Abstract
Alzheimer's disease (AD) is the leading cause of dementia among the elderly population; however, knowledge about genetic risk factors involved in disease progression is limited. We conducted a genome-wide association study (GWAS) using clinical decline as measured by changes in the Clinical Dementia Rating-sum of boxes as a quantitative trait to test for single-nucleotide polymorphisms (SNPs) that were associated with the rate of progression in 822 Caucasian subjects of amnestic mild cognitive impairment (MCI). There was no significant association with disease progress for any of the recently identified disease susceptibility variants in CLU, CR1, PICALM, BIN1, EPHA1, MS4A6A, MS4A4E or CD33 following multiple testing correction. We did, however, identify multiple novel loci that reached genome-wide significance at the 0.01 level. These top variants (rs7840202 at chr8 in UBR5: P=4.27 × 10−14; rs11637611 with a cluster of SNPs at chr15q23 close to the Tay–Sachs disease locus: P=1.07 × 10−15; and rs12752888 at chr1: P=3.08 × 10−11) were also associated with a significant decline in cognition as well as the conversion of subjects with MCI to a diagnosis of AD. Taken together, these variants define approximately 16.6% of the MCI sub-population with a faster rate of decline independent of the other known disease risk factors. In addition to providing new insights into protein pathways that may be involved with the progress to AD in MCI subjects, these variants if further validated may enable the identification of a more homogeneous population of subjects at an earlier stage of disease for testing novel hypotheses and/or therapies in the clinical setting.
Keywords: Alzheimer's disease, disease progression, GWAS, MCI
Introduction
Alzheimer's disease (AD) is a complex neurodegenerative disease with a strong genetic component. Mutations in the amyloid precursor protein, presenlin 1 and 2 genes, have been associated with the early-onset familial form of AD. However, greater than 95% of AD patients are diagnosed after the age of 65 (late-onset AD) and estimates of the heritability for late-onset AD range from 60 to 80%.1 The ɛ4 allele of the APOE gene was identified as a primary genetic risk factor for late-onset AD.2 Genome-wide association studies (GWAS) have focused on disease susceptibility analysis3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15 by comparing allele frequencies in subjects with disease vs non-disease controls and have identified robust genetic loci close to genes CLU, PICALM, CR1, BIN1, EPHA1, MS4A6A, MS4A4E and CD2AP at the genome-wide significance level7, 8, 12, 14, 15 and confirmed the previously reported putative locus at CD33.3, 12
Comparing with successes in studies of disease susceptibility, we have limited knowledge for the genetic loci that are associated with disease progression beyond the APOE ɛ4 locus.16 Candidate gene studies have suggested that a variant in the TOMM40 loci may predict AD age-of-onset in APOE ɛ3 carriers17 and variants in PPP3R118 associated with cerebrospinal fluid (CSF) tau phosphorylated at threonine 181 levels may also predict disease progression. To date, studies using age of onset for AD diagnosis9 or hippocampal atrophy measured on magnetic resonance imaging10 as end points for GWAS analysis failed to identify any genetic markers other than APOE or the close-by TOMM40 loci that reached genome-wide significance likely due to limitations in statistical power.9 Recently, a GWAS study using samples from the Alzheimer's Disease Neuroimaging Initiative (ADNI) consortium identified a novel variant close to the EPC2 loci that was associated with increased CSF total-tau levels. The analysis used a combined set of 374 subjects with AD, normal cognition or MCI, and it is unknown whether this novel variant is predictive of the conversion of MCI to AD.19
Before the current clinical diagnosis, AD subjects usually experience a transitional period of mild cognitive impairment. Amnestic MCI by the Petersen's criteria defines subjects with a primary memory component who have an elevated risk of developing dementia of the AD type.20 The annual rate of diagnosis of AD in amnestic MCI subjects is approximately 10–15%, which is substantially higher when compared with the rate of AD's diagnosis that is observed in healthy elderly individuals (1–2%). The recently proposed new diagnostic guidelines in AD formalizes the diagnosis of MCI and establish a framework to potentially incorporate additional biomarker data to the diagnosis of MCI due to AD.21 Besides the APOE ɛ4 allele,22 there are no genetic factors that have been identified to be robustly associated with cognitive decline and progression to AD in MCI subjects. Recent studies have demonstrated that changes in the Clinical Dementia Rating-sum of boxes (CDR-SB) can, with reasonable accuracy, discriminate between patients with dementia of AD and those with MCI.23 To identify genetic markers that may be associated with progression to AD in MCI subjects, we conducted a GWAS using decline in clinical status as measured in the CDR-SB as a quantitative trait in a cohort of 822 amnestic MCI subjects for which we had data measurements up to a 48-month period.
Materials and methods
Subjects
In all, 535 MCI subjects from the Vitamin E trial conducted by the Alzheimer's Disease Cooperative Study24 were genotyped in the study. All study protocols were approved by institutional review board and informed consent was obtained from participating subjects before sample collection. The ADNI study includes approximately 200 AD, 300 MCI and 200 normal controls.10, 25 Top findings from the discovery analysis of the MCI data (rs7840202, rs12752888 or rs11637611) were contrasted with analyses from an independent set of AD data. The GenADA set contains 801 AD patients and 776 controls.9 In total, 773 AD subjects from the GenADA collection were included in the association test for age of onset.
Genotyping
All genomic DNA samples for the Vitamin E trial were extracted from blood and quantified using Picogreen (Invitrogen, Carlsbad, CA, USA) before they were genotyped at Genizon Biosciences (Montreal, Quebec, Canada) using the Illumina 610Quad array. The final Vitamin E data set includes 499, 134 markers after excluding markers that failed the initial quality control (<99% call rate for all single-nucleotide polymorphisms (SNPs) and all samples, gender discrepancies and Mendelian errors for additional quality control (QC) samples). The ADNI genetic data set was downloaded from the ADNI website.
