Abstract
The relationship between major depressive disorder (MDD) and bipolar disorder (BD) remains controversial. Previous research has reported differences and similarities in risk factors for MDD and BD, such as predisposing personality traits. For example, high neuroticism is related to both disorders, whereas openness to experience is specific for BD. This study examined the genetic association between personality and MDD and BD by applying polygenic scores for neuroticism, extraversion, openness to experience, agreeableness and conscientiousness to both disorders. Polygenic scores reflect the weighted sum of multiple single-nucleotide polymorphism alleles associated with the trait for an individual and were based on a meta-analysis of genome-wide association studies for personality traits including 13 835 subjects. Polygenic scores were tested for MDD in the combined Genetic Association Information Network (GAIN-MDD) and MDD2000+ samples (N=8921) and for BD in the combined Systematic Treatment Enhancement Program for Bipolar Disorder and Wellcome Trust Case–Control Consortium samples (N=6329) using logistic regression analyses. At the phenotypic level, personality dimensions were associated with MDD and BD. Polygenic neuroticism scores were significantly positively associated with MDD, whereas polygenic extraversion scores were significantly positively associated with BD. The explained variance of MDD and BD, ∼0.1%, was highly comparable to the variance explained by the polygenic personality scores in the corresponding personality traits themselves (between 0.1 and 0.4%). This indicates that the proportions of variance explained in mood disorders are at the upper limit of what could have been expected. This study suggests shared genetic risk factors for neuroticism and MDD on the one hand and for extraversion and BD on the other.
Keywords: bipolar disorder, genetic correlation, genome-wide association, polygenic score analysis, personality-major depression
Introduction
The relationship between major depressive disorder (MDD) and bipolar disorder (BD) remains controversial. Both are characterized by depressive episodes, whereas BD is, in addition, characterized by manic episodes.1 Research into risk factors for MDD and BD reports similarities and differences.
This also applies to studies investigating the association between personality and MDD or BD. Most of these studies have focused on the personality traits neuroticism/negative emotionality (N), extraversion/positive emotionality (E)2, 3 and, to a lesser extent, agreeableness (A), conscientiousness (C) and openness to experience (O).2 ‘N' is commonly defined as a tendency toward emotional instability. ‘E' is characterized by a disposition toward positive emotions, gregariousness and the tendency to be active, seek out stimulation and enjoy the company of others. ‘O' involves active imagination, aesthetic attentiveness, variety preference and intellectual curiosity. ‘A' can be defined as the tendency to be cooperative and compassionate rather than suspicious and antagonistic toward others. Finally, the dimension ‘C' reflects traits of self-discipline, carefulness, thoroughness, organization, deliberation and achievement.2, 3
In a recent meta-analysis on the relationship between personality and MDD, MDD was significantly associated with higher N and with lower C with a Cohen's d of 1.33 for N and −0.90 for C.4 The association with C became weaker (Cohen's d −0.59) after controlling for N, but remained significant. Although a negative link between E and MDD has often been reported, the effect was modest and not significant in the meta-analysis (Cohen's d −0.62). The associations with O and A were not significant either.
Studies on the association between personality and BD are sparser, but have consistently shown higher levels on N and O and lower levels of C compared with normal controls.5, 6, 7, 8, 9 This suggests that subjects with MDD and BD are similar regarding N and C and differ regarding O. This is supported by studies directly comparing personality profiles for MDD and BD.5, 6, 8, 9 All studies showed the same trend with higher O in BD subjects than in MDD subjects. This was significant in only one of these studies,6 but the other samples included far fewer subjects and probably did not have the power to detect the effect (<100 BD subjects versus ∼1000 subjects).5, 8, 9 Most of these studies have been performed in MDD or BD subjects in an euthymic phase; thus, the results do not reflect a state effect of mood on personality.
Mood disorders and personality traits are partly influenced by genetic risk factors. Heritability estimates are ∼40% for MDD, 50% for personality traits and between 60 and 90% for BD.10, 11, 12, 13, 14, 15, 16 This raises the question whether associations between personality and mood disorders are explained by shared genetic risk factors. So far, this has only been investigated for MDD. Twin studies have provided considerable support for overlapping genetic risk factors influencing N and MDD (reviewed in Middeldorp et al.17). Fewer twin studies have investigated the association with other personality traits and MDD suggesting a smaller, but significant genetic correlation between C and O and MDD,18 but not between E and MDD.19, 20, 21, 22
Genome-wide association (GWA) data also provide an opportunity to investigate whether traits are influenced by overlapping genetic risk factors. On the basis of GWA results for one trait, for instance, neuroticism, performed in one sample (the discovery sample), a polygenic score is calculated for each individual in another sample (the target sample). These polygenic scores are obtained by taking a set of top single-nucleotide polymorphisms (SNPs), for example, all SNPs with P-values below 0.1 and multiplying the individual's genotypic score (0, 1 or 2) by the effect of the SNP. If the polygenic scores are significantly related to a second trait, for instance, MDD, in the target sample, this indicates that the two traits, namely neuroticism and MDD, are influenced by overlapping genetic risk factors. In this manner, a genetic relationship was observed for schizophrenia and BD23 and for MDD and anxiety.24 In the former study, polygenic scores based on a GWA study in schizophrenia explained between 1 and 2% in BD. In the latter study, polygenic scores based on a MDD GWA study explained ∼2% of the variance in anxiety disorders.
This study investigates the genetic association between the five personality traits N, E, O, A and C and MDD, as well as BD. On the basis of the results of a GWA meta-analysis of N, E, O, A and C in >13 000 subjects,25 individual polygenic scores were calculated and tested for their effect on case–control status in 2 combined target MDD samples and in 2 combined target BD samples totaling 8921 and 6329 subjects, respectively. We first asked whether the genetic association between N and MDD as found in the twin studies is confirmed using polygenic score analysis and to what extent the other personality traits are genetically associated with MDD. Second, we investigated the genetic relationship between personality and BD. Finally, we asked what the differences are between the genetically mediated personality profiles underlying BD and MDD.
