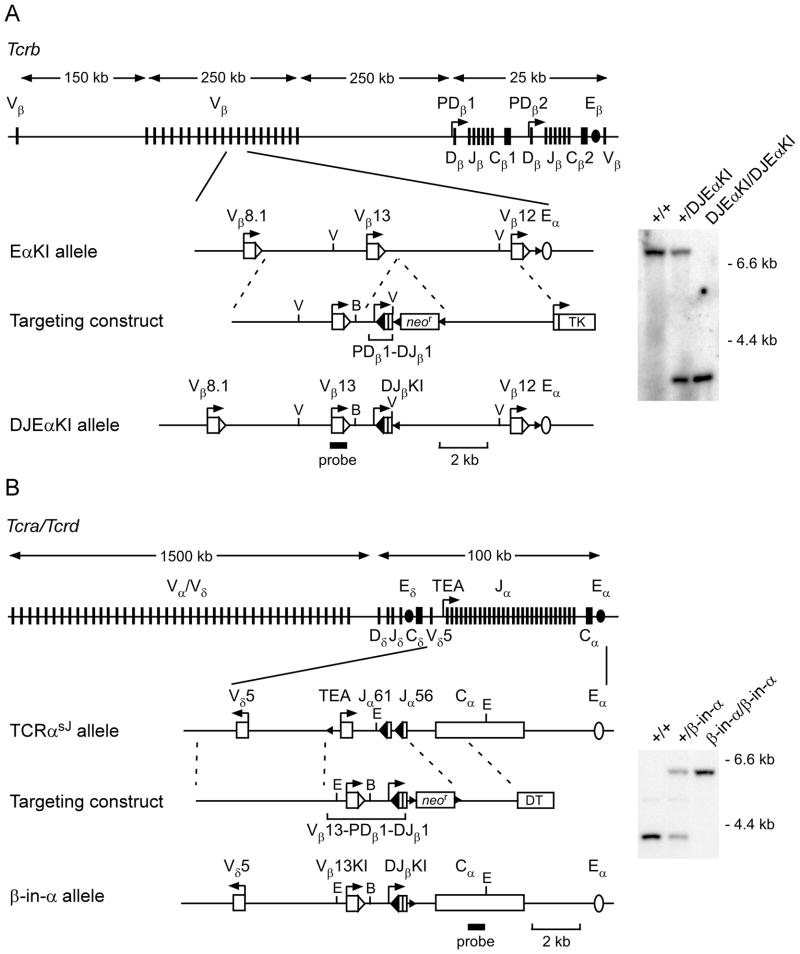
DJEαKI mice were generated as follows: DJβKI and homology arms were PCR amplified using Pfu Turbo (Stratagene) and cloned using a TOPO Cloning kit (Invitrogen, Carlsbad, CA). DJβKI was PCR amplified from a plasmid carrying PDβ1 and a Dβ1-Jβ1.1 rearrangement (5′ end at nucleotide 152528 and 3′ end at nucleotide 154066 of Genbank file MMAE000665). 5′ and 3′ homology arms extended from nucleotides 2531 to 8110 and nucleotides 8115 to 12960 of Genbank file MMAE000664. We introduced a BamHI site 90 bp 3′ of the Vβ13 RSS at nucleotide 7425. Cloned fragments were introduced into the pLNTK vector containing a phosphoglycerate kinase (PGK) promoter-driven loxP flanked neomycin-resistance (neor) cassette and a thymidine kinase selection marker. The 5′ arm and DJβKI were cloned into the XhoI site and the 3′ arm was cloned into the SalI site. ES cells derived from EαKI mice (21) were used for homologous recombination which was verified by Southern blot of SacI-digested genomic DNA analyzed with a 5′ Tcrb probe and of EcoRI-digested genomic DNA analyzed with a 3′ Tcrb probe (Supplementary Table 1). The neor cassette was removed by transient transfection of ES cells with Cre recombinase.
β-in-α mice were generated as follows: The Tcrb substrate was amplified from the DJEαKI targeting construct and extends from nucleotide 5794 of GenBank file MMAE000664 (upstream of the Vβ13 promoter) to nucleotide 154066 of GenBank file MMAE000665 (downstream of Jβ1.1). 5′ and 3′ homology arms extended from nucleotides 12960 to 18291 and 84861 to 87477 of GenBank file M64239. Homology arms and the Tcrb substrate were cloned into PGKneolox2.DTA (http://www.addgene.org/pgvec1?f=c&identifier=13449&cmd=findpl&attag=c). The 5′ arm and Tcrb substrate were cloned between the NotI and XmaI sites. The 3′ arm was cloned between the NheI and the SalI sites. ES cells derived from TCRαsJ/TCRαsJ mice (24) were used for homologous recombination, which was verified by Southern blot of EcoRI-digested genomic DNA analyzed with a 3′ TCRα (Cα) probe and using KpnI and BamHI-digested genomic DNA analyzed with a 5′ TCRα probe (Supplementary Table 1). The neor cassette was removed by transient transfection of ES cells with Cre recombinase.
3D-FISH
Bacterial artificial chromosome clones 75P5 (5′Tcrb probe) and 203H5 (3′Tcrb probe) were directly labeled and used for 3D-FISH as previously described (25). Probe-to-probe distances were calculated as previously described (25). Only nuclei with two distinguishable signals for both alleles were analyzed. Statistical tests were performed using Prism 3.0 (GraphPad Software, Inc.).
Cell sorting
DN3 thymocytes were isolated as previously described (26). Cells were stained with the following antibodies for 30 minutes on ice: Cy5-conjugated anti-CD3ε (clone 145-2C11), anti-CD4 (clone GK1.5), and anti-CD8 (clone 53-6.7); biotinylated anti-CD24 (clone M1-69), PE-conjugated anti-CD44 (clone 1M7), and FITC-conjugated anti-CD25 (clone 7D4). After washing, cells were stained with Texas Red-conjugated streptavidin. Cells were collected from the CD24+CD3−CD4−CD8− and CD25+CD44+ gates. DP thymocytes were isolated as previously described (26). Cells were stained with FITC-conjugated anti-CD4 (clone GK1.5) and PE-conjugated anti-CD8 (clone 53-6.7) for 30 minutes on ice. Cells were collected from the CD4+CD8+ gate. All antibodies were purchased from eBioscience. Samples were sorted to at least 95% purity using a DiVa cell sorter (Becton Dickinson); analysis was with CellQuest software.
Chromatin immunoprecipitation
Chromatin was prepared from primary thymocytes of 2–3 week old mice by small scale micrococcal nuclease digestion as previously described (27). Immunoprecipitations were performed using anti-H3K4me3 (Millipore, 04–745, clone MC315) and normal rabbit IgG (R&D Systems, AB-105-C ). All samples were resuspended in 200 μl of 10 mM Tris-HCl pH 8, 0.1 mM EDTA; input samples were further diluted 1:50. Bound and input samples were quantified using 2 μl of sample by SYBR Green real-time PCR using a LightCycler 480 (Roche). All PCR amplifications used a touchdown strategy (TD-qPCR) in which annealing temperature was reduced gradually from 65°C to 58°C over 10 cycles, followed by 35 cycles at 58°C. Primers used are listed in Supplementary Table 1.