Genotype data QC
Most QC procedures were performed using the genetic analysis package PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/).26 Similar criteria were applied for all Illumina data sets: SNPs with a minor allele frequency ≥1% and SNP and sample call rate ≥99% were retained; SNPs out of Hardy–Weinberg equilibrium in the control population (−log 10(P-value)>5) were excluded; duplicated samples and samples with any possible identification errors were excluded; and the analysis only included Caucasian samples identified via multi-dimensional scaling after merging with the HapMap data. No imputation was carried out for the GWAS analysis.
We adapted the QC procedure from the original GenADA sample set to accommodate a different genotyping platform (Affymetrix 550K). GenADA genotype data (after QC) were imputed by Mach (http://www.sph.umich.edu/csg/abecasis/mach/) using reference haplotypes from HapMap III phased data (release 2). A two-step imputation was performed for GenADA to investigate the top two variants for association with age of onset. Variant rs7840202 had an imputation quality score of 0.90 and an r2 value of 0.84 in GenADA data set. Variant rs11637611 had a quality score and r2 value of 1 in GenADA data set.
Statistical analysis
CDR-SB data were available for up to 48 months for 822 genotyped MCI Caucasian patients from Vitamin E trial and ADNI (78% subjects have data at 24 months). The statistical model used for progression was a repeated-measures analysis of variance with covariates of study, baseline age, baseline Mini-Mental State Examination (MMSE), baseline CDR sum of boxes, gender and APOE ɛ4 carrier status (+/−). The response variable was the change from baseline of CDR-SB, and the primary factors of interest were months and genotype, with their interaction used to assess the effect of genotype on progression. Because progression of cognitive defects may be nonlinear, a more general approach was taken, with time points treated as nominal to allow for nonlinear or piecewise linear changes in CDR-SB over time. A significant genotype × time interaction implies that the changes in CDR-SB over time are different depending on genotype. This model is used frequently at Pfizer for assessing treatment effects in clinical trials; here, ‘treatment' is replaced with ‘genotype'. Data at a maximum of five time points (6, 12, 18, 24 and 30 months) were used.
The random subject effect was assumed to have compound symmetric variance–covariance matrix. This is similar to the model used by Cruchaga et al.,18 modified to allow for variations in slope over time by treating time (months) as categorical.
The genotype × time interaction was the factor of primary interest. A significant genotype × time interaction implies a different time course of progression depending on genotype, in contrast to a significant main genotype effect, which implies a different mean change in CDR-SB over the time course depending on genotype. The reported P-value is based on the type III sum of squares for the interaction term, which assesses the effect of the interaction between genotype and time after all the main effects (including time and genotype) have been accounted for. These P-values were adjusted for genome-wide significance based on the null distribution of the interaction test statistic. To assess the null distribution of the interaction test statistic for this data set, a series of >300 000 random univariate genotypes based on empirical genotype distributions (that is, sampled from the observed genotype frequencies) were applied to the subjects and over 300 000 instances of the interaction effect were computed. The first percentile of the distribution of interaction P-values was 0.000378. Applying the Bonferroni correction for 1 million SNPs to this percentile gives a cutoff for the interaction P-value of 3.78 × 10−10 to have a family-wise type I error rate of no more than 1%. This is a stricter cutoff than the usual GWAS criterion, and was necessary since the overall progression of the cohort could inflate the significance of the interaction effect if the correct null distribution is not applied.
To identify effects that could be clinically meaningful, a prospective filter on the results was applied. SNPs showing statistically significant genotype × time interactions based on the model and criterion described above were selected for further consideration if they passed all three rules for the filter. SNPs were filtered using the following rules: (1) a single genotype must have the maximum (minimum) progression (numerical change from baseline) at each of the time points of 12, 18, 24 and 30 months. The time point of 6 months was considered too early to require a consistent signal that soon. (2) The change from baseline at 24 months for that genotype must be statistically different from at least one of the other two genotypes at a level of 0.05. (3) The difference in least-square means between the two genotypes must be at least 1 point on the CDR-SB scale, to assure clinical relevance.
Those top SNPs that passed genome-wide significance level and also passed the filters were further assessed for consistency of results by employing another cognitive measure ADAS-13 (AD Assessment Scale 13 item) as the end point (the model is similar to the one described as above, except replacing CDR-SB with ADAS-13). We also analyzed the time to conversion from MCI to AD, based on diagnostic changes as recorded by the investigators in each study. The first date when the diagnosis changed from MCI to probable AD was noted. Subjects without conversion were considered to be censored at their last recorded visit date that still had a diagnosis available. The Cox proportional hazards model was used. For the age-of-onset analysis in the GenADA set, which does not contain longitudinal cognition data, we conducted analysis of covariance analysis including APOE ɛ4 status (+/−) and gender. The models compared cases homozygous for the minor allele to the other two genotypes for each of the three variants.
CSF biomarkers were log 10 transformed and assessed in a recessive analysis of covariance model that adjusted for age, sex and baseline clinical diagnosis (AD, MCI, control).
We combined rs7840202, rs12752888 and rs11637611 to define a FastP (fast progression) status. Patients with any of the minor homozygous genotypes from the top three markers were defined as FastP group and the other patients without any of the minor homozygous genotypes were non-FastP group. Disease progression and conversion analysis were described as above.