Materials and methods
Subjects, measurement instruments and genotyping
Discovery samples for personality
The GWA meta-analyses were performed on personality data collected from nine samples: SardiNIA–Italy,26, 27 Erasmus Rucphen Family study (ERF)–The Netherlands,28 Study of Addiction: Genetics and Environment (SAGE)–United States of America,29 Helsinki Birth Cohort Study (HBCS)–Finland,30, 31, 32 Nicotin Addiction Genetics Study/Interactive Research Project Grants (NAG/IRPG) study–Australia,33, 34 Queensland Institute of Medical Research (QIMR) adolescent study–Australia,35, 36 Lothian Birth Cohort 36 (LBC36)–United Kingdom,37 Baltimore Longitudinal Study of Aging (BLSA)–United States of America38 and Estonian Genome Project of University of Tartu (EGPUT)–Estonia.39 For a detailed description of these samples, we refer to de Moor et al.25 The total number of subjects available for the meta-analyses was 13 835. Sample sizes ranged from 600 to 3972 individuals. Mean age ranged from 19 to 70 years. In 5 studies, the mean age was between 40 and 50 years, in 1 study the mean age was 19 years, and in 3 studies, the mean age was between 60 and 70 years. It must be noted that the meta-analysis as described in de Moor et al.25 also included the GAIN-MDD sample. This sample was excluded in the personality traits meta-analyses for this study as the GAIN-MDD set served as one of the target samples (see below for the description of the sample).
Personality scores were assessed with NEO Personality Inventory–Revised (NEO-PI-R), NEO-PI-3 or the NEO Five-Factor Inventory.2, 40 In each study, scores for the 5 factors N, E, O, A and C were based on the 60 items of the NEO Five-Factor Inventory (12 items per phenotype).2 Summed scores were computed for all five personality dimensions.
DNA was extracted from blood samples. Genotyping was performed on Illumina platforms (Illumina, San Diego, CA, USA) in all studies, except in SardiNIA in which an Affymetrix platform (Affymetrix, Santa Clara, CA, USA) was used. Genotype data were checked in each study independently, using slightly different inclusion criteria. Among the basic checks that were performed are checks for European ancestry, Mendelian errors, gender inconsistencies and high genome-wide homozygosity. Genotype data were further cleaned based on Hardy–Weinberg equilibrium, minor allele frequencies, SNP call rate (% of subjects with missing genotypes per SNP) and sample call rate (% of missing SNPs per subject). Imputation to ∼2.5 common SNPs included in HapMap was performed using the HapMap phase II CEU data as the reference sample (NCBI build 36/UCSC hg18, Bethesda, MD, USA). Imputation was carried out using IMPUTE for SAGE and EGPUT samples.41 For the other samples, genotype data were imputed using MACH software.42
Target samples for MDD and BD
Polygenic scores were tested in two MDD case–control samples: GAIN-MDD and MDD2000+ and in two BD case–control samples: Wellcome Trust Case–Control Consortium (WTCCC) and Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD). The four samples have been described in detail elsewhere.43, 44, 45, 46, 47, 48 MDD and BD diagnoses were assessed with commonly used standardized interviews.
GAIN-MDD
This sample consisted of subjects from two large-scale longitudinal studies:43, 46 the NESDA (Netherlands Study of Depression and Anxiety)49 and the NTR (Netherlands Twin Register).50 The mean ages of cases (N=1738) and controls (N=1802) were 43 and 45 years, respectively. Inclusion criteria for MDD cases were a lifetime diagnosis of Diagnostic and Statistical Manual of Mental Disorders 4th Edition MDD,1 age of 18–65 years and self-reported Western European ancestry. Inclusion criteria for control subjects were no report of MDD at any measurement occasion and low genetic liability for MDD based on survey data measuring MDD-related traits. In addition, controls and their parents were required to have been born in the Netherlands or Western Europe. Only one control per family was selected.
Individual genotyping was conducted by Perlegen Sciences (Mountain View, CA, USA) using a set of four proprietary, high-density oligonucleotide arrays. Imputation was carried out using IMPUTE software41 with the HapMap phase II CEU data as the reference sample using NCBI build 36 (UCSC hg18).
MDD2000+
The second MDD target sample consisted of 2101 cases and 3280 screened controls, a subset of the MDD2000+ sample after excluding samples that overlapped with the discovery and GAIN-MDD samples.48 Samples were provided by the Queensland Institute of Medical Research (QIMR, Brisbane, QLD, Australia), NESDA, NTR, the University of Edinburgh (UoE, Scotland, UK) and the MGS (Molecular Genetics of Schizophrenia) study (controls only, United States). Control subjects from NTR who also participated in the GAIN-MDD study (N=223) were excluded, as well as cases and controls from the NAG/IPRG study that had been included in the personality traits meta-analysis (N=500). Mean ages of cases and controls were 41 and 48 years, respectively. MGS controls completed an online questionnaire including the short-form CIDI, supplemented by questions about schizophrenia, psychosis and BD. Controls were required to never have met the criteria for these disorders or MDD.
Genotyping was conducted on different Illumina and Affymetrix platforms. Imputation was conducted in four analysis sets (I317, I370, I610 and A6.0) to a common set of SNPs present in HapMap3 CEU/TSI, using Beagle 3.04.51, 52
Wellcome Trust Case–Control Consortium
The sample comprises 1868 BD cases.47 They were over the age of 16 years and of European descent. Individuals who had been in contact with mental health services were recruited if they suffered from a major mood disorder in which clinically significant episodes of elevated mood had occurred, such as bipolar I disorder (71% cases), schizoaffective disorder bipolar type (15% cases), bipolar II disorder (9% cases) and manic disorder (5% cases).
