Abstract
The tendency to seek stimulating activities and intense sensations define excitement-seeking, a personality trait akin to some aspects of sensation-seeking. This trait is a central feature of extraversion and is a component of the multifaceted impulsivity construct. Those who score high on measures of excitement-seeking are more likely to smoke, use other drugs, gamble, drive recklessly, have unsafe/unprotected sex and engage in other risky behaviors of clinical and social relevance. To identify common genetic variants associated with the Excitement-Seeking scale of the Revised NEO Personality Inventory, we performed genome-wide association studies in six samples of European ancestry (N=7860), and combined the results in a meta-analysis. We identified a genome-wide significant association between the Excitement-Seeking scale and rs7600563 (P=2 × 10−8). This single-nucleotide polymorphism maps within the catenin cadherin-associated protein, alpha 2 (CTNNA2) gene, which encodes for a brain-expressed α-catenin critical for synaptic contact. The effect of rs7600563 was in the same direction in all six samples, but did not replicate in additional samples (N=5105). The results provide insight into the genetics of excitement-seeking and risk-taking, and are relevant to hyperactivity, substance use, antisocial and bipolar disorders.
Keywords: catenin cadherin-associated protein alpha 2 (CTNNA2), excitement-seeking, genome-wide association (GWA), impulsivity, sensation-seeking
Introduction
Genome-wide association studies (GWAS) have reliably identified common genetic variants associated with complex traits and diseases.1, 2, 3, 4 For personality traits, as for other psychological and psychiatric conditions, finding genome-wide significant associations has been difficult,5, 6, 7, 8, 9 presumably because of relatively small effects and sample sizes. By combining results from multiple cohorts, however, a meta-analysis based on the NEO five-factor inventory found genome-wide significant associations for Openness to Experience and Conscientiousness, two of the five major dimensions of personality.10 In addition to the five broad factors, recent studies have examined more narrowly defined personality facets, with promising results.11, 12 Using a meta-analytic approach, in this article we report a genome-wide significant association with excitement-seeking, a facet of personality with clinical and health implications.
Excitement-seeking is a personality trait that reflects an individual's tendency to crave stimulation and thrills.13, 14 This trait, akin to some aspects of sensation seeking, is a major component of the multifaceted impulsivity construct and is a core defining feature of extraversion.13, 14, 15, 16, 17 Excitement-seeking is also conceptually related to the approach-oriented motivation system and the behavioral activation system.15, 18, 19, 20, 21 Those who score high on measures of excitement-seeking tend to engage in exciting activities, crave intense sensations, pursue a high level of arousal and are generally attracted by bright colors and crowded environments. They are more likely to take unnecessary risks for thrills (for example, skydiving) and engage in risky behaviors and dangerous activities just for fun. The behavioral consequences of this trait are more common among adolescents and young adults, who compared with older adults are generally more likely to seek excitement.22, 23 In addition, male subjects tend to score higher on measures of excitement-seeking compared with female subjects, a pattern observed in countries around the world.23, 24 This trait is of interest because it accounts for individual differences in basic psychological processes, and because it is linked to many risky behaviors, including cigarette smoking, alcohol and other drug use, reckless driving, gambling and unsafe/unprotected sex.14, 17, 25, 26, 27, 28, 29, 30 These excitement-seeking-related behaviors do not necessarily lead to psychiatric conditions or antisocial behaviors.17, 29, 30 Still, the above links suggest that excitement-seeking, especially in combination with other impulsivity-related traits, may increase risk for externalizing disorders. Twin studies point to a shared genetic vulnerability of externalizing disorders, and a large portion of the common genetic liability is accounted for by impulsivity-related personality constructs, such as behavioral undercontrol and disinhibited personality.31, 32, 33 Furthermore, excitement-seeking behaviors are common in attention deficit/hyperactivity disorders and are heightened during manic states in patients with bipolar spectrum disorders.34, 35, 36, 37
According to behavioral genetic studies, about 50% of the variance in excitement-seeking is heritable.38, 39, 40 Yet, little is known about the actual genetic variants that contribute to this and other impulsivity-related traits.41 To identify common genetic variants associated with excitement-seeking, we meta-analyzed results of GWAS from six cohorts of European ancestry, all assessed with the Excitement-Seeking scale of the Revised NEO Personality Inventory (NEO-PI-R).13 We further tested whether any single-nucleotide polymorphisms (SNPs) significantly associated with the Excitement-Seeking scale in the meta-analysis replicated in additional samples.
Materials and methods
Samples description
Each study was reviewed and approved by local institutional review boards and informed consent was obtained in all studies. The samples contributing GWAS results for the meta-analysis are briefly described below in order of sample size, followed by the description of replication samples.