Results
Sample/data set characteristics
We conducted a GWAS study in 489 MCI Caucasian cases collected through the Vitamin E MCI trial (Vitamin E MCI)18 combined with 333 Caucasian MCI patients from the ADNI data set (ADNI MCI)10, 25 for which longitudinal cognitive measures were available for up to 48 months (Table 1). Both studies included amnestic MCI subjects according to the Peterson's criteria27 without any other neurological diseases or any psychiatric disorder as described in DSM-IV (Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition). Characteristics of these two sample sets were similar for baseline scores (MMSE, CDR-SB) as well as age distribution (Table 1). A majority of subjects in the combined data set had longitudinal CDR-SB scores evaluated up to 24 (78.3%) and 36 months (52.7%). The ADNI MCI cohort included more male subjects (64.3 vs 54.0%) and slightly fewer subjects with the APOE ɛ4 (54.5 vs 58.5%) allele when compared with subjects from the Vitamin E trial.
Table 1. Summary of baseline sample characteristicsa.
| Sample set (N) | Age (mean±s.d.) | Baseline CDR-SB (mean±s.d.a) | Baseline MMSE (mean±s.d.a) | Gender (% male) | APOE status (% ɛ4 carriers)b |
|---|---|---|---|---|---|
| ADNI (333 MCI) | 75.0±7.14 | 1.6±0.88 | 27.1±1.75 | 64.3 | 54.5 |
| Vitamin E Trial (489 MCI) | 73.0±7.11 | 1.8±0.78 | 27.4±1.75 | 54.0 | 58.5 |
| Combined (822 MCI) | 73.9±7.18 | 1.7±0.82 | 27.3±1.76 | 58.2 | 57.3 |
| GenADA (783 AD) | 72.2±8.49 | NA | NA | 42.5 | 62.8 |
Abbreviations: AD, Alzheimer's disease; ADNI, Alzheimer's Disease Neuroimaging Initiative; CDR-SB, Clinical Dementia Rating-sum of boxes; MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination; NA, not applicable; s.d., standard deviation.
Only Caucasian subjects are included in the analysis and reported here.
APOE ɛ4 carriers include subjects with one or two copies of ɛ4 allele at the APOE locus.
All genotyping data obtained from the Illumina 610 platform for the combined data sets were processed through a strict quality control process (see Materials and methods for details). A linear repeated measures model was used to examine the contributions of genetic variants on disease progression adjusted for study (Vitamin E or ADNI), baseline age, gender, baseline MMSE, baseline CDR-SB and APOE ɛ4 status. A similar model has been previously used to model disease progression.18 Our model treats time (months) as categorical rather than continuous, to allow for the possibility that the progression rate was not constant over time. We observed that study (P<0.01), baseline MMSE scores (P<10−9), baseline age (P<0.001) and APOE ɛ4 status (P<10−4) significantly contributed to the progression rate as measured by the change in clinical status (CDR-SB). These observations were consistent for all of the variants tested in our analysis.
Pre-specified candidate variants involved in disease susceptibility are not strongly associated with disease progression in subjects with amnestic MCI
We first tested the association of cognitive decline with the pre-specified candidate variants in CLU (rs11136000), PICALM (rs3851179), CR1 (rs3818361) and BIN1 (rs744373) that had been identified in previous disease susceptibility studies.7, 8, 12 None of these candidate variants were significantly associated with the rate of disease progression after multiple testing corrections in our data set (Table 2). The PICALM variant (rs3851179) showed a nominal association (P=0.027, corrected P=0.108) with the AA carriers having a slower rate of decline when compared to the AG/GG carriers. This result is consistent with the previously reported protective effect of the A allele for AD susceptibility.7
Table 2. Candidate variants identified from recent GWAS scans for AD susceptibilitya.
| SNP | Gene | Chr. | Position (b.p.) | Minor allele frequency | Genotype × time interaction nominal P-value | Bonferroni corrected P-valueb |
|---|---|---|---|---|---|---|
| Pre-specified | ||||||
| rs3818361 | CLU | 8 | 27 520 436 | 0.38 | 0.948 | 1 |
| rs3851179 | PICALM | 11 | 85 546 288 | 0.34 | 0.027 | 0.108 |
| rs3818361 | CR1 | 1 | 205 851 591 | 0.22 | 0.933 | 1 |
| rs744373 | BIN1 | 2 | 127 611 085 | 0.3 | 0.566 | 1 |
| Post hoc | ||||||
| rs11767557 | EPHA1 | 7 | 142 819 261 | 0.19 | 0.013 | 0.065 |
| rs3865444 | CD33 | 19 | 56 419 774 | 0.29 | 0.439 | 1 |
| rs3764650 | ABCA7 | 19 | 997 520 | 0.10 | 0.804 | 1 |
| rs610932 | MS4A6A | 11 | 59 695 883 | 0.44 | 0.840 | 1 |
| rs670139 | MS4A6A/MS4A4E | 11 | 59 728 371 | 0.41 | 0.592 | 1 |
| rs9349407 | CD2AP | 6 | 47 561 337 | NA | NA | NA |
Abbreviations: AD, Alzheimer's disease; CDR-SB, Clinical Dementia Rating-sum of boxes; MMSE, Mini-Mental State Examination; GWAS, genome-wide association studies; NA, not applicable; SNP, single-nucleotide polymorphism.
The analysis was performed with experimentally obtained genotype data using change of CDR-SB as end point and a repeated mixed model to adjust for study, baseline age, gender, baseline MMSE, baseline CDR-SB and APOE ɛ4 status (+/−).
Variants in CLU, PICALM, CR1 and BIN1 were pre-specified, whereas the other recently reported variants were investigated after the primary analysis.