Half of the 3000 controls came from the 1958 British Birth cohort (58C) and were between 44 and 45 years of age at the time of DNA collection. The other half was selected from blood donors. Age ranged from 18 to 69 years. Analyses were carried out on observed genotypes. Ancestry principal components were available for 3919 subjects (1452 cases and 2467 controls), and these subjects were included in the current analyses.
Systematic Treatment Enhancement Program for Bipolar Disorder
STEP-BD was a US national, longitudinal public health initiative designed to examine the effectiveness of treatments and their impact on the course of BD.44, 45 Over a 7-year period, 4361 participants were enrolled across 20 sites and followed for up to 2 years. From the parent STEP-BD study, 2089 individuals were enrolled in a genetic substudy. Mean age was 43 years. Only non-Hispanic Caucasian individuals with European ancestry based on self-reported race and ethnicity information were included. Controls were used from the NIMH (National Institute of Mental Health) Genetics Repository and were MGS controls. This study included 1507 cases with bipolar I and bipolar II disorder and 903 controls.
Genotyping was performed using the Affymetrix GeneChip Human Mapping 500K Array Set. Beagle, version 3.1.1 was used to impute missing genotypes, with HapMap2 (Centre d'Etude du Polymorphisme Humain from Utah population, release 23, forward strand) as the reference panel.
Statistical analyses
GWA meta-analyses of personality
Genome-wide association meta-analyses were carried out as described in de Moor et al.25 without inclusion of the GAIN-MDD data. Meta-analyses were based on GWA analyses for the five personality traits conducted in each study with linear regression (under an additive model) and included sex and age as covariates. Meta-analyses of the results were conducted by the weighted inverse variance method as implemented in METAL (http://www.sph.umich.edu/csg/a becasis/metal/index.html),53 which computes a pooled effect estimate (ln(beta)), its s.e. and its P-value by weighing the effect estimates of individual samples by the inverse of its variance and by taking into account the direction of effect.
Polygenic score analyses
The genetic associations of the five personality scales with MDD and BD were analyzed for each personality scale separately in the combined samples for MDD and in the combined samples for BD. Polygenic scores were calculated and tested for their effect according to Purcell et al.23 Five sets of SNPs were selected from the GWA results for each of the five NEO personality scales meta-analyses. This selection of SNPs was based on their nominal P-value (Pdiscovery) for association with the NEO-personality scale in the discovery sample. For each of the five personality scales, five sets of SNPs were selected, one with Pdiscovery threshold 0.1, one with threshold 0.2, one with threshold 0.3, one with threshold 0.4 and one with threshold 0.5. These sets of SNPs were used to calculate the polygenic scores for subjects in the four target samples by multiplying the dosage score of risk allele count per SNP from 0 to 2 by the beta, summed over all SNPs in the considered set of SNPs and divided by the total number of SNPs.23 In short: ∑dosageScore*beta/N(SNPs). Dosage scores were used to account for the uncertainty in the imputed genotypes. We calculated individual scores for each set of SNPs using the PLINK software.54
The association of the polygenic scores with MDD was evaluated by logistic regression. Ancestry principal components were included as covariates. The explained variance was based on Nagelkerke's R2. To analyze polygenic scores in the combined samples (GAIN-MDD+MDD2000+ and WTCCC+STEP-BD), polygenic scores were corrected for ancestry principal components. Next, a logistic regression with sample as covariate was performed with standardized residuals of polygenic scores as independent variables.
Results
To illustrate the association between personality dimensions and MDD or BD, Table 1 shows the scores for five personality traits measured with the NEO Five-Factor Inventory2 in GAIN-MDD controls, GAIN-MDD cases and STEP-BD cases at entry to the study. Four of the five personality scales were significantly different in MDD patients compared with controls: N scores were higher, whereas A, E and C scores were lower in depressed subjects (all P-values <0.0001 in a t-test). As described in Barnett et al.,6 BD patients at a euthymic phase significantly differed from the population norms for all scales exhibiting higher levels of N and O and lower levels of E, C and A.
Table 1. Mean (s.d.) neuroticism (N), extraversion (E), agreeableness (A), openess to experience (O) and conscientiousness scores (C) in GAIN-MDD controls and GAIN-MDD cases and STEP-BD cases.
| Controls | MDD | BD | |
|---|---|---|---|
| N | 14.6 (5.9) | 27.3 (8.1)* | 30.9 (8.6)** |
| E | 30.5 (5.7) | 23.1 (7.0)* | 23.2 (7.5)** |
| A | 33.4 (4.7) | 31.4 (5.4)* | 29.9 (6.9)** |
| O | 25.0 (5.7) | 24.8 (5.6) | 29.5 (7.0)** |
| C | 33.5 (4.8) | 25.6 (6.1)* | 26.8 (8.0)** |
Abbreviations: BD, bipolar disorder; MDD, major depressive disorder; STEP-BD, Systematic Treatment Enhancement Program for Bipolar Disorder.
*P<0.00001 compared with GAIN-MDD controls.
**P<0.001 compared with population norms.6
The sets of SNPs with P-values below 0.1, 0.2, 0.3, 0.4 and 0.5 based on the results in the discovery sample included the number of SNPs as expected for these P-values, that is, ∼200 k, 400 k, 600 k, 800 k and 1 million SNPs, respectively. The overlap with SNPs used to calculate polygenic scores in the target samples was 100% for the GAIN-MDD sample, 40% for the MDD2000+ sample, 20% for the WTCCC sample and 90% for the STEP-BD sample. The lower overlap is due to the use of HapMap 3 as a reference set in the MDD2000+ sample and to the use of observed genotypes only in the WTCCC sample.