SardiNIA
The SardiNIA project is a prospective community-based cohort study that recruited over 62% of the population from four towns in the Ogliastra province of Sardinia, Italy.42 All subjects are of European ancestry and they derive from one of the oldest and largest founder populations.43 A total of 4375 subjects had valid scores on the NEO-PI-R and were genotyped using Affymetrix mapping array sets as described in previous studies.7, 11, 42 The sample includes 57% women and ranged in age from 14 to 94 years (M=42.5, s.d.=17). Additional information on the sample has been reported elsewhere.7, 11, 42
Estonian Genome Center of University of Tartu
The Estonian sample comes from the population-based biobank of the Estonian Genome Center of University of Tartu (EGCUT). A total of 51 515 individuals aged 18 years or older were part of this cohort (33% male, 67% female). Subjects were randomly recruited by general practitioners and physicians in hospitals. This study is based on 1031 randomly selected subjects with data available from both the Illumina HumanCNV370 genotype and the NEO-PI-3 questionnaire,44 which is a more readable version of the NEO-PI-R. The items for excitement-seeking are identical across the two measures, except for one. This item slightly improves internal consistency, but does not affect the mean level of the scale.45 The sample was 53.7% women and the age range was 18–87 years (M=43.0, s.d.=16.5). Additional information on the sample has been reported elsewhere.46
Baltimore Longitudinal Study of Aging
The Baltimore Longitudinal Study of Aging (BLSA) is an ongoing multidisciplinary study of community-dwelling volunteers mainly from the east coast of the United States.11, 22, 47 The GWAS analysis was restricted to subjects of European ancestry to reduce population stratification biases. A total of 840 subjects (46% women) were successfully genotyped (using the Illumina 550K array) and completed the personality questionnaire at least once. In this sample, age at first assessment ranged from 20 to 93 (M=58.5; s.d.=17). In the BLSA sample, multiple personality assessments were available for most participants. Although individual differences on personality traits are generally stable over time,22, 47 we combined all available assessments for each individual, for a total of 3879 assessment points. By averaging across multiple time points, we reduce variability due to temporary effects and random error, thereby obtaining more reliable and robust personality score estimate.
Queensland Institute of Medical Research
NEO-PI-R data from the Australian adolescent and young adult population sample, aged 17–27 years (M=20.7, s.d.=2.3), were collected as part of a mail and phone study examining health and well-being conducted between 2002 and 200348 and as part of a borderline personality disorder study conducted between 2003 and 2006.49 Both phenotypes and genotypes were available for 782 individuals (59.5% female) from 437 families and included 101 monozygotic twin pairs. Genotyping for this sample is described in ref.50 (see Project5: ADOL deCODE).
Germany
A total of 2420 healthy control participants were randomly selected from the general population of Munich, Germany, and contacted by mail. We included 476 individuals with GWA data (Illumina HumanHap300 chip) in this study. Several screenings were conducted to exclude subjects with current or lifetime history of neuropsychiatric disorders or with first-degree relatives with psychiatric disorders. Only participants with German descent (all four grandparents German) were included. A large battery of personality questionnaires was administered, including the NEO-PI-R. The sample (56% females) mean age was 46 years (s.d.=15).
Cilento
The Cilento study is a population-based study that includes 2137 individuals from three isolated populations in South Italy. Of these individuals, 859 were genotyped on the 370K SNP map from Illumina. Personality data were available for 343 genotyped subjects representing the final sample. Of this sample, 65.6% were women, and the mean age of all participants was 58.9 years (s.d.=19).
EGCUT: replication sample
A first replication sample consisted of 608 subjects from the EGCUT Estonia cohort described above. The subjects in the replication sample were not included in the GWAS. This sample was assessed with the NEO-PI-3 and genotyped using TaqMan method (Applied Biosystems by Life Technologies). The sample included 63% women and ranged in age from 18 to 87 years (M=41.6, s.d.=16.3).
Helsinki Birth Cohort Study: replication sample
The Helsinki Birth Cohort Study (HBCS) is composed of 8760 individuals born between 1934 and 1944 in one of the two main maternity hospitals in Helsinki, Finland. Between 2001 and 2003, a randomly selected sample of 928 men and 1075 women participated in a clinical follow-up study, with a focus on cardiovascular, metabolic and reproductive health, cognitive function and depressive symptoms. In 2004, the participants filled-in the NEO-PI.51 There are eight items in the Excitement-Seeking scale of the NEO-PI, with four items that are identical to the NEO-PI-R. There were 1454 subjects with both valid phenotype and genotype data (57.3% women). The mean age of the subjects was 63.4 years (s.d.=2.9).
DNA was extracted from the blood samples and genotyping was performed with the modified Illumina 610K chip by the Wellcome Trust Sanger Institute (Cambridge, UK). Detailed information on the selection of the HBCS participants and on the study design can be found elsewhere.52, 53, 54
Netherlands Twin Register: replication sample
A third replication sample consist of 3043 (62% female; age: M=39.6, s.d.=14) genotyped subjects from the Netherlands Twin Register study,55, 56 who completed Zuckerman's Sensation-Seeking scales18, 57 at multiple time points (in 1991, 1993, 1997, 2000, 2002; the available average scores were used in the analyses). The Sensation-Seeking questionnaire consists of four scales: Thrill and Adventure-Seeking, Experience-Seeking, Disinhibition and Boredom Susceptibility. Except for Boredom Susceptibility, the Sensation-Seeking scales are significantly correlated with the NEO-PI-R Excitement-Seeking scale (r=0.53 with Thrill- and Adventure-Seeking; r=0.36 with Experience-Seeking; r=0.49 with Disinhibition),58 but these scales share <30% of the variance. We examined whether any common genetic variant significantly associated with the NEO-PI-R Excitement-Seeking scale is also associated with the Zuckerman's Thrill- and Adventure-Seeking, Experience-Seeking and Disinhibition scales.