We also conducted a post-hoc analysis of the recently identified variants in EPHA1, MS4A6A, MS4A4E and CD33 (Table 2). Genotype data for the top variant in CD2AP (rs9349407) were not available in our data set. The EPHA1 variant (rs11767557) showed a nominal association (P=0.013, corrected P=0.065) with the rate of decline. This association is primarily driven by the CC carriers having a slower progression toward the end of the study period, but is not consistent over the full course of observation. None of the other previously identified variants contribute to disease progression in our sample set (Table 2).
GWAS analysis identified multiple novel loci associated with the rate of progression in subjects of MCI
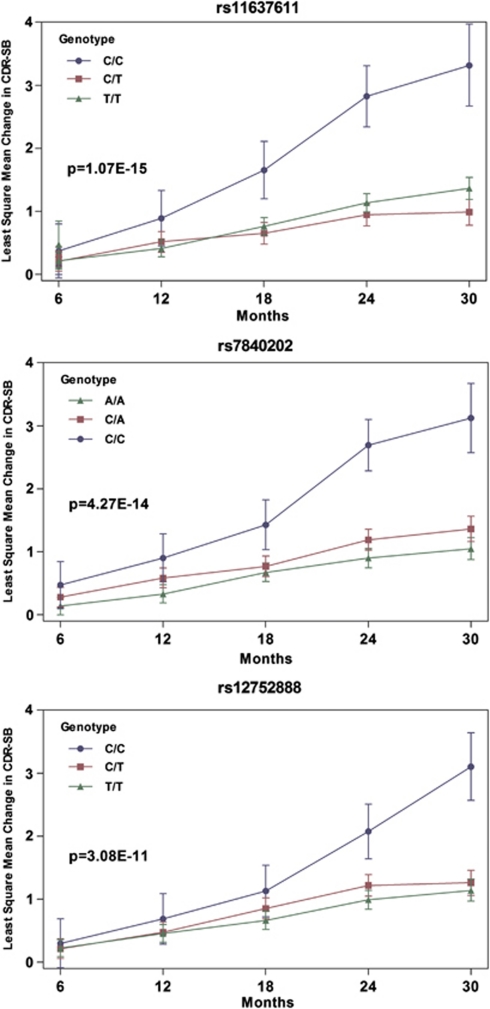
We next conducted an unbiased GWAS analysis in the sample set. SNPs with rare genotype counts (<5% for any individual genotype group) were removed to avoid technical artifacts (233 603 SNPs were included in the final analysis). Our GWAS analysis (QQ plot was presented in Supplementary Figure S2) identified 23 variants that reached genome-wide significance level at the false-positive rate of 0.01 (Bonferroni correction for 1 million SNPs based on simulated P-value null distribution, P-value <3.78 × 10−10). In total, 11 variants were selected, which demonstrated a consistent genotype effect on disease progression at each of the time points examined, with the possible exception of the 6-month time point that produced an inconsistent signal owing to the variability at this early time point (Supplementary Table S1). These variants were also selected based on potential clinical utility, defined as at least 1 point difference in estimated CDR-SB scores between the fastest and slowest progressing genotype groups at 24 months. None of the SNPs deviated from Hardy–Weinberg equilibrium (Supplementary Table S1). The most significant SNPs that reached genome-wide significance level included: rs7840202 on chr8, rs12752888 on chr1 and nine SNPs on chr15. The SNPs on chr15 appeared to have very similar allele frequencies and are in high linkage disequilibrium (pair-wise correlation coefficient r2⩾0.64; Supplementary Figure S1). We selected rs11637611 (with the smallest P-value) to represent the chr15 region.
All the top variants were found to have consistent effects in each of the independent sample sets that we probed (Table 3). Two SNPs reached genome-wide significance in the Vitamin E MCI trial alone (P-values for rs1275288 and rs11637611 passed a false-positive rate of 0.05 after adjusting for 1 million SNPs; P<8.31 × 10−9). The other SNP (rs7840202, P=5.71 × 10−7) did not reach genome-wide significance level in the Vitamin E MCI trial alone, but was highly significant when combined with the ADNI sample set (Table 3). For these variants, the minor homozygous genotype group had greater disease progression over the study period (Figure 1). Importantly, the effect of the risk genotypes is independent of and additive to other factors such as baseline age, gender and APOE ɛ4 status (+/−) that are known to influence progression.
Table 3. Top markers associated with the rate of progression from the GWAS analysis in MCI subjectsa.
| SNP |
UCSD |
ADNI |
Combined |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Geno | Least-square mean estimate (24 months) | Genotype × time interaction P-valueb | Geno | Least-square mean estimate (24 months) | Genotype × time interaction P-valueb | Geno | Least-square mean estimate (24 months) | Genotype × time interaction P-valueb | |
| rs7840202 | CC | 2.59 | 5.71E-07 | CC | 2.82 | 2.48E-05 | CC | 2.692 | 4.27E-14 |
| CA | 0.94 | CA | 1.55 | CA | 1.192 | ||||
| AA | 0.66 | AA | 1.25 | AA | 0.899 | ||||
| rs11637611c | CC | 2.92 | 9.38E-10 | CC | 2.7 | 4.80E-06 | CC | 2.826 | 1.07E-15 |
| CT | 0.78 | CT | 1.19 | CT | 0.947 | ||||
| TT | 0.79 | TT | 1.64 | TT | 1.138 | ||||
| rs12752888 | CC | 2.18 | 2.48E-10 | CC | 1.97 | 3.30E-01 | CC | 2.076 | 3.08E-11 |
| CT | 0.92 | CT | 1.64 | CT | 1.221 | ||||
| TT | 0.69 | TT | 1.39 | TT | 0.992 | ||||
Abbreviations: ADNI, Alzheimer's Disease Neuroimaging Initiative; CDR-SB, Clinical Dementia Rating-sum of boxes; Geno, genotype; GWAS, genome-wide association studies; MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination; SNP, single-nucleotide polymorphism.