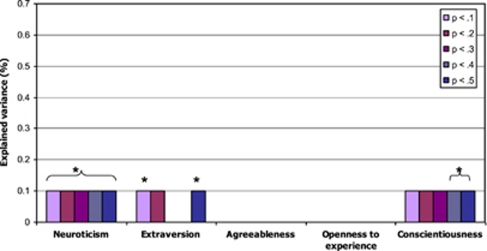
Figure 1 presents the results of the logistic regression analysis for MDD with the personality polygenic scores based on the GWA meta-analysis as independent variables. Significant positive associations were found between MDD and polygenic N scores for all sets of SNPs. In addition, significant negative associations were found between MDD and polygenic E scores based on the sets of SNPs with P-values <0.1 and <0.5 and polygenic C scores based on the sets of SNPs with P-values <0.3 to <0.5. These effects explained 0.1% of the variance in MDD (P-value <0.05).
Figure 1.
Explained variance by the five personality traits polygenic scores in MDD in GAIN-MDD + MDD2000+, *P<0.05. MDD, major depressive disorder.
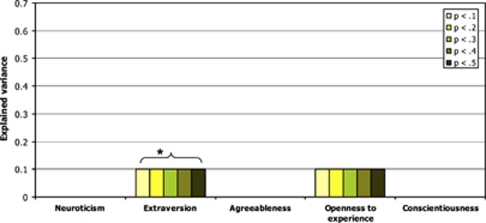
Figure 2 shows results of the logistic regression analysis for BD. Polygenic E scores were significantly positively related to BD, explaining 0.1% of the variance (P-value <0.05).
Figure 2.
Explained variance by the five personality traits polygenic scores in BD STEP-BD+WTCCC, *P<0.05. BD, bipolar disorder; STEP-BD, Systematic Treatment Enhancement Program for Bipolar Disorder; WTCCC, Wellcome Trust Case–Control Consortium.
The analyses were also carried out in the four samples separately to check for heterogeneity in the results across studies. None of the polygenic personality scores were significantly associated with MDD in the MDD2000+ study with the proportion of explained variance always below 0.1%. This is in contrast to the analyses of the GAIN-MDD sample in which polygenic N scores based were significantly higher in MDD cases than in controls, explaining between 0.2 and 0.4% of the variance (P-values <0.005) (see Supplementary Figure 1a). Moreover, polygenic C scores, based on the sets of SNPs with P-values <0.3 and <0.5, were lower in MDD cases explaining 0.2 and 0.3% of the variance (P-values <0.05). Thus, the significant results for MDD are mostly driven by the GAIN-MDD study.
The picture looks different for BD (see Supplementary Figures 1b and 1c). In the STEP-BD sample, polygenic E scores based on the sets of SNPs with P-values <0.4 and 0.5 and polygenic C scores based on the sets of SNPs with P-values <0.1 showed a significant positive and negative association, respectively, with BD. There were no significant effects detected in WTCCC, but the proportion of explained variance by polygenic E scores is similar as in the STEP-BD study. This suggests that, although separate studies are sometimes underpowered to detect the small effects of polygenic E scores, they both contribute to the significant effects as found in the analyses of the combined samples.
Given the relatively low proportion of variance explained by the polygenic personality scores for mood disorders, additional regression analyses were performed to detect the proportion of variance explained by the polygenic personality scores in the personality trait scores themselves. It was tested whether the polygenic N, E, O, A and C scores, as used in the analyses above, significantly predicted the respective N, E, O, A and C trait scores as measured in GAIN-MDD subjects. The polygenic scores explained between 0.1 and 0.4% of the corresponding personality trait scores (P-values between 0.0003 and 0.2) with the best results for polygenic O scores predicting O and the worst results for polygenic A scores predicting A.
Discussion
This study suggests that MDD is influenced by similar genetic risk factors as neuroticism (N) and that BD is influenced by similar genetic risk factors as extraversion (E). Both associations are positive: that is, genetic risk factors related to a higher N score increase the risk for MDD and genetic risk factors related to a higher E score increase the risk for BD. Moreover, genetic risk factors influencing conscientiousness (C) and E might be negatively associated with MDD, but results are less consistent.
The results for MDD agree with earlier genetic epidemiological research. Several studies have already suggested an overlap in genetic risk factors for neuroticism and MDD as reviewed by Middeldorp et al.17 A negative genetic correlation between MDD and C has also been reported.18
The genetic relationship between personality and BD has not been investigated before. With the exception of E, the absence of a genetic association between personality traits and BD is surprising given the phenotypic relationships between BD and the five personality traits found in the STEP-BD sample (Table 1) and other studies.5, 6, 7, 8, 9 Furthermore, the genetic association with E was positive, whereas the phenotypic association found in the total sample of STEP-BD subjects was negative. However, Barnett et al.6 showed in the same study sample that high N was related to a depression-prone BD course, whereas high E was related to a manic-prone BD course. Similar findings were reported by Quilty et al.55
The explained variance in MDD and BD by the polygenic scores was very modest. However, this was also true for prediction of the personality traits themselves with comparable proportions of explained variance (up to 0.4%) and P-values. Thus, the proportions of explained variance for mood disorders are at the upper limit of what could have been expected. Still, these numbers are lower than the explained variance of ∼2% reported for polygenic schizophrenia scores predicting BD and polygenic MDD scores predicting anxiety disorders.23, 24 This can be partly explained by a lack of power. Although the discovery sample was large with >13 000 subjects, only a few SNPs reached genome-wide significance and the deviation of the line of observed P-values from the line of expected P-values in the QQ plots is also modest.25 As the individual effect sizes of SNPs are small, the error of the estimates is relatively large. Therefore, the explained variance in a prediction analysis is low.56 This is comparable to the results of the analyses of GWA data for intelligence, which showed that the proportion of variance explained by all SNPs varied between 40 and 51%, whereas prediction analyses only explained 1% of the variance in intelligence.57
An additional explanation could be the large age differences in the studies included in the discovery set, resulting in top hits in the meta-analysis that reflect genetic variants associated with stability in personality traits from adolescence through older age. It is possible that genetic risk factors influencing stability over time are less related to MDD and BD than genetic risk factors for personality that are mostly important around early adulthood, the period of onset of MDD and BD. However, longitudinal twin studies suggest that genetic influences on personality are for the largest part stable, thus without much change in genetic risk factors across time (see, for example, Kandler et al.58 for a study in adults and Gillespie et al.59 and Hopwood et al.60 for studies in adolescents and young adults). A strength of our study, on the other hand, is that polygenic scores were determined from GWA study results in a sample of individuals without mood disorders and are not confounded by mood state in the cases.