Germline transcription
Approximately 2 × 107 primary thymocytes were resuspended in 1 ml of Trizol (Invitrogen) and RNA was isolated according to manufacturer’s instructions. cDNA was synthesized using the Super Script III kit (Invitrogen) using up to 2 μg of purified RNA. Transcripts were quantified by SYBR Green real-time PCR using the TD-qPCR program. Actb was amplified for 40 cycles at 62°C. Primers are listed in Supplementary Table 1.
Coding joints
Genomic DNA isolated from sorted DN3 and DP thymocytes was amplified by touchdown PCR (TD-PCR) as follows: 5 minutes at 94°C, 31–35 cycles of 30 seconds at 94°C, 30 seconds at annealing temperature, and 30–60 seconds at 72°C. Annealing temperature was held at 68°C, 66°C and 64°C for 5 cycles each and at 62°C for the remaining cycles. Amplicons were resolved on a 2.0% agarose gel, were transferred onto a nylon membrane, and were detected by hybridization with γ 32P-labeled oligonucleotide probes. Cd14 was amplified for 23 cycles at 62°C. Primers and probes are listed in Supplementary Table 1.
RSS retention
DP thymocyte genomic DNA was amplified by TD-qPCR as described above. Primers are listed in Supplementary Table 1.
Genomic Southern blot
Whole thymus genomic DNA was digested with restriction enzyme, subjected to 0.7% agarose gel electrophoresis and transferred to a nylon membrane. Blots were hybridized with α32P-labeled probes listed in Supplementary Table 1.
Signal ends
Genomic DNA (1.5 μg) of DN3 and DP thymocytes was analyzed by LM-PCR as previously described (21, 28). Amplification by TD-PCR and detection of amplicons was as described above. Supplementary Table 1 lists linker sequences, primers and probes.
Results
EαKI locus conformation
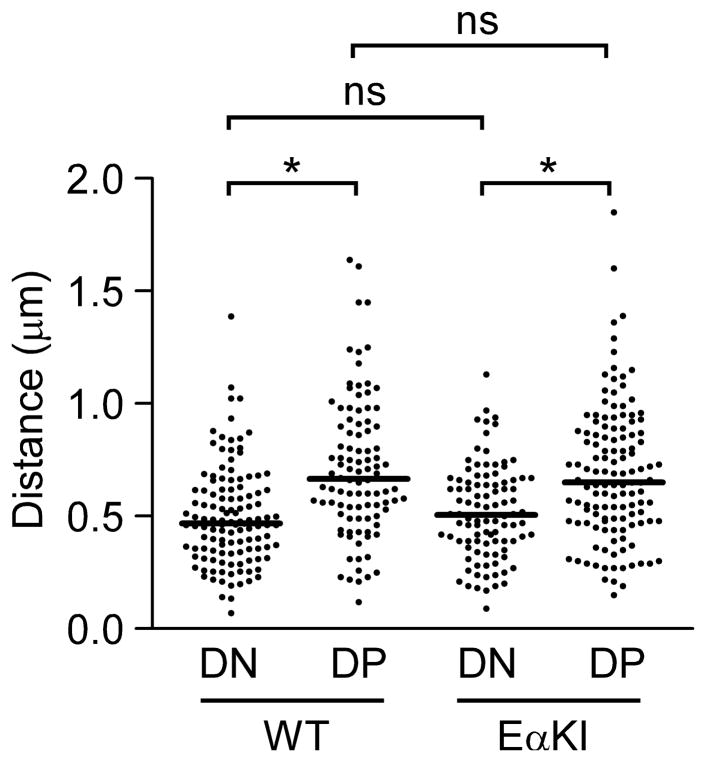
Previous studies of mice carrying Tcrb alleles with an introduced Eα (EαKI; Fig 1A) indicated that elevation of Vβ accessibility, by itself, could not subvert feedback inhibition of Vβ-to-DJβ recombination in DP thymocytes. We hypothesized that recombination might remain suppressed on accessible EαKI alleles if, like wild-type alleles, they were decontracted in DP thymocytes. To assess this, we used three-dimensional fluorescence in situ hybridization (3D-FISH) to measure the distance between the 5′ and 3′ ends of wild-type and EαKI Tcrb alleles in recombinase-deficient DN and DP thymocytes (Fig 2). Consistent with previous experiments (16), we found that, on average, wild-type alleles were contracted in DN thymocytes and decontracted in DP thymocytes. The behavior of EαKI alleles was indistinguishable from wild-type, indicating that they decontract in DP thymocytes despite the presence of Eα and an accessible chromatin configuration. Therefore, to formally test whether the distance between accessible Vβ and DJβ gene segments limited Vβ-to-DJβ recombination in DP thymocytes, we generated and characterized two novel TCR locus alleles that approximated accessible Vβ and DJβ gene segments in DP thymocytes.
FIGURE 1.
TCR loci and gene targeting strategies. (A) Generation of the DJEαKI allele. Top, Tcrb locus, including relative positions of and distances between the V, D, J and C gene segments and cis-elements. Below, EαKI allele, targeting construct and DJEαKI allele. Right, Southern blot analysis of genomic DNA from wild-type (+/+), heterozygous (+/DJEαKI) and homozygous (DJEαKI/DJEαKI) mice. DNA was digested with EcoRV and hybridized with a Vβ13 probe. Expected wild-type and DJEαKI fragments are 7.1 kb and 3.5 kb, respectively. (B) Generation of the β-in-α allele. Top, Tcra/d locus, including relative positions of and distances between the V, D, J and C gene segments and cis-elements. Below, TCRαsJ allele, targeting construct and β-inα allele. Right, Southern blot of genomic DNA from wild-type (+/+), heterozygous (+/βin-α) and homozygous (β-in-α/β-in-α) mice. DNA was digested with EcoRI and hybridized with a 3′ TCRα (Cα) probe. Expected wild-type and β-in-α fragments are 3.8 kb and 5.9 kb, respectively. V, EcoRV; E, EcoRI, B, BamHI; DT, diphtheria toxin; bent arrow, promoter; open and filled large triangles, 23 bp and 12 bp RSSs, respectively; small triangles, loxP sites.
FIGURE 2.
Conformation of wild-type and EαKI alleles. 3D-FISH was performed using probes to the 5′ and 3′ ends of the Tcrb locus. Scatter plots display distances between centers of probe hybridization in Rag2−/− DN thymocytes (130 alleles from 3 slides), EαKI Rag2−/− DN thymocytes (90 alleles from 3 slides), DP thymocytes from Rag2−/− mice treated with an anti-CD3ε (90 alleles from 3 slides), and DP thymocytes from EαKI Rag2−/− mice treated with anti-CD3ε (130 alleles from 3 slides). Median values are indicated by horizontal lines. *, P<0.0001; ns, not statistically significant. Data were accumulated from two independent experiments for each cell type.