GWAS analysis was performed with experimentally obtained genotype data using change of CDR-SB as end point and a repeated mixed model to adjust for study, baseline age, gender, baseline MMSE, baseline CDR-SB and APOE ɛ4 status (+/−).
Nominal P<3.78 × 10−10 reflects genome-wide correction (1 million SNPs) for family-wide error rate of no >0.01 based on 300 000 simulations from the null distribution.
Other SNPs in the region (listed in Supplementary Table S1) are also strongly associated with disease progression.
Figure 1.
rs11637611, rs7840202 and rs12752888 define sub-populations with different progression rates in 822 subjects of mild cognitive impairment (MCI). Least-square means were estimated for Clinical Dementia Rating (CDR)-sum of boxes scores over time by genotype groups. Ninety-five percent confidence intervals were shown for each time point. At least 78% of patients contributed data to 6, 12, 18 and 24 months. Close to half of the patients contributed data at 30 months.
rs7840202, rs12752888 or rs11637611 are also associated with the rate of decline in cognition and conversion of amnestic MCI to a diagnosis of AD
We further tested the association of the variants that were identified with a faster rate of increase in disease severity with related, but different cognitive endpoints to confirm our finding. All three variants that were associated with a faster rate of disease progression as measured by a significant increase in the CDR-SB demonstrated significant associations with changes in the ADAS-13 from baseline after adjusting for baseline age, gender, APOE, baseline MMSE and baseline ADAS-13 scores (genotype × months interaction P-value—rs7840202: 2.09 × 10−13; rs11637611: 1.5 × 10−3; rs12752888: 7.8 × 10−3). Furthermore, the effect sizes from the genotype groups are consistent for the two measures (Table S2).
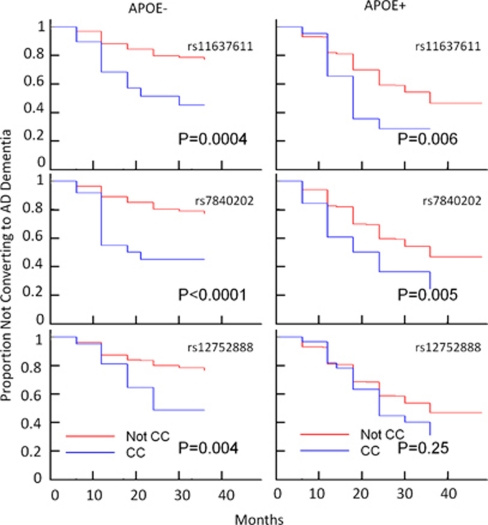
Next, we examined whether or not the top three variants (rs11637611, rs7840202 and rs12752888) that we had identified to be associated with disease progression were associated with the time to develop possible or probably AD according to the criteria defined by NINCDS-ADRDA (National Institute of Neurological and Communicative Diseases and Stroke and the Alzheimer's Disease and Related Disorder Association). The conversion of subjects from MCI to AD was based on diagnostic changes as recorded by the investigators in each study (Figure 2). For all of the variants that we identified, the minor homozygous genotype groups had an increased probability of converting to AD (rs7840202, hazard ratio=2.26, 95% confidence interval [1.56, 3.26] rs12752888, hazard ratio=1.78, 95% confidence interval [1.21, 2.64] rs11637611, hazard ratio=2.15, 95% confidence interval [1.40, 3.31]) when compared with the other genotype groups. The increased probability of converting to AD for all of the minor allele variants that we discovered were of a similar magnitude as that which has been previously reported for MCI subjects who are APOE ɛ4 carriers (hazard ratio=2.25, 95% confidence interval [1.73, 2.9]). Furthermore, a stratified analysis according to APOE ɛ4 status indicated that the effect of rs7840202, rs11637611 or rs1275288 for conversion to AD is observed in both APOE ɛ4 carriers and non-carriers (Figure 2).
Figure 2.
Kaplan–Meier estimates of the rate of conversion from mild cognitive impairment (MCI) to Alzheimer's disease (AD) for rs7840202, rs11637611 or rs12752888. Panels showed the effect of genotypes (CC is the minor homozygous genotype group vs non-CC groups) on the rate of conversion from MCI to AD diagnosis in APOE ɛ4 carriers (+) and non-carriers (−). The Cox proportional hazards model was used.
rs7840202 and rs11637611 are associated with an earlier age-of-onset in an independent AD set
MCI subjects with a faster rate of disease progression may, in theory, have an earlier age of AD onset. We therefore investigated whether the novel loci that we had identified to be associated with a more rapid rate of disease progression (rs11637611 and rs7840202) were associated with an earlier age of disease onset in an independent data set. This analysis comprised 772 AD subjects from the GenADA sample set who met the ADRDA/NINCDS criteria for diagnosis of probable AD and whose age-of-onset information had been collected.9 Genotype data from the rs12752888 SNP were not available in this data set. Both the rs7840202 and rs11637611 SNPs were modestly associated with an earlier age of disease onset in AD subjects that was independent of APOE ɛ4 carrier status and gender (rs7840202: P=0.04; rs11637611: P=0.05). On average, carriers for the CC genotype were diagnosed with AD at a younger age (rs7840202: age difference estimate=−1.93 years; rs11637611: age difference estimate=−2.5) when compared with the age of disease diagnosis in individuals representing the other genotypes. This earlier age of disease onset (approximately 2 years) in AD subjects whose genotype is the minor homozygous allele groups in rs7840202 and rs11637611, represents a larger effect when compared with the effect by APOE ɛ4 carrier status, which is approximately −1.04 years in the same data set. Note that longitudinal clinical decline is a more accurate assessment compared with age of onset, which may not be consistently measured in different sample sets and may show varying results even for the effect of the APOE ɛ4 variant. Nevertheless, the results support the hypothesis that these variants potentially define a group of MCI subjects with a faster cognitive decline that might also be associated with an earlier age of AD onset.