Excitement-seeking assessment
The excitement-seeking personality trait was assessed with the NEO-PI-R.13, 23 The scale consists of eight items answered on a five-point Likert scale, from strongly disagree to strongly agree. After reversing two negatively keyed items, the summed scores were standardized (M=50, s.d.=10) in all samples using American combined gender norms.13 The mean Excitement-Seeking scores were 47.7 (s.d.=10) for SardiNIA, 50.1 (s.d.=10) for EGCUT, 47.1 (s.d.=9) for BLSA, 59.3 (s.d.=8.7) for Queensland Institute of Medical Research (QIMR), 43.6 (s.d.=9.7) for RUJ-Germany, 43.1 (s.d.=8) for Cilento and 51.7 (s.d.=13) for the EGCUT replication sample. The QIMR adolescent sample had the highest average score on this scale, which is consistent with the universal maturational decline of excitement-seeking with age reported in longitudinal and cross-sectional studies.22, 23 Despite a decline in excitement-seeking with age, which we control in each study by using age as a covariate, the rank-order of individual differences on the NEO-PI-R excitement-seeking is highly stable. For example, in a BLSA sample of 676 adults followed for an average of 10 years, the uncorrected test–retest correlation for excitement-seeking was rtt=0.74. Correcting this value for attenuation due to measurement error would give a stability estimate of about 0.90.38, 47
Genotyping and imputation
DNA was extracted from blood in all samples. Genotyping was performed using Illumina or Affymetrix mapping array sets, as noted above for each sample. Extensive quality control analyses were performed in each cohort and at the meta-analysis stage. The standard quality screening conducted independently in each sample included SNP and sample call rates (⩾95%), minor allele frequencies (⩾1%), Hardy–Weinberg equilibrium (P<10−6), Mendelian errors, gender inconsistencies and checks for population stratification.
To allow for meta-analysis across different marker sets, all studies used their genotype data to impute the autosomal HapMap phase II CEU SNPs. The NCBI build 36 was used as reference. The imputation was performed using the MACH (http://www.sph.umich.edu/csg/abecasis/MACH/) or IMPUTE (https://www.mathgen.stats.ox.ac.uk/impute/impute.html) software.
Statistical analyses
Each study independently performed single marker association analyses with excitement-seeking using an additive genetic model implemented in MACH2DAT (http://www.genome.sph.umich.edu/wiki/Mach), SNPTEST (http://www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html), MERLIN (http://www.sph.umich.edu/csg/abecasis/Merlin/) or ABEL (http://www.mga.bionet.nsc.ru/~yurii/ABEL/). These imputation and association analyses software have been used successfully in numerous other GWAS. Age and sex were used as covariates in each study. Additional study-specific covariates were added as needed, such as principal components to account for population admixture. For the BLSA sample, we aggregated data across multiple assessments; therefore, the mean age was used as covariate. For the SardiNIA sample, an additional covariate was used to control for differences related to test administration (subjects who required a tester to read and report answers to the questionnaire).
We ran the meta-analysis using the program METAL (http://www.sph.umich.edu/csg/abecasis/Metal/). We used the inverse variance method, which is based on regression coefficients (β values) and standard errors (s.e.) from each individual study. Before the meta-analysis, we filtered out SNPs with imputation quality ≤0.30 (r2-hat from MACH) or ≤0.40 (proper_info from IMPUTE), and genomic control (λ) correction was applied to the results from each study. Before correction, the genomic inflation factors λ were 1.021 for SardiNIA, 1.011 for EGCUT, 1.058 for BLSA, 1.005 for QIMR, 1.010 RUJ-Germany and 1.022 for Cilento, providing little evidence of an increased false-positive rate due to stratification. As further quality control, we tracked differences in allele frequency among the cohorts for the most strongly associated SNPs. The meta-analysis was independently run by two analysts at two sites (United States and Estonia). The reported P-values were from two-tailed tests, unless otherwise specified.
Subjects in cohorts such as the SardiNIA and BLSA are characterized on hundreds of traits and disorders. Assessing multiple phenotypes in the same genotyped cohort is a cost-effective strategy,59 but it might raise the issue of correction for multiple independent tests. To balance the risk of false positives against the risk of false negatives,60, 61 a corrected P-value <5 × 10−8 (the standard 0.05 divided by 1 million independent SNP tested) was considered genome-wide significant. P-values <5 × 10−8 are generally recognized as significant in GWAS, irrespective of the exact number of SNPs and phenotypes tested in a particular study.