Regulation of the DJEαKI allele
We used homologous recombination to introduce a cassette containing the Dβ1 promoter (PDβ1) and a rearranged Dβ1Jβ1.1 (DJβKI) approximately 1.0 kb 3′ of the Vβ13 RSS on the EαKI allele (DJEαKI allele; Fig 1A). Mice homozygous for the DJEαKI allele displayed normal thymocyte development as assessed by cell number and expression of cell surface markers CD4, CD8, CD25 and CD44 (data not shown).
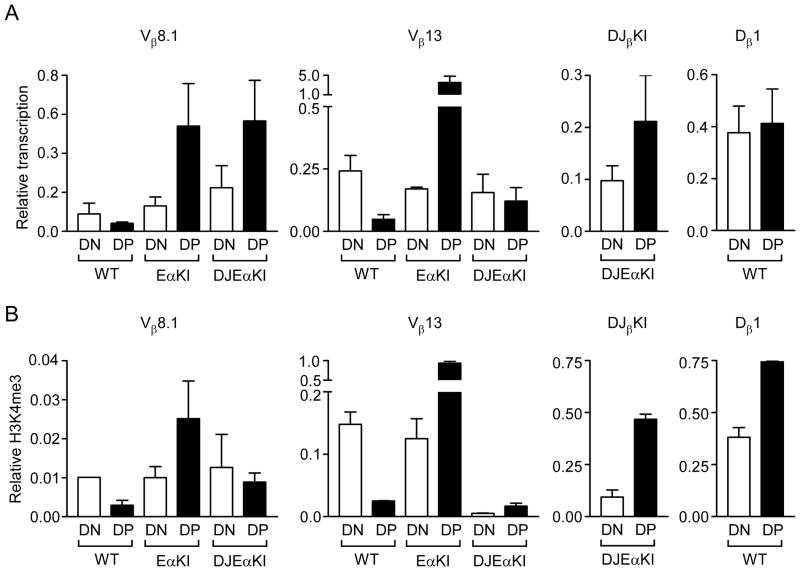
We addressed chromatin accessibility on the DJEαKI allele by introducing it onto Rag2-deficient (Rag2−/−) and Rag2−/− xTcrb transgene (tg) backgrounds for analysis of steady state germline transcripts and histone modifications in DN and DP thymocytes. Like Dβ1 transcripts on wild-type alleles, DJβKI transcripts on DJEαKI alleles were of comparable abundance in DN and DP thymocytes (Fig 3A, right). Moreover, like nucleosomes at the Dβ1 RSS on wild-type alleles, those at the DJβKI RSS on DJEαKI alleles were H3K4me3-modified in DN thymocytes and displayed increased H3K4me3 in DP thymocytes (Fig 3B, right). Hence, the DJβKI RSS appears to reside in accessible chromatin in both DN and DP thymocytes of DJ EαKI mice.
FIGURE 3.
Chromatin accessibility of DJEαKI alleles. (A) Germline transcription in wild-type (WT), EαKI and DJEαKI DN (all Rag2−/−) and DP (all Rag2−/− x Tcrb tg) thymocytes, analyzed by quantitative real-time PCR. Results were normalized to those for Actb and represent the mean ± s.e.m. of 2–6 independent experiments, using cDNA diluted 1:10 for Dβ1 and Actb PCRs and undiluted cDNA for Vβ13 and Vβ8.1 PCRs. (B) Chromatin immunoprecipitation of H3K4me3-modified nucleosomes of wild-type, EαKI and DJEαKI DN (all Rag2−/−) and DP (all Rag2−/− x Tcrb tg) thymocytes. Ratios of bound to input were normalized to those for β2–microglobulin (B2m) and represent the mean ± s.e.m. of 2–4 independent experiments.
As documented previously (21), germline transcription of Vβ13 is downregulated on transition from DN to DP on wild-type alleles but is upregulated on EαKI alleles (Fig 3A, middle). Unexpectedly, we found that introduction of the DJβKI blunted the effect of Eα on Vβ13 in DJEαKI DP thymocytes, such that Vβ13 transcripts were upregulated as compared to wild-type DP thymocytes, but were no more abundant in DJEαKI DP than in DJEαKI DN thymocytes. This might reflect a suppression of transcription due to competition between PDβ1 and the Vβ13 promoter, or an effect of the DJβKI on Vβ13 transcript stability. The DJβKI also unexpectedly suppressed H3K4me3 at the Vβ13 RSS in both DN and DP thymocytes (Fig. 3B, center). Given these results, we also analyzed accessibility at Vβ8.1, which lies 5 kb upstream of Vβ13. Unlike Vβ13, the upregulation of Vβ8.1 transcription in EαKI DP thymocytes was maintained in DJEαKI DP thymocytes (Fig. 3A, left). Moreover, the upregulation of Vβ8.1 H3K4me3 in EαKI DP thymocytes was only partly suppressed by the DJβKI (Fig. 3B, left). Taken together, the transcription and chromatin data suggest that Vβ13 is moderately accessible and that Vβ8.1 is highly accessible in DJEαKI DN and DP thymocytes.
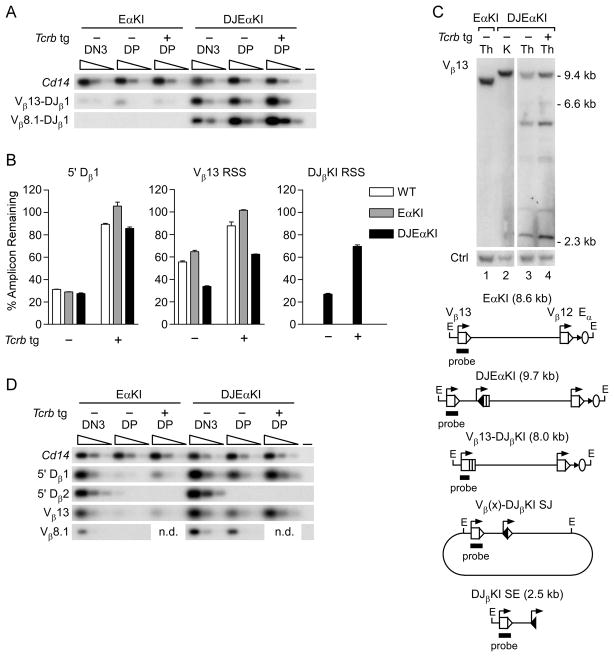
To analyze Vβ-to-DJβ recombination, we prepared genomic DNA from purified DN3 and DP thymocytes of EαKI and DJEαKI mice, amplified with Vβ13 and Jβ1.1 primers, and distinguished DJβKI from endogenous DJβ1.1 rearrangement using a DJβKI-specific probe (Fig. 4A). Vβ13-to-DJβKI rearrangement was readily detected in sorted DN3 and DP thymocytes of DJEαKI mice but not in EαKI controls. To measure the frequency of DJβKI recombination, we quantified residual unrearranged Vβ13 and DJβKI RSSs in DP thymocyte genomic DNA (Fig. 4B, center and right). We found that approximately 70% of the DJβKI and Vβ13 RSSs were lost in DJEαKI DP thymocytes. These losses likely reflect recombination to DJβKI as well as recombination to the endogenous Dβ gene segments that would delete Vβ13 and DJβKI.