Combining rs7840202, rs12752888 or rs11637611 information (FastP) defines a larger population of FastP that may be additive to CSF--biomarkers for patient selection
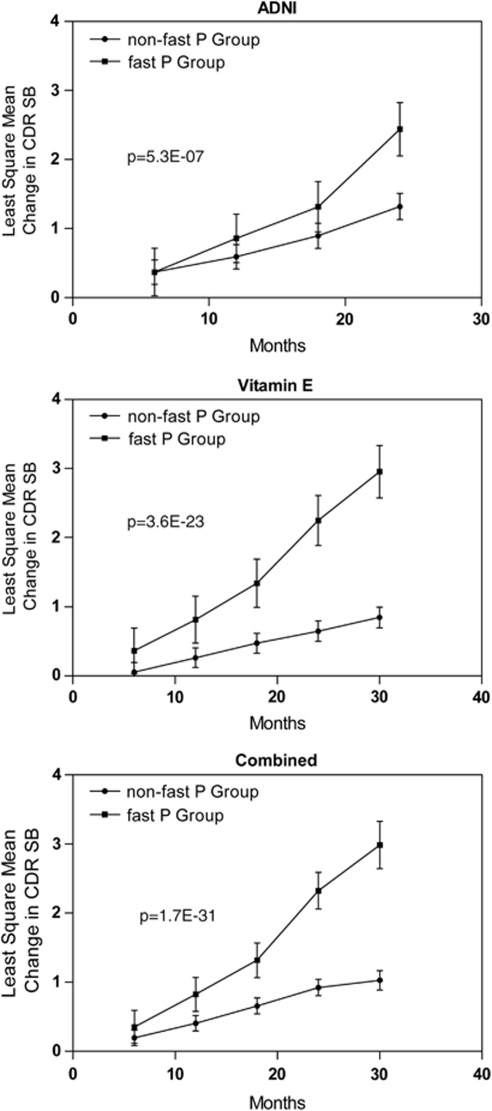
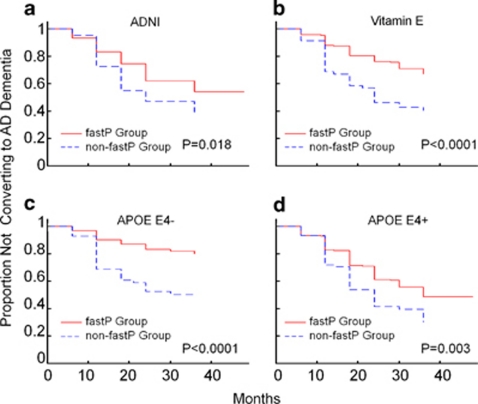
Although each of the top variants is strongly associated with disease progression in MCI subjects alone, each variant only defines a relatively small group (5–7%), therefore limiting the clinical utility for these markers individually. However, there is minimal correlation among the genetic variants and it is possible to combine them to define a larger MCI population (approximately 16.6%). MCI subjects can be defined by a FastP status by the presence of the minor homozygous genotype for at least one of the three variants that we identified (FastP group). When compared with MCI subjects where the minor homozygous genotype is completely absent (non-FastP group), there is a significant difference in the disease progression profile (genotype × time interaction: P=1.66 × 10−31; Figure 3). The CDR-SB change from baseline least-square mean estimate at 24 months is 2.33 for FastP group and only 0.92 for non-FastP group. The effect of FastP status on disease progression was similar for each study cohort (2.44 for FastP group and 1.32 for non-FastP group in the ADNI MCI subjects; 2.25 for FastP group and 0.65 for non-FastP group in the Vitamin E MCI cohort). Next, we probed whether or not FastP status would influence the conversion to AD dementia in MCI subjects. Although a higher percentage of MCI subjects are projected to convert to AD in the Vitamin MCI trial vs ADNI MCI, FastP group consistently had a significant increased rate of conversion to AD independent of APOE ɛ4 carrier status when compared with non-FastP group in each of the two independent sample sets that we analyzed (Figure 4).
Figure 3.
Fast progression (FastP) group with any of the risk genotypes in rs7840202, rs11637611 or rs12752888 define different rate of progression compared with non-FastP group without any of the risk genotypes. Panels a–c showed the least-square mean estimates for Clinical Dementia Rating-sum of boxes (CDR-SB): (a) for Alzheimer's Disease Neuroimaging Initiative (ADNI) mild cognitive impairment (MCI) subjects; (b) for Vitamin E MCI subjects; and (c) for the combined sample set.
Figure 4.
Kaplan–Meier estimates of the rate of conversion from mild cognitive impairment (MCI) to Alzheimer's disease (AD) dementia for the fast progression (FastP) group and non-FastP group. Panels a–d showed the Kaplan–Meier estimates for the conversion of MCI to AD. Panels showed results (a) for Alzheimer's Disease Neuroimaging Initiative (ADNI) MCI subjects; (b) for Vitamin early MCI (E MCI) subjects; (c) and (d) for the stratified analysis according to APOE ɛ4 status (+/−) in the combined MCI sample set.