Results
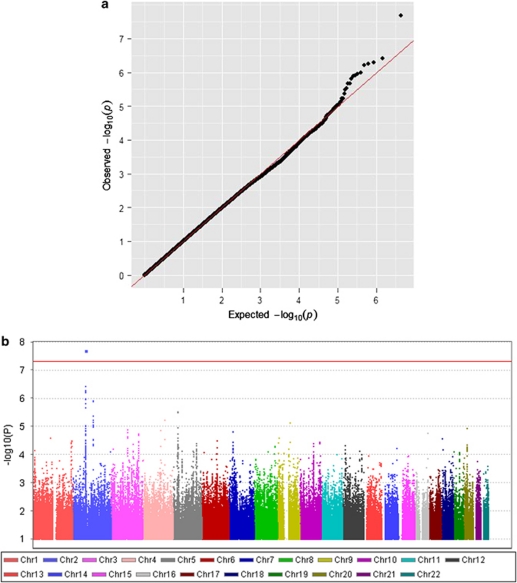
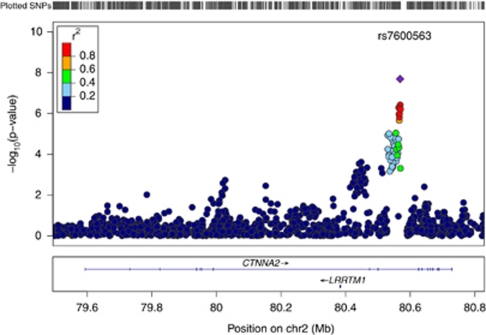
We meta-analyzed the results from six GWAS using over two million genotyped or imputed SNPs and the Excitement-Seeking personality scale. The six samples included 7860 subjects, all of European ancestry. The genomic inflation factor for the meta-analyzed results (λ=0.995), and the quantile–quantile plot (Q–Q plot; Figure 1a) showed no evidence of inflated test statistics, with all SNPs falling on the diagonal line of the observed vs expected P-values, with the exception of the few SNPs on the extreme tail of the distribution with a strong association signal. As reported in Table 1 (see Supplementary Table S1 for SNPs with P-values <10−3) and shown in the Manhattan and regional plots (Figures 1b and 2), the meta-analysis revealed a genome-wide significant association (rs7600563; P=2 × 10−8) on chromosome 2, within an intron of the gene catenin (cadherin-associated protein), alpha 2 (CTNNA2). CTNNA2 is a brain-expressed gene that plays a major role in synaptic plasticity, and has been associated with bipolar disorder,62 attention deficit/hyperactivity disorders63 and schizophrenia.64 As depicted in Figure 2, a gene within 200 kb of rs7600563 is the leucine-rich repeat transmembrane neuronal 1 (LRRTM1). LRRTM1 is thought to be involved in neuronal differentiation and connectivity, and it has been associated with handedness and schizophrenia/schizoaffective disorder.65
Figure 1.
(a) Quantile–quantile (Q–Q) plots of the meta-analyses of genome-wide association studies (GWAS) results for excitement-seeking. (b) Manhattan plot summarizing the meta-analysis results for excitement-seeking in six samples. Notes: Each dot represents a single-nucleotide polymorphism (SNP), with the chromosomal position on the x axis and the P-value on the y axis.
Table 1. SNPs with the strongest association with excitement-seeking from the meta-analysis of GWAS results.
| SNP | Chr. | Pos | Gene | AL1 | AL2 | Freq | β | s.e. | Direction | P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| rs7600563 | 2 | 80.57 | CTNNA2 | t | g | 69 (58–73) | 0.94 | 0.17 | ++++++ | 2.04 × 10−8 |
| rs17019225 | 2 | 80.57 | CTNNA2 | a | t | 30 (27–42) | −0.90 | 0.18 | −−−−?− | 3.80 × 10−7 |
| rs6547311 | 2 | 80.56 | CTNNA2 | c | g | 28 (24–33) | −0.89 | 0.18 | −−−−−− | 5.16 × 10−7 |
| rs6547313 | 2 | 80.57 | CTNNA2 | a | g | 69 (58–73) | 0.87 | 0.17 | ++++?+ | 5.48 × 10−7 |
| rs6742029 | 2 | 80.57 | CTNNA2 | a | g | 33 (30–45) | −0.82 | 0.16 | −−−−−− | 6.16 × 10−7 |
| rs1971117 | 2 | 80.57 | CTNNA2 | c | g | 69 (57–72) | 0.82 | 0.17 | ++++++ | 1.01 × 10−6 |
| rs11902138 | 2 | 80.57 | CTNNA2 | t | c | 25 (19–31) | −0.90 | 0.18 | −−−−−− | 1.10 × 10−6 |
| rs2445234 | 2 | 129.01 | — | a | g | 11 (06–12) | 1.25 | 0.26 | ++−+?+ | 1.24 × 10−6 |
| rs841086 | 2 | 129.00 | — | a | g | 11 (05–12) | 1.21 | 0.25 | ++−+++ | 1.28 × 10−6 |
| rs11126769 | 2 | 80.57 | CTNNA2 | t | c | 69 (57–72) | 0.80 | 0.17 | ++++++ | 1.52 × 10−6 |
| rs1897771 | 2 | 80.57 | CTNNA2 | a | g | 31 (28–45) | −0.79 | 0.17 | −−−−−− | 2.17 × 10−6 |
| rs1434073 | 2 | 80.57 | CTNNA2 | a | t | 69 (58–73) | 0.83 | 0.18 | ++++?+ | 2.17 × 10−6 |
| rs4421076 | 5 | 27.48 | — | a | g | 41 (36–44) | −0.73 | 0.16 | −−−−−− | 3.02 × 10−6 |
| rs10039598 | 5 | 27.48 | — | a | g | 41 (36–44) | −0.73 | 0.16 | −−−−−− | 3.24 × 10−6 |
Abbreviations: N>7000 (range: 7384–7861); Minor Allele Frequency⩾1%, chr, chromosome; POS, chromosomal position; AL1, effect allele; AL2, other allele; Freq, average and (range) of frequency of AL1 across the six samples; SNP, single nucleotide polymorphism; GWAS, genome-wide association studies; CTNNA2, catenin cadherin-associated protein alpha 2.