FIGURE 4.
Recombination of DJEαKI alleles. (A) Coding joint analysis. Genomic DNAs from sorted EαKI and DJEαKI DN3 and DP thymocytes without (−) or with (+) a Tcrb tg were serially three-fold diluted (wedges) and analyzed by PCR. Blots of PCRs using Vβ and Jβ1.1 primers were hybridized with a DJβKI-specific probe. Cd14 amplification was used to control for DNA loading. –, no DNA. Data are representative of two independent experiments. (B) RSS usage. Genomic DNAs from DJEαKI kidney and from sorted wild-type (WT), EαKI and DJEαKI DP thymocytes, without (−) or with (+) a Tcrb tg, were analyzed by quantitative real-time PCR. Percent amplicon remaining was calculated as [(experimental amplicon in thymus/experimental amplicon in kidney)/(B2m in thymus/B2m in kidney)] x 100. Data are the mean ± s.e.m. of 2–3 samples for each genotype. (C) Genomic Southern blot. Top, unfractionated thymus (Th) and kidney (K) genomic DNAs were digested with EcoRI and analyzed by Southern blot using a Vβ13 probe. DNA loading was assessed using a control (Ctrl) trypsinogen probe. Below, schematic of expected EcoRI fragments, including a diagram of predicted excision circles containing Vβ(x)-DJβKI signal joint (SJ) recombination products. (D) SE analysis. Thymocyte genomic DNA samples were linker-ligated, serially three-fold diluted (wedges) and analyzed by PCR. Cd14 amplification was used to control for DNA loading. –, no DNA. The data are representative of two independent experiments. n.d., not determined.
We also analyzed DJβKI recombination by genomic Southern blot of EcoRI-digested whole thymus DNA (Fig. 4C). As compared to DJEαKI kidney (lane 2), a Vβ13 probe detected substantial loss of DNA carrying unrearranged Vβ13 and DJβKI, and detected two major and several minor rearranged fragments (lane 3). However, the predicted 8.0 kb fragment representing Vβ13-DJβKI rearrangement was not detected. The additional rearranged fragments may represent excision circles carrying signal joints generated by rearrangement of upstream Vβ segments to DJβKI (eg., Vβ8.1 = 6.7 kb, Vβ8.2 = 5.5 kb Vβ8.3 = 3.0 kb, Vβ5.1 = 4.2 kb and, Vβ5.2 = 4.6 kb), as well as DJβKI signal end (SE) recombination intermediates (2.5 kb), all of which would hybridize to the Vβ13 probe. Consistent with the former, we detected Vβ8.1-to-DJβKI recombination using a PCR strategy (Fig. 4A). Excision circles and SE intermediates generated in DN thymocytes should be undetectable by genomic Southern blot of whole thymus because they would be diluted by the proliferative burst that accompanies the DN to DP transition. The apparent abundance of Vβ13-containing excision circles and DJβKI SE intermediates in DJEαKI thymocytes suggested that they were generated by recombination events occurring in DP rather than in DN thymocytes.
To test directly for Vβ and DJβKI recombination in DP thymocytes, we used ligation-mediated-PCR (LM-PCR) to detect SE recombination intermediates in sorted thymocyte subpopulations (Fig. 4D). Because this assay cannot distinguish DJβKI from endogenous Dβ1 SEs, we evaluated DJβKI SEs by comparison of DJEαKI to EαKI samples. As expected, 5′ Dβ1, Vβ13 and Vβ8.1 SEs were readily detected in EαKI DN thymocytes but were barely detected in EαKI DP thymocytes. However these SE intermediates were all readily detected in DJEαKI DP thymocytes (Fig. 4D). In contrast, control 5′ Dβ2 SEs were undetectable in DJEαKI DP thymocytes, indicating a selective loss of feedback inhibition involving DJβKI and upstream Vβ gene segments.
To formally demonstrate that DJβKI rearrangements in DP thymocytes occurred chromosomally rather than on excision circles generated by Vβ-to-endogenous Dβ recombination in DN thymocytes, we analyzed Vβ-to-DJβKI recombination in thymocytes of DJEαKI mice that express a Tcrb tg. Feedback inhibition by such transgenes specifically suppresses Vβ-to-DJβ recombination and the excision circles generated by these recombination events (21). Indeed, increased retention of a DNA segment situated 5′ of Dβ1, normally lost during Vβ-to-endogenous DJβ recombination, was apparent in wild-type, EαKI and DJEαKI Tcrb tg DP thymocytes (Fig. 4B, left). However, suppression of Vβ13 and DJβKI RSS loss was only partial in DJEαKI Tcrb tg DP thymocytes and for Vβ13 was much diminished as compared to the complete suppression in EaKI Tcrb tg DP thymocytes (Fig. 4B, center and right). This indicates continued chromosomal recombination of Vβ13 and upstream Vβs to DJβKI in DP thymocytes, despite feedback inhibition of endogenous Vβ-to-DJβ recombination by the Tcrb tg. Consistent with this interpretation, recombination events detected by the Vβ13 probe were more abundant on Southern blots of DJEαKI Tcrb tg thymus DNA (Fig. 4C, compare lanes 3 and 4) and Vβ13 and DJβKI SE intermediates were more abundant in DJEαKI Tcrb tg thymus DNA (Fig. 4D). We conclude that by reducing the distance between accessible gene segments, the DJβKI promotes chromosomal Vβ recombination in DP thymocytes and thereby subverts the process of feedback inhibition.
Regulation of theβ-in-α allele
To further assess constraints on Tcrb gene segment recombination in DP thymocytes, we introduced a Tcrb recombination substrate into the Tcra locus, since this locus normally undergoes recombination in DP thymocytes (Tcrb-in-Tcra; β-in-α allele). The Tcrb substrate contained the same DJβKI as in the DJEαKI allele, with the Vβ13 promoter and gene segment (Vβ13KI) situated just upstream (Fig. 1B). A BamHI site introduced approximately 90 bp 3′ of the Vβ13 RSS was used in some experiments to distinguish the Vβ13KI from the endogenous Vβ13. We used homologous recombination to introduce this Tcrb recombination substrate into a previously generated Tcra allele (TCRαsJ, which contains only the Jα61 and Jα56 gene segments (24)), such that it replaces the TEA promoter and the entire Jα array of the wild-type Tcra locus. In this way, the Tcrb recombination substrate carries Vβ and DJβ segments that are in close physical proximity and that will be accessible in DP thymocytes due to the activity of the endogenous Eα. We generated heterozygous β-in-α mice which were then intercrossed to produce β-in-α homozygous mice (Fig. 1B). These mice displayed normal DN thymocyte development and efficient differentiation to the DP stage, but were blocked in their development beyond the DP stage (data not shown). We presume that the chimeric TCRα proteins encoded by β-in-α alleles (which would include Dβ and Jβ, rather than Jα sequences) are either unstable, cannot assemble with TCRβ proteins, or cannot create a TCRαβ complex that can support positive selection.