Biomarker data such as baseline CSF Aβ and tau levels have been previously shown to be associated with the diagnosis of AD in MCI subjects.28 We also investigated whether the FastP signature can be additive to the other CSF biomarkers. The analysis was limited to the subjects in the ADNI cohort with both CSF and genotype data (n=173) as CSF biomarker data were not collected for the Vitamin E MCI subjects. Subjects with CSF amyloid-β 1–42 peptide (Aβ1−42) levels ⩽192 pg ml−1 in the ADNI set had been shown to decline more rapidly than subjects above the cutoff.29 In that subset of subjects (n=133), the FastP group had a significantly faster rate of decline when compared with the non-FastP group (change in CDR-SB, P=0.028), with an increasing effect at the 24-month time point.
Discussion
To date, the majority of GWAS studies have focused on genes associated with AD disease status. In this study, we undertook a GWAS that utilized disease progression as defined by a change in the CDR-SB score over time as a quantitative trait in Caucasian subjects with MCI. By using this approach, we identified several novel variants that appear to be associated with disease progression in MCI subjects and that are independent of other genetic loci that affect disease susceptibility.7, 8, 12 None of the candidate variants identified from the recent GWAS susceptibility study seem to have large effect in disease progression in our sample set. We identified several novel variants in which the minor homozygous genotype groups are associated with a significantly faster rate of disease progression as measured by increase in the CDR-SB and ADAS-13. In addition, inheritance of the minor homozygous genotype for each of these novel variants (rs7840202, rs12752888 or rs11637611) was also associated with a significantly increased likelihood of conversion from MCI status to AD. We validated our findings of an association for the minor homozygous genotype carriers for rs7840202 and rs11637611 with an earlier age of disease onset in an independent cross-sectional set of AD subjects, consistent with the expectation that the minor homozygous genotype group is associated with a faster rate of disease progression. Note that changes in cognitive decline is a more sensitive measure compared with age of onset, which may not be accurately assessed and could potentially introduce inconsistencies across data sets. To our knowledge, these novel variants are the first loci that are robustly associated with the progression of disease that reached genome-wide significance level in the combined set.
Although each individual variant alone only captures a relatively small fraction of the MCI population that has a faster rate of disease progression, in aggregate they define approximately 16.6% of the MCI population (FastP group). Moreover, the estimated difference in the changes in the CDR-SB at 24 months between the FastP group and non-FastP group is 1.40 and highly significant (P=1.87 × 10−21). This large effect is independent of APOE ɛ4 carrier status and can therefore be potentially utilized to model disease progression in clinical trials. We observed a significant study effect for the variants, but the estimates for mean change of CDR-SB at 24 months are similar for these variants and consistent for the two independent studies (Table 3).
CSF biomarker data were available for only a small subset of the ADNI set (n=174 with baseline data). Combined with the low frequency of the genotypes for each variant (5–7%), this limits the statistical power to detect association of the variants with either CSF Aβ1−42 or total-tau levels. We observed a borderline association with phosphorylated-tau (tau phosphorylated at threonine181 (p-tau181p)) for rs7840202 (Supplementary Table S3). Subjects with CSF Aβ1−42 levels ⩽192 pg ml−1 in the ADNI set declined more rapidly than subjects above the cutoff.29 After combining subjects with any of the fast progress genotypes to a FastP status, we observed a moderately significant contribution of the FastP signature in a subset of MCI patients with lower baseline CSF-Aβ1−42 levels (<192 ng ml−1), suggesting that the signature may be additive to the known CSF biomarkers such as CSF Aβ1−42.
In addition to the potential of utilizing FastP group for testing clinical candidates, the novel variants that we have identified that are associated with a more rapid rate of disease progression over time may provide insights into the mechanistic basis for disease progression. We defined a linkage disequilibrium region for each locus by identifying all variants with pair-wise r2⩾0.5 in the HapMap Caucasian subjects and extended the furthest SNPs into the haplotype block boundaries (see Supplementary Table S4 for all region boundaries). The region for rs1275288 contains a single gene: ACOT11 (a member of the acyl-CoA thioesterase family that catalyzes the conversion of activated fatty acids); the region for rs7840202 includes two genes: UBR5 (ubiquitin protein ligase E3 component n-recognin 5) and RRM2B (ribonucleotide reductase M2 B); and the variants on chr15q23 (top variant rs11637611) define a large genomic region containing multiple genes (MYO9A, SENP8, GRAMD2, PKM2, CELF6, PARP6, HEXA and ARIH1), including the HEXA gene, which had been identified in a lysosomal storage disease, Tay–Sachs disease. We employed various in silico techniques to prioritize the gene candidates based on the potential regulatory effect of the variants on gene expression (eQTL) and also non-random connections among these gene candidates through word-usage in PubMed abstracts (GRAIL) (Supplementary Figure S3). The eQTL browser (http://www.eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/) did not reveal any strong cis-effects for the variants, but this may be largely limited by the availability of the variants/transcripts on the tested platforms and we cannot also exclude the possibility that these variants may be specific to any activity-induced expression patterns. We expanded the GRAIL analysis to include additional loci associated with rate of decline below genome-wide significance level in the GWAS analysis (rs1412451, rs582420, rs7678888 and rs1958399), which did not pass our selection filter owing to lower potential for clinical utility (the estimated differences between the fastest and slowest genotype groups for CDR-SB scores were less than 1 point at 24 months) (Supplementary Table S4). The GRAIL analysis (Supplementary Figure S3) suggested connections between PTPRM (rs582420) and EPHA5 (rs7678888), which are associated with tyrosine phosphorylation and axon guidance. The EPHA5 variant associated with disease progression is independent of the EPHA1 variant for the disease susceptibility (correlation coefficient: r2=0.017) and together support the roles of ephrin receptors for AD etiology.30 It also highlighted connections among UBR5, ARIH1 and SENP8 (Supplementary Figure S3), which are associated with the ubiquitination process. Ubiquitination was reported to play a role in synaptic plasticity as well as other neurological diseases such as UBE3A in Angelman's syndrome31 and PARK2 gene (parkin) in familial Parkinson's disease.31 A previous report32 indicated that UBR5 expression is elevated in pyramidal cells from multiple regions of post-mortem AD brain including hippocampus as compared with the same regions from aged normal brain tissue. We also observed a similar increase in UBR5 mRNA levels in samples analyzed from AD vs control (Supplementary Figure S3), supporting the need for further elucidation of the role of UBR5 and ubiquitin ligases in AD biology.