The STEP-BD sample and the MDD2000+ sample both included MGS controls. That is no problem as the discovery sample and target samples were not overlapping.
Analyses of the effects of the polygenic personality scores on mood disorders in the four separate studies indicated that for BD, results were consistent over studies, whereas for MDD, results were mainly driven by the GAIN-MDD study. There does not seem to be an obvious explanation for the absence of effect in the MDD2000+ sample. The overlap in SNPs used to calculate polygenic scores was far lower in the MDD2000+ sample than in the GAIN-MDD sample because of the use of different reference sets for the imputation, HapMap3 in MDD2000+ and HapMap2 in MDD-GAIN. HapMap3 includes less SNPs but is based on more subjects. However, the overlap in SNPs was also lower in the WTCCC sample, whereas their results were similar to the results in the STEP-BD sample, which had a high overlap in SNPs. Therefore, the low overlap in SNPs does not seem to explain the difference in results between the MDD2000+ and the MDD-GAIN sample. Given the repeatedly found genetic correlation between N and MDD, it seems most likely that the finding in MDD2000+ is a false negative finding.
Despite the low explained variance, the results of this study indicate some interesting issues regarding the etiology of MDD and especially BD. Although studies investigating the phenotypic association between personality and MDD, as well as BD suggest that both disorders are related to high N and that, in addition, BD is related to high O, the genetic association shows a different picture of shared genetic risk factors for N and MDD on the one hand and for E and BD on the other. As previous studies have already pointed to differences in personality profiles between depression-prone and manic-prone BD patients,6, 55 these results imply that BD is a heterogeneous disorder with different expressions related to different, genetically influenced, personality profiles. This view is supported by the finding that an association with polymorphisms in the GABA receptor β1 subunit gene is most significant in cases fulfilling the Research Diagnostic Criteria for schizo-affective disorder. Cases fulfilling the Research Diagnostic Criteria for Bipolar Disorder type II showed a similar allele frequency as did controls.61 Future studies in larger samples, for example, the Psychiatric GWAS Consortium62 are suited to further investigate the complex association between personality and BD. For MDD, the genetic association with N is now well established as studies using different methods reveal similar results. This is beneficial for molecular genetic studies as statistical power can be increased by performing bivariate association analyses of neuroticism and MDD.
Acknowledgments
This study makes use of data generated by the Wellcome Trust Case–Control Consortium. A full list of the investigators who contributed to the generation of the WTCCC data is available from http://www.wtccc.org.uk. Funding for the WTCCC project was provided by the Wellcome Trust under award 076113. NESDA/NTR: Funding support was provided by the Netherlands Scientific Organization (904-61-090, 904-61-193, 480-04-004, 400-05-717, 912-100-20) Centre for Medical Systems Biology (NWO Genomics), the Neuroscience Campus Amsterdam (NCA) and the EMGO+ Institute; the European Union (EU/WLRT-2001-01254), ZonMW (Geestkracht program, 10-000-1002), NIMH (RO1 MH059160) and matching funds from participating institutes in NESDA and NTR. The NTR controls in MDD2000+ were genotyped in the Genomics platform (certified service provider (CSPro(R)) for Illumina) at the LIFE and BRAIN Center, Bonn (funded by NWO-SPI 56-464-1419). Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the NWO (480-05-003). MHM de Moor and CM Middeldorp are financially supported by the Netherlands Organization for Scientific Research (NWO) (ZonMW Addiction program, grant 31160008, VENI-016-115-035 and VENI grant 916-76-125). MDD2000+/QIMR: Funding was provided by the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496675,496739, 552485, 552498, 613608), the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254), and the US National Institutes of Health (NIH grants AA07535, AA10248, AA13320, AA13321, AA13326, AA14041, MH66206, DA12854, DA019951). A portion of the genotyping on which this study was based (Illumina 370K scans on 4300 individuals) was carried out at the Center for Inherited Disease Research, Baltimore (CIDR), through an access award to our late colleague Dr Richard Todd (Psychiatry, Washington University School of Medicine, St Louis). GW Montgomery is supported by the National Health and Medical Research Council (NHMRC) Fellowship Scheme. NRWray and DR Nyholt are supported by the Australian Research Council Future Fellowship Scheme. The ERF study was supported by grants from The Netherlands Organization for Scientific Research (NWO), Erasmus MC and the Netherlands Genomics Initiative (NGI)-sponsored Center of Medical Systems Biology (CMSB). Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative (GEI) (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392) and the Family Study of Cocaine Dependence (FSCD; R01 DA013423, R01 DA019963). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse and the NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease' (HHSN268200782096C). The Collaborative Study on the Genetics of Alcoholism (COGA) Principal Investigators: B Porjesz, V Hesselbrock, H Edenberg, L Bierut, includes 10 different centers: University of Connecticut (V Hesselbrock); Indiana University (HJ Edenberg, J Nurnberger Jr., T Foroud); University of Iowa (S Kuperman, J Kramer); SUNY Downstate (B Porjesz); Washington University in St Louis (L Bierut, A Goate, J Rice, K Bucholz); University of California at San Diego (M Schuckit); Rutgers University (J Tischfield); Southwest Foundation (L Almasy), Howard University (R Taylor) and Virginia Commonwealth University (D Dick). A Parsian and M Reilly are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P Michael Conneally, Raymond Crowe and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). The Collaborative Genetic Study of Nicotine Dependence (COGEND) project is a collaborative research group and part of the NIDA Genetics Consortium. Lead investigators directing data collection are Laura Bierut, Naomi Breslau, Dorothy Hatsukami and Eric Johnson. We thank Heidi Kromrei and Tracey Richmond for their assistance in data collection. In memory of Theodore Reich, founding Principal Investigator of COGEND, we are indebted to his leadership in the establishment and nurturing of COGEND and acknowledge with great admiration his seminal scientific contributions to the field. COGEND is supported by the NIH grant P01CA89392 from the National Cancer Institute. SardiNIA: We acknowledge support from the Intramural Research Program of the NIH, National Institute on Aging. Funding was provided by the National Institute on Aging, NIH contract NO1-AG-1-2109 to the SardiNIA (‘ProgeNIA') team. HBCS: We acknowledge financial support from the Academy of Finland (grant no. 120315 and 129287 to EW, 1129457 and 1216965 to KR, 120386 and 125876 to JGE), the European Science Foundation (EuroSTRESS), the Wellcome Trust (grant no. 89061/Z/09/Z and 089062/Z/09/Z) and the Signe and Ane Gyllenberg foundation. NAG/IRPG: This study is supported by NIH grants DA12854 (to PAFM), AA07728, AA07580, AA11998, AA13320 and AA13321 (to ACH); and grants from the Australian National Health and Medical Research Council; MLP is supported by DA019951. LBC36: We thank David Liewald and Paul Redmond for technical assistance; the study Secretary Paula Davies; Alan Gow, Michelle Taylor, Janie Corley, Caroline Brett and Caroline Cameron for data collection and data entry; nurses and staff at the Wellcome Trust Clinical Research Facility, where subjects were tested and at the genotyping was performed; staff at the Lothian Health Board and staff at the SCRE Centre, University of Glasgow. The research was supported by a program grant from Research Into Ageing. The research continues with program grants from Help the Aged/Age Concern (The Disconnected Mind). GWAS funding awarded by the Biotechnology and Biological Sciences Research Council (BBSRC) to IJD and AT. ML is a Royal Society of Edinburgh/Lloyds TSB Foundation for Scotland Personal Research Fellow. The study was conducted within the University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, supported by the (BBSRC), Engineering and Physical Sciences Research Council (EPSRC), Economic and Social Research Council (ESRC) and Medical Research Council (MRC), as part of the cross-council Lifelong Health and Wellbeing Initiative. This work has made use of the resources provided by the Edinburgh Compute and Data Facility (ECDF) (http://www.ecdf.ed.ac.uk/). The ECDF is partially supported by the eDIKT initiative (http://www.edikt.org.uk). BLSA: We acknowledge support from the Intramural Research Program of the NIH, National Institute on Aging. We thank Robert McCrae. EGPUT: AM and TE received support from FP7 grants (201413 ENGAGE, 212111 BBMRI, ECOGENE (no. 205419, EBC)). AM and TE also received targeted financing from Estonian Government SF0180142s08 and by EU through the European Regional Development Fund, in the frame of Centre of Excellence in Genomics. The genotyping of the Estonian Genome Project samples was performed in Estonian Biocentre Genotyping Core Facility, AM and TE thank Mari Nelis and Viljo Soo for their contributions. A Realo and J Allik are supported by a grant from the Estonian Ministry of Science and Education (SF0180029s08). STEP-BD: JH Barnett was funded by the Parke-Davis Exchange Fellowship. JW Smoller and LM McGrath were supported in part by NIMH grant R01 MH-079799 (Dr Smoller, PI).