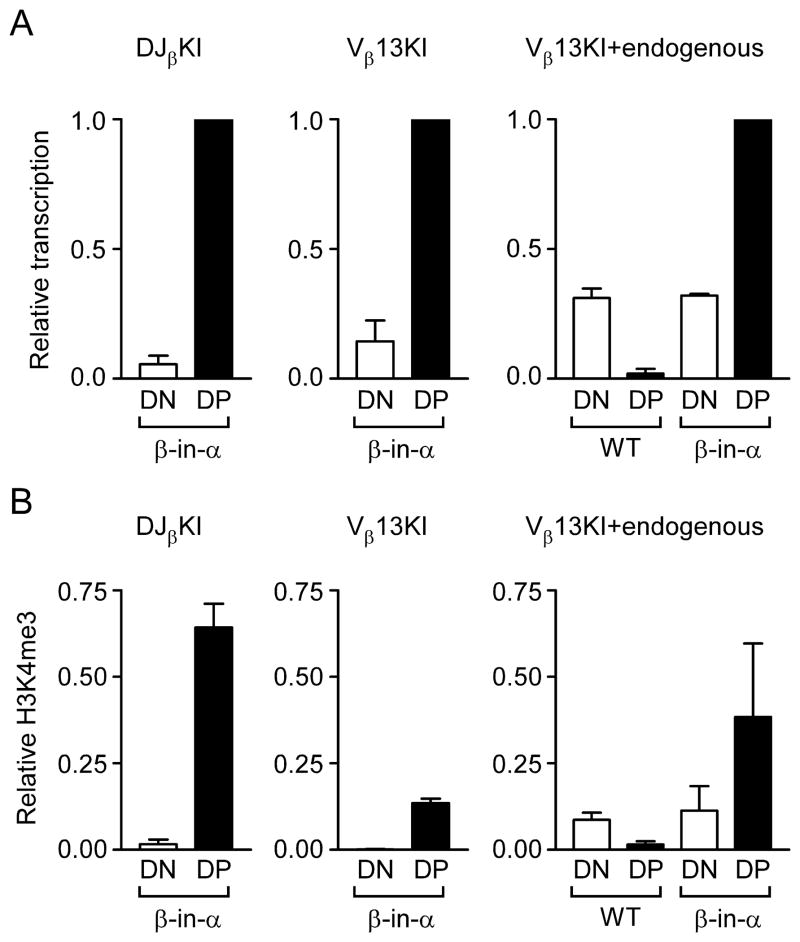
We analyzed germline transcription and H3K4me3 modified nucleosomes on the β-in-α allele after introducing it onto Rag2−/− and Rag2−/− xTcrb tg backgrounds. As expected, transcription of the DJβKI was low in DN thymocytes and was substantially upregulated in DP thymocytes (Fig. 5A, left). Specific amplification of Vβ13KI revealed it to behave similarly (Fig. 5A, center). We also directly compared Vβ13KI transcription to endogenous Vβ13 transcription using a PCR strategy that amplified both equally (Fig. 5B, right). Vβ13KI transcripts were more abundant in β-in-α DP thymocytes than were endogenous Vβ13 transcripts in wild-type DN thymocytes. Similar conclusions were drawn from analysis of H3K4me3 on β-in-α alleles (Fig. 5B). These data suggest that the Tcrb substrate, like Jα gene segments on a wild-type allele, is regulated by Eα and is highly accessible in β-in-α DP thymocytes.
FIGURE 5.
Chromatin accessibility of β-in-α alleles. (A) Germline transcription in wild-type (WT) and β-in-α DN (Rag2−/−) and DP (Rag2−/− x Tcrb tg) thymocytes was analyzed by quantitative real-time PCR. Data were normalized to Actb and then expressed as a fraction of the value inβ-in-α DP thymocytes. Results represent the mean ± s.e.m. of 2–4 independent experiments. (B) Chromatin immunoprecipitiation of H3K4me3-modified nucleosomes of wild-type (WT) and β-in-α DN (Rag2−/−) and DP (Rag2−/− x Tcrb tg) thymocytes. Ratios of bound to input were normalized to those for B2m and represent the mean ± s.e.m. of 2–3 independent experiments.