Further investigation and replication studies will be required to fully validate and elucidate the roles the novel variants that we have identified here. However, our results demonstrate that it is possible to identify genetic factors associated with the rate of decline in a cohort of MCI subjects where sufficient longitudinal data exists. Elucidation of the biological pathways that are involved with disease progression in various stages of the disease may reveal novel targets that are amenable to drug discovery efforts and in the short term may provide useful tools to identify an enriched population of fast progressors.
Acknowledgments
We acknowledge all the patients who contributed materials and Pfizer BIOBANK to allow research investigation in the study. Pfizer provided funding for generating GWAS data in the Vitamin E trial samples, which were facilitated through collaboration with Leon Thal at UCSD. We appreciate James Duerr, Philip Iredale, Michael Ehlers, Aidan Power and Bill Billing for useful discussions and assistance. Larry Altstiel reviewed the paper and provided useful comments. Data collection and sharing for ADNI was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). The genotypic and associated phenotypic data used in the GenADA study were provided by the GlaxoSmithKline, R&D Limited and the data sets were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000219.v1.p1. ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace Inc., Merck and Co. Inc., Novartis AG, Pfizer Inc., F Hoffman-La Roche, Schering-Plough, Synarc Inc., as well as non-profit partners, the Alzheimer's Association and Alzheimer's Drug Discovery Foundation, with participation from the US Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego, CA, USA. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles, CA, USA. This research was also supported by NIH Grants P30 AG010129, K01 AG030514, and the Dana Foundation.
Pfizer provided funding for the study and most of the authors (except Dr Paul Aisen) are/were employees of Pfizer.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, et al. Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer's disease. Nat Genet. 2009;41:192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J Clin Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, Morgan A, et al. Evidence for novel susceptibility genes for late-onset Alzheimer's disease from a genome-wide association study of putative functional variants. Hum Mol Genet. 2007;16:865–873. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer's disease. PLoS One. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, et al. GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers. Neuron. 2007;54:713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Pickering E, Liu YC, Hall S, Fournier H, Katz E, et al. Meta-analysis for genome-wide association study identifies multiple variants at the BIN1 locus associated with late-onset Alzheimer's disease. PLoS One. 2011;6:e16616. doi: 10.1371/journal.pone.0016616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C, Mayeux R. Use of genetic variation as biomarkers for mild cognitive impairment and progression of mild cognitive impairment to dementia. J Alzheimers Dis. 2010;19:229–251. doi: 10.3233/JAD-2010-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, Sundseth SS, et al. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease. Pharmacogenomics J. 2009;10:375–384. doi: 10.1038/tpj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchaga C, Kauwe JS, Mayo K, Spiegel N, Bertelsen S, Nowotny P, et al. SNPs associated with cerebrospinal fluid phospho-tau levels influence rate of decline in Alzheimer's disease. PLoS Genet. 2010;6:e1001101. doi: 10.1371/journal.pgen.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Swaminathan S, Shen L, Risacher SL, Nho K, Foroud T, et al. Genome-wide association study of CSF biomarkers A{beta}1-42, t-tau, and p-tau181p in the ADNI cohort. Neurology. 2010;76:69–79. doi: 10.1212/WNL.0b013e318204a397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC. Normal aging, mild cognitive mpairment, and early Alzheimer's disease. Neurologist. 1995;1:326–344. [Google Scholar]
- Albert MS, Dekosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging and Alzheimer's Association workgroup. Alzheimers Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Smith GE, Ivnik RJ, Tangalos EG, Schaid DJ, Thibodeau SN, et al. Apolipoprotein E status as a predictor of the development of Alzheimer's disease in memory-impaired individuals. JAMA. 1995;273:1274–1278. [PubMed] [Google Scholar]
- O'Bryant SE, Waring SC, Cullum CM, Hall J, Lacritz L, Massman PJ, et al. Staging dementia using Clinical Dementia Rating Scale Sum of Boxes scores: a Texas Alzheimer's research consortium study. Arch Neurol. 2008;65:1091–1095. doi: 10.1001/archneur.65.8.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Thomas RG, Grundman M, Bennett D, Doody R, Ferris S, et al. Vitamin E and donepezil for the treatment of mild cognitive impairment. N Engl J Med. 2005;352:2379–2388. doi: 10.1056/NEJMoa050151. [DOI] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, et al. Alzheimer's Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimers Dement. 2010;6:265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256:183–194. doi: 10.1111/j.1365-2796.2004.01388.x. [DOI] [PubMed] [Google Scholar]
- Blennow K, Hampel H. CSF markers for incipient Alzheimer's disease. Lancet Neurol. 2003;2:605–613. doi: 10.1016/s1474-4422(03)00530-1. [DOI] [PubMed] [Google Scholar]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, et al. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Mabb AM, Ehlers MD. Ubiquitination in postsynaptic function and plasticity. Annu Rev Cell Dev Biol. 2010;26:179–210. doi: 10.1146/annurev-cellbio-100109-104129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Walker DG, et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics. 2007;28:311–322. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.