The direction column indicates whether the sign of the β is positive or negative in each of the six samples, in the order of sample size: SardiNIA, Estonian Genome Center of University of Tartu, Baltimore Longitudinal Study of Aging, Queensland Institute of Medical Research, Germany, and Cilento. The question mark is for missing statistics.
Figure 2.
Regional plot of the association between catenin cadherin-associated protein, alpha 2 (CTNNA2) single-nucleotide polymorphisms (SNPs) and excitement-seeking. Notes: Each dot represents an SNP, with the chromosomal position on the x axis and the P-value on the y axis. The color of the dots depicts the HapMap CEU derived r2 between the SNP with the strongest signal (rs7600563) and the other SNPs in the region. Plot was generated using LocusZoom,72 http://www.csg.sph.umich.edu/locuszoom.
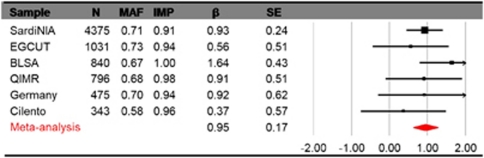
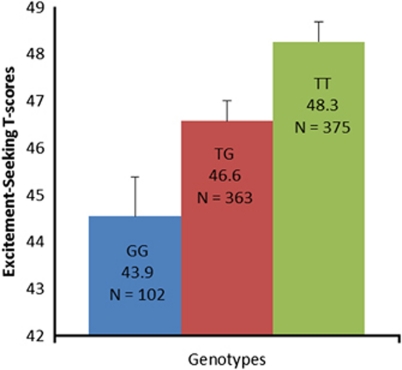
The association between excitement-seeking and rs7600563 was in the same direction in all six samples (Figure 3), and there was no evidence of heterogeneity (Q=4.2, d.f.=5, P=0.52). Each copy of the T allele (frequency 69%) was associated with almost 0.1 s.d. higher Excitement-Seeking scores. The strongest association between excitement-seeking and rs7600563 was found in the BLSA sample (Figures 3 and 4). The SNP rs7600563 was directly genotyped in the BLSA sample (Hardy–Weinberg equilibrium, χ2=0.95, P>0.05), and it was imputed in the other samples with good imputation quality (>0.90). Table 1 and Figure 2 show that a number of SNPs within CTNNA2 had a strong signal, all of them in high linkage disequilibrium with rs7600563.
Figure 3.
Association of rs7600563 and excitement-seeking in six samples and the meta-analysis. Notes: N=sample size; MAF=major allele frequency (T); IMP=imputation quality. The single-nucleotide polymorphism (SNP) was genotyped and not imputed in the Baltimore Longitudinal Study of Aging (BLSA). Plot drawn with the program Comprehensive Meta-Analysis V.2.
Figure 4.
Association of rs7600563 genotypes and excitement-seeking in the Baltimore Longitudinal Study of Aging (BLSA). Notes: N=840. Estimated means controlling for age and sex. Hardy–Weinberg Equilibrium (HWE) χ2=0.95, P>0.05.
Replication analyses
We tested whether the association between excitement-seeking and rs7600563 was replicated in three additional samples. The genotyped rs7600563 showed no association with excitement-seeking in a sample of 608 subjects from Estonia (T allele frequency=68%, β=−0.27, s.e.=0.65, P=0.67) and in a sample of 1454 subjects from the HBCS study (T allele frequency=76%, β=−0.27, s.e.=0.25, P=0.27). There was also no association between the rs7600563 (proper_info=0.87, T allele frequency 72%) and the Zuckerman's Sensation-Seeking scales in the NTR sample (N=3043). Although not significant, there was a trend for the rs7600563 T allele to be associated with higher scores on Thrill and Adventure-Seeking (β=0.36, P=0.08 one-tail), the Zuckerman's Sensation-Seeking scale most strongly associated with excitement-seeking.