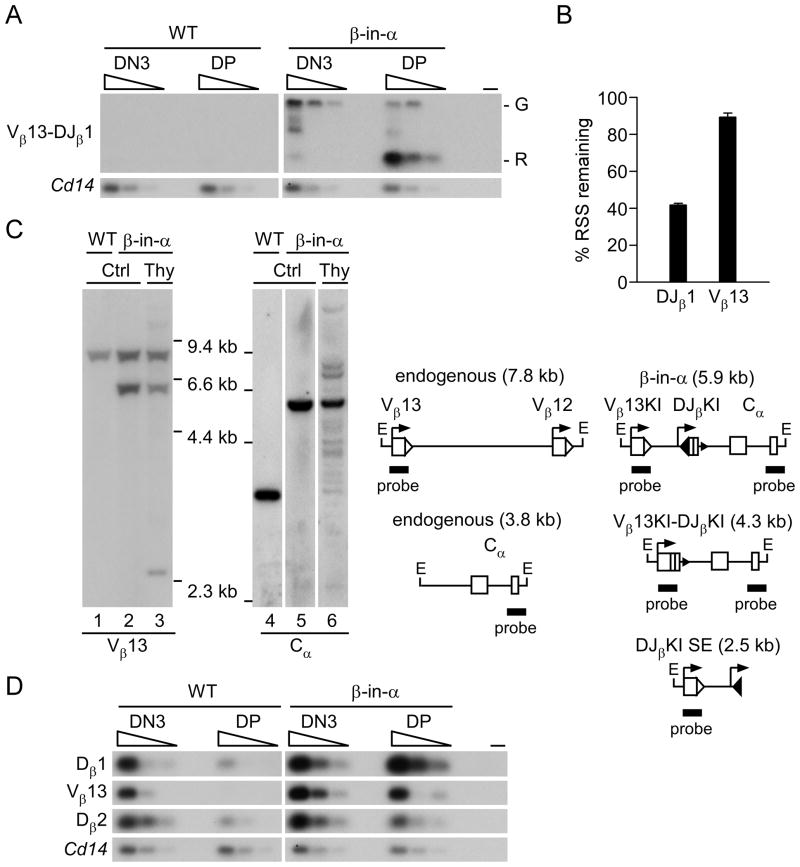
We assayed Vβ13KI-to-DJβKI recombination in wild-type and β-in-α DN3 and DP thymocytes by amplification with Vβ13 and Jβ1.1 primers followed by hybridization with a DJβKI-specific probe (Fig. 6A). This strategy detected the germline (G) substrate in βin-α DN and DP thymocytes (Fig. 6A), but detected abundant Vβ13KI-to-DJβKI rearranged (R) alleles selectively in β-in-α DP thymocytes (Fig. 6A). On the basis of germline RSS loss, fully 60% of the DJβKI had undergone recombination in β-in-α DP thymocytes (Fig. 6B). However, the same analysis indicated that only about 10% of the Vβ13 KI had undergone recombination (Fig. 6B), indicating that DJβKI could rearrange to RSSs other than Vβ13 KI. To further address this, we hybridized EcoRI-digested whole thymus genomic DNA to Vβ13 and Cα probes. In addition to unrearranged Vβ13 (7.8 kb) and unrearranged Vβ13KI (5.9 kb), the Vβ13 probe detected a potential DJβKI SE intermediate (2.5 kb) and one or two additional minor species (Fig. 6C, lane 3). However, consistent with low frequency Vβ13KI recombination, we could not detect Vβ13KI-to-DJβKI rearranged alleles (4.3 kb) (Fig 6C, lane 3). The Cα probe also detected a 5.9 kb unrearranged fragment, but in addition detected a large number of additional recombination events involving the DJβKI, including a potential 4.3 kb Vβ13KI-to-DJβKI rearrangement (Fig. 6C, lane 6). However, the majority of DJβKI rearrangements did not involve Vβ13KI, but presumably, upstream Vα gene segments instead. Thus, Vα RSSs appear to outcompete the Vβ13KI RSS for the DJβKI RSS in vivo. This result is consistent with previous studies demonstrating that proximal Vα segments are contracted and accessible in DP thymocytes (25, 29), and that Vα RSSs are generally superior to Vβ RSSs as recombinase substrates (30).
FIGURE 6.
Recombination of β-in-α alleles. (A) Coding joint analysis. Genomic DNAs from sorted wild-type (WT) and β-in-α DN3 and DP thymocytes were serially three-fold diluted (wedges) and analyzed by PCR. Blots of Vβ to Jβ1.1 PCRs were hybridized with a DJβKI-specific probe. Cd14 amplification was used to control for DNA loading. –, no DNA; G, germline Vβ13 KI-DJβKI; R, rearranged Vβ13KI-DJβKI. Data are representative of two independent experiments. (B) RSS usage. Genomic DNAs from β-in-α kidney and unfractionated thymus were analyzed by quantitative real-time PCR using DJβKI-and Vβ13KI-specific primers. Percent amplicon remaining was calculated as [(experimental amplicon in thymus/experimental amplicon in kidney)/(B2m in thymus/B2m in kidney)] x 100. Data are the mean ± s.e.m. of 2–3 samples. (C) Genomic Southern blot. Left, wild-type (WT) andβ-in-α nonrearranged control tissue (Ctrl) and β-in-α unfractionated thymus (Thy) genomic DNAs were digested with EcoRI and analyzed by Southern blot using Vβ13 or Cα probes. Right, schematic of expected EcoRI fragments. (D) SE analysis. Thymocyte genomic DNA samples were linker-ligated, serially three-fold diluted (wedges) and analyzed by PCR. Cd14 amplification was used to control for DNA loading. Vβ13 primers amplify SEs from endogenous Vβ13 and Vβ13 KI. –, no DNA. The data are representative of three independent experiments.
To confirm that β-in-α alleles undergo recombination and are not subject to feedback inhibition in DP thymocytes, we sorted DN and DP thymocytes from wild-type and β-in-α mice and detected SE recombination intermediates by LM-PCR (Fig. 6D). As previously described (21), wild-type thymocytes displayed 5′Dβ1, 5′Dβ2 and Vβ13 SEs that were abundant in DN but not in DP thymocytes. In contrast, 5′Dβ1 and Vβ13 SEs were at least as abundant in β-in-α DP thymocytes as they were in β-in-α DN thymocytes (Fig. 6D). This represents dysregulation of the β-in-α substrate rather than the endogenous gene segments because 5′Dβ2 SEs were reduced in abundance in DP as compared to DN thymocytes. We conclude that β-in-α alleles undergo both Vβ13KI-to-DJβKI and endogenous Vα-to-DJβKI recombination in DP thymocytes.
Discussion
Numerous studies have correlated reduced antigen receptor locus accessibility and an extended antigen receptor locus conformation with the feedback inhibition of V(D)J recombination that mediates allelic exclusion (2, 13, 14). We previously forced Vβ accessibility in DP thymocytes but could not overcome the inhibition of Vβ-to-DJβ recombination that normally characterizes this compartment (21). Here we found that, like wild-type alleles, those accessible EαKI alleles are extended in DP thymocytes. We therefore generated two new alleles (DJEαKI and β-in-α) to formally test whether gene segment proximity and accessibility are both critical effectors of feedback inhibition. By comparing the behavior of DJEαKI to EαKI alleles, we varied the proximity of accessible Vβ and DJβ segments; in DP thymocytes these gene segments are accessible on both alleles, but they are in physical proximity on DJEαKI alleles only. We found that DJEαKI but not EαKI alleles supported Vβ-to-DJβ recombination in DP thymocytes. By comparing β-in-α alleles in DN and DP thymocytes, we varied the accessibility of proximal Vβ and DJβ segments; these gene segments are in physical proximity in both compartments, but become accessible due to developmental activation of Eα in DP thymocytes only. We found that β-in-α alleles supported Vβ-to-DJβ recombination in DP but not in DN thymocytes. Based on the data from both models, we conclude that gene segment accessibility and gene segment proximity are both essential for chromosomal V(D)J recombination, and that feedback inhibition of Vβ-to-DJβ recombination on wild-type Tcrb alleles in DP thymocytes is normally enforced by both a loss of RSS accessibility to RAG1/2 and a decontracted locus conformation that inhibits RSS synapsis.
Pre-TCR signals initiate feedback inhibition and promote Tcrb epigenetic changes that enforce feedback inhibition, but the critical signaling pathways and downstream effector proteins are only partially understood (14). To the best of our knowledge, the only signaling pathway or downstream effector that has clearly been shown to impact Tcrb allelic exclusion through effects in DP thymocytes is the transcription factor E47. E47 supports Tcrb locus accessibility and recombination in DN thymocytes and is downregulated in response to pre-TCR signaling in DP thymocytes (31). Notably, its overexpression was shown to override feedback inhibition and to promote Vβ-to-DJβ recombination in DP thymocytes (31). However, Tcrb locus accessibility and conformation were not evaluated in E47-overexpressing DP thymocytes, leaving the basis for this override of feedback inhibition undefined. We predict that E47 must support Vβ accessibility and Tcrb locus contraction to account for the described effects on Tcrb recombination.