Dr Laura Bierut is listed as an inventor on a patent, ‘Markers of Addiction,' covering the use of certain SNPs in diagnosing, prognosing and treating addiction. Dr Bierut served as a consultant to Pfizer in 2008. Paul Costa receives royalties from the NEO Inventories. The other authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders 4th Edition4th edn.APA: Washington DC; 1994 [Google Scholar]
- Costa PT, McCrae R. Manual for the Revised NEO Personality Inventory (NEO-PI-R) and NEO Five-Factor Inventory (NEO-FFI) Psychological Assessment Resources: Odessa; 1992. [Google Scholar]
- Eysenck HJ, Eysenck SBG. Manual of the Eysenck Personality Questionnaire. Educational and Industrial Testing Service: San Diego, CA; 1975. [Google Scholar]
- Kotov R, Gamez W, Schmidt F, Watson D. Linking ‘big' personality traits to anxiety, depressive, and substance use disorders: a meta-analysis. Psychol Bull. 2010;136:768–821. doi: 10.1037/a0020327. [DOI] [PubMed] [Google Scholar]
- Bagby RM, Bindseil KD, Schuller DR, Rector NA, Young LT, Cooke RG, et al. Relationship between the five-factor model of personality and unipolar, bipolar and schizophrenic patients. Psychiatry Res. 1997;70:83–94. doi: 10.1016/s0165-1781(97)03096-5. [DOI] [PubMed] [Google Scholar]
- Barnett JH, Huang J, Perlis RH, Young MM, Rosenbaum JF, Nierenberg AA, et al. Personality and bipolar disorder: dissecting state and trait associations between mood and personality. Psychol Med. 2010;41:1593–1604. doi: 10.1017/S0033291710002333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld RM, Klerman GL, Keller MB, Andreasen NC, Clayton PJ. Personality of recovered zpatients with bipolar affective disorder. J Affect Disord. 1986;11:81–89. doi: 10.1016/0165-0327(86)90064-9. [DOI] [PubMed] [Google Scholar]
- Nowakowska C, Strong CM, Santosa CM, Wang PW, Ketter TA. Temperamental commonalities and differences in euthymic mood disorder patients, creative controls, and healthy controls. J Affect Disord. 2005;85:207–215. doi: 10.1016/j.jad.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Tackett JL, Quilty LC, Sellbom M, Rector NA, Bagby RM. Additional evidence for a quantitative hierarchical model of mood and anxiety disorders for DSM-V: the context of personality structure. J Abnorm Psychol. 2008;117:812–825. doi: 10.1037/a0013795. [DOI] [PubMed] [Google Scholar]
- Kieseppa T, Partonen T, Haukka J, Kaprio J, Lonnqvist J. High concordance of bipolar I disorder in a nationwide sample of twins. Am J Psychiatry. 2004;161:1814–1821. doi: 10.1176/ajp.161.10.1814. [DOI] [PubMed] [Google Scholar]
- Middeldorp CM, Birley AJ, Cath DC, Gillespie NA, Willemsen G, Statham DJ, et al. Familial clustering of major depression and anxiety disorders in Australian and Dutch twins and siblings. Twin Res Hum Genet. 2005;8:609–615. doi: 10.1375/183242705774860123. [DOI] [PubMed] [Google Scholar]
- Jang KL, Livesley WJ, Vernon PA. Heritability of the big five personality dimensions and their facets: a twin study. J Pers. 1996;64:577–591. doi: 10.1111/j.1467-6494.1996.tb00522.x. [DOI] [PubMed] [Google Scholar]
- Distel MA, Trull TJ, Willemsen G, Vink JM, Derom CA, Lynskey M, et al. The five-factor model of personality and borderline personality disorder: a genetic analysis of comorbidity. Biol Psychiatry. 2009;66:1131–1138. doi: 10.1016/j.biopsych.2009.07.017. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry. 2003;60:497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- Middeldorp CM, Cath DC, Van Dyck R, Boomsma DI. The co-morbidity of anxiety and depression in the perspective of genetic epidemiology. A review of twin and family studies. Psychol Med. 2005;35:611–624. doi: 10.1017/s003329170400412x. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Myers J. The genetic and environmental relationship between major depression and the five-factor model of personality. Psychol Med. 2010;40:801–806. doi: 10.1017/S0033291709991140. [DOI] [PubMed] [Google Scholar]
- Fanous AH, Neale MC, Aggen SH, Kendler KS. A longitudinal study of personality and major depression in a population-based sample of male twins. Psychol Med. 2007;37:1163–1172. doi: 10.1017/S0033291707000244. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. A longitudinal twin study of personality and major depression in women. Arch Gen Psychiatry. 1993;50:853–862. doi: 10.1001/archpsyc.1993.01820230023002. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Gatz M, Gardner CO, Pedersen NL. Personality and major depression—A Swedish longitudinal, population-based twin study. Arch Gen Psychiatry. 2006;63:1113–1120. doi: 10.1001/archpsyc.63.10.1113. [DOI] [PubMed] [Google Scholar]
- Pakkala I, Read S, Kaprio J, Koskenvuo M, Kauppinen M, Rantanen T. Genetic contribution to the relationship between personality and depressive symptoms among older women. Psychol Med. 2010;40:1357–1366. doi: 10.1017/S0033291709991401. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirkan A, Penninx BW, Hek K, Wray NR, Amin N, Aulchenko YS, et al. Genetic risk profiles for depression and anxiety in adult and elderly cohorts. Mol Psychiatry. 2011;16:773–783. doi: 10.1038/mp.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor MH, Costa PT, Terracciano A, Krueger RF, de Geus EJ, Toshiko T, et al. Meta-analysis of genome-wide association studies for personality Mol Psychiatry 2010 10.1038/mp.2010.128(e-pub ahead of print). [DOI] [PMC free article] [PubMed]
- Pilia G, Chen WM, Scuteri A, Orru M, Albai G, Dei M, et al. Heritability of cardiovascular and personality traits in 6148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Sanna S, Uda M, Deiana B, Usala G, Busonero F, et al. Genome-wide association scan for five major dimensions of personality. Mol Psychiatry. 2010;15:647–656. doi: 10.1038/mp.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo LM, MacKay I, Oostra B, van Duijn CM, Aulchenko YS. The effect of genetic drift in a young genetically isolated population. Ann Hum Genet. 2005;69:288–295. doi: 10.1046/j.1529-8817.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Osmond C, Forsen TJ, Kajantie E, Eriksson JG. Trajectories of growth among children who have coronary events as adults. N Engl J Med. 2005;353:1802–1809. doi: 10.1056/NEJMoa044160. [DOI] [PubMed] [Google Scholar]