Discussion
Across six studies, with a total of 7860 subjects, we found a genome-wide significant association between rs7600563, a common variant in CTNNA2 and the Revised NEO Personality Inventory Excitement-Seeking scale, a trait measure of thrill-seeking and risk-taking behaviors (P=2 × 10−8). Although the association was not replicated in two additional samples, it showed a trend (P=0.08) with a related sensation-seeking scale. Excitement-seeking was one of many traits that we tested, so the likelihood of finding a significant association may be inflated by multiple testing. Even a modest correction would render the effect nonsignificant. Still, this association points to a gene with clear biological relevance for brain structure and function, and can provide insight into the biological bases of individual differences in excitement-seeking and impulsive tendencies.
CTNNA2 is a large (∼1 Mb) gene on chromosome 2 that is conserved across species. Microarray expression data (see http://www.genome.ucsc.edu) indicate that CTNNA2 is strongly expressed in the central nervous system, especially in the prefrontal cortex, temporal lobe, cingulate cortex, hypothalamus and amygdala. These structures are responsible for executive function, memory, learning, energy balance and emotion-related processes that are likely to be involved in the regulation of excitement-seeking tendencies. Outside the brain, there is little expression of CTNNA2 in other tissues, except for the testis. CTNNA2 encodes for a cell-adhesion protein known as αN-catenin, which plays an important role as regulator of synaptic plasticity: it binds cadherins and the actin cytoskeleton, and as such is essential for maintaining the stability of dendritic spines and synaptic contact.66, 67 Deletion of the homologous gene in mice causes cerebellar and hippocampal lamination defects, axon migration defects and other deficiencies in brain morphogenesis.68, 69 Mice with deletion of Catna2 show a number of abnormal behaviors, including cerebella ataxia, impaired startle modulation and deficits in fear conditioning. These mice had stronger acoustic startle responses to intense/abrupt auditory stimuli and compromised startle modulation following prepulse inhibition or fear conditioning.68 Furthermore, the expression of a Catna2 transgene in mice with partially deleted Catna2 restored brain morphology and behavioral performance,68 confirming the role of Catna2 and catenin–cadherin cell–adhesion complexes in the brain structure and behaviors. The mouse model, knowledge of the protein function and the gene expression profile all point to potential neurophysiological mechanisms underlying the association we identified with a bottom–up molecular genetic approach.
The search for exciting and stimulating activities and environments are common human behaviors, but in the extreme such behaviors can be symptoms of psychiatric disorders. In that regard, it is worth noting that a common variant in CTNNA2 has been associated with bipolar disorder in a meta-analysis of GWAS that included 3683 cases and 14 507 controls.62 Similarly, a GWAS of pooled DNA also reported evidence that CTNNA2 is involved in attention deficit/hyperactivity disorders.63 CTNNA2 has been considered a susceptibility gene for schizophrenia, based on microarray analyses and qPCR-confirmed differences in gene expression in post-mortem brain tissue of schizophrenic patients compared with controls.64 Although these findings need further replication, they may suggest a pleiotropic action of CTNNA2, and supports the notion that genes that harbor common variants associated with personality traits might harbor additional variants associated with more extreme behavioral, emotional and cognitive phenotypes that fall under psychiatric nosology. Other GWAS have pointed to shared genetic susceptibility for a number of psychiatric disorders.11, 63, 70, 71
In conclusion, our systematic search across the genome for variants associated with the Excitement-Seeking scale identified a locus within CTNNA2, an α-catenin widely expressed in the central nervous system and critical for brain morphogenesis and synaptic plasticity. Furthermore, this study provides association results for a large number of additional SNPs across the genome that showed nominal associations with excitement-seeking (see Supplementary Table S1), which may be a valuable resource for future research. Identifying genetic factors associated with excitement-seeking is a valuable step forward to pinpoint biological mechanisms underlying risk-taking behaviors. Furthermore, this association is relevant to a number of related psychiatric conditions, and for dangerous behaviors that have serious medical, psychological and societal consequences.