Although modulation of gene segment proximity appears to represent an important component of the feedback inhibition program, the mechanisms of locus contraction and decontraction are poorly understood. Recent studies have implicated architectural proteins cohesin and CTCF as regulators of long-distance interactions and V(D)J recombination at the Tcra and Igh loci (32–34) but it is not known whether these proteins regulate overall locus conformation. Eμ (35) and transcription factors Pax5 (11, 36), YY1(37) and Ikaros (38) have all been implicated in Igh locus contraction, but whether and how they might trigger Igh locus decontraction is uncertain. Much less is known about the roles of architectural proteins and transcriptional regulators in Tcrb locus contraction and decontraction events. This will certainly be an important avenue for future studies.
Our data argue that gene segment proximity and accessibility are critical determinants of the Tcrb locus feedback inhibition program. Moreover, our results suggest that there are not likely any additional constraints imposed on the rearrangement of most Vβ gene segments to Dβ1 in DP thymocytes, for example, specific factors that regulate the usage of Vβ and 5′Dβ1 RSSs. Were such constraints to exist, they should have been unperturbed by our genetic manipulations, and feedback would have remained intact on both the DJEαKI and β-in-α alleles. We caution that we cannot formally eliminate the possibility that what we interpret to result from a change in physical distance could actually reflect the loss of an intervening regulatory element that is intrinsically inhibitory to Vβ-to-DJβ recombination. The identity of that element would be a matter of speculation. However we imagine that it would function, like a change in physical proximity, to limit synapsis of Vβ and Dβ1 RSSs.
Despite the conclusions outlined above, additional layers of regulation may be required to explain the suppression of certain types of Tcrb locus recombination events in DP thymocytes. A particularly vexing issue is Vβ14-to-DJβ recombination, since, unlike all other Vβ gene segments, Vβ14 is located near Dβ and Jβ gene segments and its accessibility is not downregulated by pre-TCR signaling and is apparently high in DP thymocytes (17, 39–41). Since Dβ and Jβ segments are also accessible and support RAG1/2 binding in DP thymocytes (42), the suppression of Vβ14 rearrangement may depend on unique features of inversional rearrangement (40) or of the Vβ14 RSS (41).
A second issue is the problem of secondary rearrangements. Reduced accessibility and locus decontraction can account for inhibition of Vβ-to-DJβ1 or -DJβ2 rearrangement on a Tcrb allele that had not yet undergone Vβ rearrangement. However, because the Vβ segments immediately upstream of a rearranged Vβ are accessible in DP thymocytes (43, 44) and are proximal to accessible downstream Dβ2 and Jβ2 segments (42) it is not clear what would suppress secondary Vβ-to-DJβ2 rearrangement on an allele that had already undergone primary Vβ-to-DJβ1 rearrangement. Recent work has demonstrated that secondary rearrangements can occur on these alleles, and that they can replace even an in-frame VDJβ1 rearrangement (18), but there was no indication that this occurred in DP as opposed to DN thymocytes. Indeed, analysis of DP thymocytes failed to detect SE intermediates at the accessible Vβ segments upstream of a rearranged Vβ gene segment (43). Moreover, SEs at 5′Dβ2 RSS are strongly suppressed in DP thymocytes (21) (and this study). Thus, DP thymocytes appear not to be permissive for secondary Tcrb recombination.
Because we found that accessible Vβs can rearrange to the DJβKI in DJEαKI DP thymocytes, it seems unlikely that any additional mechanism that might be required to suppress secondary recombination would be directed at Vβ RSSs. However, it remains possible that there is a specific regulatory mechanism directed at the 5′Dβ2 RSS. Indeed, Dβ2 regulation appears to be unusually complex, with promoters both upstream and downstream of Dβ2 (45, 46). The downstream promoter is preferentially active on unrearranged alleles and presumably directs Dβ2-to-Jβ2 rearrangement; the upstream promoter only becomes active once the downstream promoter is eliminated by Dβ2-to-Jβ2 rearrangement and is likely important to direct Vβ-to-DJβ2 rearrangement. Activity of the 5′ promoter suggests that the 5′Dβ2 RSS resides in accessible chromatin on Dβ2-to-Jβ2 rearranged alleles in DP thymocytes. However, it is unclear whether these alleles support RAG1/2 binding, since the only assays of RAG1/2 binding at Dβ2 in DP thymocytes were conducted on alleles that were in germline configuration and in which only the downstream promoter should have been active (42). Therefore it is not known whether RAG1/2 can bind to the 5′Dβ2 RSS in DP thymocytes, and it remains possible that secondary rearrangements could be suppressed in DP thymocytes by a specific mechanism that occludes RAG1/2 binding to the 5′Dβ2 RSS. Our results demonstrate conclusively that, for most Vβ gene segments, accessibility and conformational constraints alone can fully account for the suppression of Vβ-to-DJβ1 recombination in DP thymocytes. However additional work will be required to clarify the mechanisms, beyond accessibility and conformational constraints, which impart feedback inhibition to Vβ14 recombination and to secondary recombination events involving DJβ2.
Supplementary Material
Acknowledgments
We thank L. Martinek of the Duke University Cancer Flow Cytometry Facility for help with cell sorting and analysis; Zanchun Huang for technical assistance; and Dr. Eugene Oltz for critical review of the manuscript.
Footnotes
This work was supported by National Institutes of Health Grant AI049934 (to M.S.K.).
Abbreviations used in this paper: 3D-FISH, three-dimensional fluorescence in situ hybridization; DN, double negative; DP, double positive; Eα, Tcra enhancer; ES, embryonal stem; H3K4me3; histone H3 lysine 4 trimethylation; LM-PCR, ligation-mediated-polymerase chain reaction; PDβ1, promoter Dβ1; RSS, recombination signal sequence; SE, signal end; tg, transgene.