- Eriksson JG, Osmond C, Kajantie E, Forsen TJ, Barker DJ. Patterns of growth among children who later develop type 2 diabetes or its risk factors. Diabetologia. 2006;49:2853–2858. doi: 10.1007/s00125-006-0459-1. [DOI] [PubMed] [Google Scholar]
- Rikkonen K, Pesonen AK, Heinonen K, Lahti J, Kajantie E, Forsen T, et al. Infant growth and hostility in adult life. Psychosom Med. 2008;70:306–313. doi: 10.1097/PSY.0b013e3181651638. [DOI] [PubMed] [Google Scholar]
- Pergadia ML, Agrawal A, Loukola A, Montgomery GW, Broms U, Saccone SF, et al. Genetic linkage findings for DSM-IV nicotine withdrawal in two populations. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:950–959. doi: 10.1002/ajmg.b.30924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Pergadia ML, Loukola A, Broms U, Montgomery GW, Wang JC, et al. Genetic linkage to chromosome 22q12 for a heavy-smoking quantitative trait in two independent samples. Am J Hum Genet. 2007;80:856–866. doi: 10.1086/513703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel MA, Trull TJ, Derom CA, Thiery EW, Grimmer MA, Martin NG, et al. Heritability of borderline personality disorder features is similar across three countries. Psychol Med. 2008;38:1219–1229. doi: 10.1017/S0033291707002024. [DOI] [PubMed] [Google Scholar]
- Wright MJ, Martin NG. Brisbane adolescent twin study: outline of study methods and research projects. Aust J Psychol. 2004;56:65–78. [Google Scholar]
- Deary IJ, Gow AJ, Taylor MD, Corley J, Brett C, Wilson V, et al. The Lothian Birth Cohort 1936: a study to examine influences on cognitive ageing from age 11 to age 70 and beyond. BMC Geriatr. 2007;7:28. doi: 10.1186/1471-2318-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock N, Greulich RC, Andres R, Arenberg D, Costa PT, Lakatta EG, et al. Normal Human Aging: The Baltimore Longitudinal Study of Aging. US Government Printing Office: Washington, DC; 1984. [Google Scholar]
- Metspalu A. The Estonian Genome Project. Drug Develop Res. 2004;62:97–101. [Google Scholar]
- Mccrae RR, Costa PT, Jr, Martin TA. The NEO-PI-3: a more readable revised NEO Personality Inventory. J Pers Assess. 2005;84:261–270. doi: 10.1207/s15327752jpa8403_05. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Ann Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma DI, Sullivan PF, de Geus EJ, Heutink P, Meijer P, Sondervan D, et al. Genome-wide association of major depression: description of samples for the GAIN major depressive disorder study: NTR and NESDA Biobank Projects. Eur J Hum Genet. 2008;16:335–342. doi: 10.1038/sj.ejhg.5201979. [DOI] [PubMed] [Google Scholar]
- Huang J, Perlis RH, Lee PH, Rush AJ, Fava M, Sachs GS, et al. Cross-disorder genomewide analysis of schizophrenia, bipolar disorder, and depression. Am J Psychiatry. 2010;167:1254–1263. doi: 10.1176/appi.ajp.2010.09091335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs GS, Thase ME, Otto MW, Bauer M, Miklowitz D, Wisniewski SR, et al. Rationale, design, and methods of the systematic treatment enhancement program for bipolar disorder (STEP-BD) Biol Psychiatry. 2003;53:1028–1042. doi: 10.1016/s0006-3223(03)00165-3. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, de Geus EJ, Willemsen G, James MR, Smit JH, Zandbelt T, et al. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcome Trust Case–Control Consortium Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, Nyholt DR, et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned Mol Psychiatry 2010 10.1038/mp.2010.109(e-pub ahead of print). [DOI] [PMC free article] [PubMed]
- Penninx BW, Beekman AT, Smit JH, Zitman FG, Nolen WA, Spinhoven P, et al. The Netherlands Study of Depression and Anxiety (NESDA): rationale, objectives and methods. Int J Methods Psychiatr Res. 2008;17:121–140. doi: 10.1002/mpr.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma DI, de Geus EJ, Vink JM, Stubbe JH, Distel MA, Hottenga JJ, et al. Netherlands Twin Register: from twins to twin families. Twin Res Hum Genet. 2006;9:849–857. doi: 10.1375/183242706779462426. [DOI] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilty LC, Sellbom M, Tackett JL, Bagby RM. Personality trait predictors of bipolar disorder symptoms. Psychiatry Res. 2009;169:159–163. doi: 10.1016/j.psychres.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Yang J, Goddard ME. A commentary on ‘Common SNPs explain a large proportion of the heritability for human height' by Yang et al. (2010) Twin Res Hum Genet. 2010;13:517–524. doi: 10.1375/twin.13.6.517. [DOI] [PubMed] [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic Mol Psychiatry 2011 10.1038/mp.2011.85(e-pub ahead of print). [DOI] [PMC free article] [PubMed]
- Kandler C, Bleidorn W, Riemann R, Spinath FM, Thiel W, Angleitner A. Sources of cumulative continuity in personality: a longitudinal multiple-rater twin study. J Pers Soc Psychol. 2010;98:995–1008. doi: 10.1037/a0019558. [DOI] [PubMed] [Google Scholar]
- Gillespie NA, Evans DE, Wright MM, Martin NG. Genetic simplex modeling of Eysenck's dimensions of personality in a sample of young Australian twins. Twin Res. 2004;7:637–648. doi: 10.1375/1369052042663814. [DOI] [PubMed] [Google Scholar]
- Hopwood CJ, Donnellan MB, Blonigen DM, Krueger RF, McGue M, Iacono WG, et al. Genetic and environmental influences on personality trait stability and growth during the transition to adulthood: a three-wave longitudinal study. J Pers Soc Psychol. 2011;100:545–556. doi: 10.1037/a0022409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, Jones L, Jones IR, Kirov G, Green EK, Grozeva D, et al. Strong genetic evidence for a selective influence of GABAA receptors on a component of the bipolar disorder phenotype. Mol Psychiatry. 2010;15:146–153. doi: 10.1038/mp.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry. 2009;14:10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.