Acknowledgments
We thank study subjects for their participation as well as everybody involved in each of the participating studies. EGCUT authors want to acknowledge EGCUT personnel, especially Merli Hass, Viljo Soo and Mari Nelis. EGCUT data analyzes were carried out in part in the High Performance Computing Center of University of Tartu. Cilento authors acknowledge Dr Maria Enza Amendola for the test administration and thank the personnel working in the organization of the study in the villages. QIMR thanks study participants and acknowledges Nathan Gillespie for discussion on study design related to NEO collection; Marisa Grimmer, Romana Leisser and Kim Eldridge for overseeing data collection; Anjali Henders, Megan Campbell, Lisa Bowdler, Steven Crooks and staff of the Molecular Epidemiology Laboratory for sample processing and preparation; and Harry Beeby, David Smyth and Daniel Park for IT support. We acknowledge Drs Dale R Nyholt and Scott Gordon for their substantial efforts involving the QC and preparation of the GWA data sets and Dr Sarah Medland for undertaking the imputation of the GWAS data and subsequent preparation for analyses. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. Anu Realo and Jüri Allik were supported by a grant from the Estonian Ministry of Science and Education (SF0180029s08) and by a Primus Grant (3-8.2/60) from the European Social Fund. TE and AM received support from FP7 Grants (201413, 212111, 245536). TE and AM also received targeted financing from Estonian Government SF0180142s08 and from the European Union through the European Regional Development Fund, in the frame of Centre of Excellence in Genomics. QIMR received support from Beyond Blue and the Borderline Personality Disorder Research Foundation. Genotyping was funded by the National Health and Medical Research Council (Medical Bioinformatics Genomics Proteomics Program, 389891). Helsinki Birth Cohort Study has been supported by grants from the Academy of Finland, the Finnish Diabetes Research Society, Folkhälsan Research Foundation, Novo Nordisk Foundation, Finska Läkaresällskapet, Signe and Ane Gyllenberg Foundation, University of Helsinki, European Science Foundation (EUROSTRESS), Ministry of Education, Ahokas Foundation and Emil Aaltonen Foundation.
Paul T Costa Jr received royalties from the Revised NEO-PI. The other authors declare that they have no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
This work was performed by NIH researchers (US government employees), and as such the work is “in the public domain”.
Supplementary Material
References
- Wellcome Trust Case Control Consortium Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Bhomra A, Smiley S, Wray NR, James MR, Martin NG, et al. A whole genome association study of neuroticism using DNA pooling. Mol Psychiatry. 2008;13:302–312. doi: 10.1038/sj.mp.4002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Oord EJCG, Kuo P-H, Hartmann AM, Webb BT, Moller H-J, Hettema JM, et al. Genomewide association analysis followed by a replication study implicates a novel candidate gene for neuroticism. Arch Gen Psychiatry. 2008;65:1062–1071. doi: 10.1001/archpsyc.65.9.1062. [DOI] [PubMed] [Google Scholar]
- Terracciano A, Sanna S, Uda M, Deiana B, Usala G, Busonero F, et al. Genome-wide association scan for five major dimensions of personality. Mol Psychiatry. 2010;15:647–656. doi: 10.1038/mp.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calboli FC, Tozzi F, Galwey NW, Antoniades A, Mooser V, Preisig M, et al. A genome-wide association study of neuroticism in a population-based sample. PLoS One. 2010;5:e11504. doi: 10.1371/journal.pone.0011504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij KJ, Zietsch BP, Medland SE, Gordon SD, Benyamin B, Nyholt DR, et al. A genome-wide association study of Cloninger's temperament scales: implications for the evolutionary genetics of personality. Biol Psychol. 2010;85:306–317. doi: 10.1016/j.biopsycho.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor MH, Costa PT, Terracciano A, Krueger RF, de Geus EJ, Toshiko T, et al. Meta-analysis of genome-wide association studies for personality Mol Psychiatry 2010(in press). [DOI] [PMC free article] [PubMed]
- Terracciano A, Tanaka T, Sutin AR, Sanna S, Deiana B, Lai S, et al. Genome-wide association scan of trait depression. Biol Psychiatry. 2010;68:811–817. doi: 10.1016/j.biopsych.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merjonen P, Keltikangas-Järvinen L, Jokela M, Seppälä I, Lyytikäinen L-P, Pulkki-Råback L, et al. Hostility in adolescents and adults: a genome-wide association study of the Young Finns. Translational Psychiatry. 2011;1:e11. doi: 10.1038/tp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PT, Jr, McCrae RR. Revised NEO Personality Inventory (NEO-PI-R) and NEO Five-Factor Inventory (NEO-FFI) professional manual. Psychological Assessment Resources: Odessa, FL; 1992. [Google Scholar]
- Zuckerman M, Kuhlman DM. Personality and risk-taking: common biosocial factors. J Pers. 2000;68:999–1029. doi: 10.1111/1467-6494.00124. [DOI] [PubMed] [Google Scholar]