References
- 1.Schatz DG, Spanopoulou E. Biochemistry of V(D)J recombination. Curr Top Microbiol Immunol. 2005;290:49–85. doi: 10.1007/3-540-26363-2_4. [DOI] [PubMed] [Google Scholar]
- 2.Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- 3.Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009;21:133–139. doi: 10.1016/j.coi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 5.Ji Y, Little AJ, Banerjee JK, Hao B, Oltz EM, Krangel MS, Schatz DG. Promoters, enhancers, and transcription target RAG1 binding during V(D)J recombination. J Exp Med. 2010;207:2809–2816. doi: 10.1084/jem.20101136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimazaki N, Tsai AG, Lieber MR. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol Cell. 2009;34:535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jhunjhunwala S, van Zelm MC, Peak MM, Murre C. Chromatin architecture and the generation of antigen receptor diversity. Cell. 2009;138:435–448. doi: 10.1016/j.cell.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mostoslavsky R, Alt FW, Rajewsky K. The lingering enigma of the allelic exclusion mechanism. Cell. 2004;118:539–544. doi: 10.1016/j.cell.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 13.Jackson AM, Krangel MS. Turning T-cell receptor β recombination on and off: more questions than answers. Immunol Rev. 2006;209:129–141. doi: 10.1111/j.0105-2896.2006.00342.x. [DOI] [PubMed] [Google Scholar]
- 14.Brady BL, Steinel NC, Bassing CH. Antigen receptor allelic exclusion: an update and reappraisal. J Immunol. 2010;185:3801–3808. doi: 10.4049/jimmunol.1001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roldan E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M, Skok JA. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat Immunol. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skok JA, Gisler R, Novatchkova M, Farmer D, de Laat W, Busslinger M. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nat Immunol. 2007;8:378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- 17.Senoo M, Wang L, Suzuki D, Takeda N, Shinkai Y, Habu S. Increase of TCR Vβ accessibility within Eβ regulatory region influences its recombination frequency but not allelic exclusion. J Immunol. 2003;171:829–835. doi: 10.4049/jimmunol.171.2.829. [DOI] [PubMed] [Google Scholar]
- 18.Brady BL, Oropallo MA, Yang-Iott KS, Serwold T, Hochedlinger K, Jaenisch R, Weissman IL, Bassing CH. Position-dependent silencing of germline Vβ segments on TCRβ alleles containing preassembled VβDJβCβ1 genes. J Immunol. 2010;185:3564–3573. doi: 10.4049/jimmunol.0903098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sieh P, Chen J. Distinct control of the frequency and allelic exclusion of the Vβ gene rearrangement at the TCR β locus. J Immunol. 2001;167:2121–2129. doi: 10.4049/jimmunol.167.4.2121. [DOI] [PubMed] [Google Scholar]
- 20.Bates JG, Cado D, Nolla H, Schlissel MS. Chromosomal position of a VH gene segment determines its activation and inactivation as a substrate for V(D)J recombination. J Exp Med. 2007;204:3247–3256. doi: 10.1084/jem.20071787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson A, Kondilis HD, Khor B, Sleckman BP, Krangel MS. Regulation of T cell receptor β allelic exclusion at a level beyond accessibility. Nat Immunol. 2005;6:189–197. doi: 10.1038/ni1157. [DOI] [PubMed] [Google Scholar]
- 22.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 23.Shinkai Y, Koyasu S, Nakayama K, Murphy KM, Loh DY, Reinherz EL, Alt FW. Restoration of T cell development in RAG-2-deficient mice by functional TCR transgenes. Science. 1993;259:822–825. doi: 10.1126/science.8430336. [DOI] [PubMed] [Google Scholar]
- 24.Huang CY, Sleckman BP, Kanagawa O. Revision of T cell receptor α chain genes is required for normal T lymphocyte development. Proc Natl Acad Sci U S A. 2005;102:14356–14361. doi: 10.1073/pnas.0505564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shih HY, Krangel MS. Distinct contracted conformations of the Tcra/Tcrd locus during Tcra and Tcrd recombination. J Exp Med. 2010;207:1835–1841. doi: 10.1084/jem.20100772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts JL, Lauzurica P, Krangel MS. Developmental regulation of VDJ recombination by the core fragment of the T cell receptor α enhancer. J Exp Med. 1997;185:131–140. doi: 10.1084/jem.185.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carabana J, Watanabe A, Hao B, Krangel MS. A barrier-type insulator forms a boundary between active and inactive chromatin at the murine TCRβ locus. J Immunol. 2011;186:3556–3562. doi: 10.4049/jimmunol.1003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMurry MT, Hernandez-Munain C, Lauzurica P, Krangel MS. Enhancer control of local accessibility to V(D)J recombinase. Mol Cell Biol. 1997;17:4553–4561. doi: 10.1128/mcb.17.8.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawwari A, Krangel MS. Regulation of TCR δ and α repertoires by local and long-distance control of variable gene segment chromatin structure. J Exp Med. 2005;202:467–472. doi: 10.1084/jem.20050680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang HE, Hsu LY, Cado D, Cowell LG, Kelsoe G, Schlissel MS. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 2002;17:639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- 31.Agata Y, Tamaki N, Sakamoto S, Ikawa T, Masuda K, Kawamoto H, Murre C. Regulation of T cell receptor β gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity. 2007;27:871–884. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 32.Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, Bates JG, Richards N, Myers D, Patel H, Gallagher M, Schlissel MS, Murre C, Busslinger M, Giallourakis CC, Alt FW. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen R. Two Forms of Loops Generate the Chromatin Conformation of the Immunoglobulin Heavy-Chain Gene Locus. Cell. 2011 doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M. The distal VH gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reynaud D, I, Demarco A, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chattopadhyay S, Whitehurst CE, Schwenk F, Chen J. Biochemical and functional analyses of chromatin changes at the TCR-β gene locus during CD4−CD8− to CD4+CD8+ thymocyte differentiation. J Immunol. 1998;160:1256–1267. [PubMed] [Google Scholar]
- 40.Mathieu N, Spicuglia S, Gorbatch S, Cabaud O, Fernex C, Verthuy C, Hempel WM, Hueber AO, Ferrier P. Assessing the role of the T cell receptor β gene enhancer in regulating coding joint formation during V(D)J recombination. J Biol Chem. 2003;278:18101–18109. doi: 10.1074/jbc.M212647200. [DOI] [PubMed] [Google Scholar]
- 41.Yang-Iott KS, Carpenter AC, Rowh MA, Steinel N, Brady BL, Hochedlinger K, Jaenisch R, Bassing CH. TCR β feedback signals inhibit the coupling of recombinationally accessible Vβ14 segments with DJβ complexes. J Immunol. 2010;184:1369–1378. doi: 10.4049/jimmunol.0900723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson AM, Krangel MS. Allele-specific regulation of TCR β variable gene segment chromatin structure. J Immunol. 2005;175:5186–5191. doi: 10.4049/jimmunol.175.8.5186. [DOI] [PubMed] [Google Scholar]
- 44.Jia J, Kondo M, Zhuang Y. Germline transcription from T-cell receptor Vβ gene is uncoupled from allelic exclusion. EMBO J. 2007;26:2387–2399. doi: 10.1038/sj.emboj.7601671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMillan RE, Sikes ML. Differential activation of dual promoters alters Dβ2 germline transcription during thymocyte development. J Immunol. 2008;180:3218–3228. doi: 10.4049/jimmunol.180.5.3218. [DOI] [PubMed] [Google Scholar]
- 46.McMillan RE, Sikes ML. Promoter activity 5′ of Dβ2 is coordinated by E47, Runx1, and GATA-3. Mol Immunol. 2009;46:3009–3017. doi: 10.1016/j.molimm.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.