- Eysenck HJ, Eysenck SBG. On the unitary nature of extraversion. Acta Psychol. 1967;26:383–390. doi: 10.1016/0001-6918(67)90034-0. [DOI] [PubMed] [Google Scholar]
- Whiteside SP, Lynam DR. The Five Factor Model and impulsivity: using a structural model of personality to understand impulsivity. Person Individ Differ. 2001;30:669–689. [Google Scholar]
- Smith GT, Fischer S, Cyders MA, Annus AM, Spillane NS, McCarthy DM. On the validity and utility of discriminating among impulsivity-like traits. Assessment. 2007;14:155–170. doi: 10.1177/1073191106295527. [DOI] [PubMed] [Google Scholar]
- Zuckerman M. Sensation Seeking: Beyond the Optimal Level of Arousal. Lawrence Erlbaum Associates: Hillsdale, NJ; 1979. [Google Scholar]
- Gray JA. The psychophysiological basis of introversion–extroversion. Behav Res Ther. 1970;8:249–266. doi: 10.1016/0005-7967(70)90069-0. [DOI] [PubMed] [Google Scholar]
- Cloninger CR, Svrakic DM, Przybeck TR. A psychobiological model of temperament and character. Arch Gen Psychiatry. 1993;50:975–990. doi: 10.1001/archpsyc.1993.01820240059008. [DOI] [PubMed] [Google Scholar]
- Elliot AJ, Thrash TM. Approach–avoidance motivation in personality: approach and avoidance temperaments and goals. J Pers Soc Psychol. 2002;82:804–818. doi: 10.1037//0022-3514.82.5.804. [DOI] [PubMed] [Google Scholar]
- Terracciano A, McCrae RR, Brant LJ, Costa PT., Jr Hierarchical linear modeling analyses of NEO-PI-R scales in the Baltimore Longitudinal Study of Aging. Psychology Aging. 2005;20:493–506. doi: 10.1037/0882-7974.20.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrae RR, Terracciano A, Members of the Personality Profiles of Cultures Project Universal features of personality traits from the observer's perspective: data from 50 cultures. J Person Soc Psychol. 2005;88:547–561. doi: 10.1037/0022-3514.88.3.547. [DOI] [PubMed] [Google Scholar]
- Costa PT, Jr, Terracciano A, McCrae RR. Gender differences in personality traits across cultures: robust and surprising findings. J Person Social Psychol. 2001;81:322–331. doi: 10.1037/0022-3514.81.2.322. [DOI] [PubMed] [Google Scholar]
- Terracciano A, Lockenhoff CE, Crum RM, Bienvenu OJ, Costa PT., Jr Five-Factor Model personality profiles of drug users. BMC Psychiatry. 2008;8:22. doi: 10.1186/1471-244X-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucidi F, Giannini AM, Sgalla R, Mallia L, Devoto A, Reichmann S. Young novice driver subtypes: relationship to driving violations, errors and lapses. Accid Anal Prev. 2010;42:1689–1696. doi: 10.1016/j.aap.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Samuels J, Bienvenu OJ, Cullen B, Costa PT, Jr, Eaton WW, Nestadt G. Personality dimensions and criminal arrest. Compr Psychiatry. 2004;45:275–280. doi: 10.1016/j.comppsych.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Ball SA, Carroll KM, Rounsaville BJ. Sensation seeking, substance abuse, and psychopathology in treatment-seeking and community cocaine abusers. J Consult Clin Psychol. 1994;62:1053–1057. doi: 10.1037//0022-006x.62.5.1053. [DOI] [PubMed] [Google Scholar]
- Bagby RM, Vachon DD, Bulmash EL, Toneatto T, Quilty LC, Costa PT. Pathological gambling and the five-factor model of personality. Person Individ Differ. 2007;43:873–880. [Google Scholar]
- Fischer S, Smith GT. Binge eating, problem drinking, and pathological gambling: Linking behavior to shared traits and social learning. Person Individ Differ. 2008;44:789–800. [Google Scholar]
- Krueger RF, Hicks BM, Patrick CJ, Carlson SR, Iacono WG, McGue M. Etiologic connections among substance dependence, antisocial behavior, and personality: modeling the externalizing spectrum. J Abnorm Psychol. 2002;111:411–424. [PubMed] [Google Scholar]
- Young SE, Stallings MC, Corley RP, Krauter KS, Hewitt JK. Genetic and environmental influences on behavioral disinhibition. Am J Med Genet. 2000;96:684–695. [PubMed] [Google Scholar]
- Zietsch BP, Verweij KJ, Bailey JM, Wright MJ, Martin NG. Genetic and environmental influences on risky sexual behaviour and its relationship with personality. Behav Genet. 2010;40:12–21. doi: 10.1007/s10519-009-9300-1. [DOI] [PubMed] [Google Scholar]
- Maremmani I, Pacini M, Perugi G. Addictive disorders, bipolar spectrum and the impulsive link: the psychopathology of a self-regenerating pathway. Heroin Addict Relat Clin Problems. 2005;7:33–45. [Google Scholar]
- Hegerl U, Himmerich H, Engmann B, Hensch T. Mania and attention-deficit/hyperactivity disorder: common symptomatology, common pathophysiology and common treatment. Curr Opin Psychiatry. 2010;23:1–7. doi: 10.1097/YCO.0b013e328331f694. [DOI] [PubMed] [Google Scholar]
- McElroy SL, Pope HG, Jr, Keck PE, Jr, Hudson JI, Phillips KA, Strakowski SM. Are impulse-control disorders related to bipolar disorder. Compr Psychiatry. 1996;37:229–240. doi: 10.1016/s0010-440x(96)90001-2. [DOI] [PubMed] [Google Scholar]
- Moeller FG, Barratt ES, Dougherty DM, Schmitz JM, Swann AC. Psychiatric aspects of impulsivity. Am J Psychiatry. 2001;158:1783–1793. doi: 10.1176/appi.ajp.158.11.1783. [DOI] [PubMed] [Google Scholar]
- McCrae RR, Kurtz JE, Yamagata S, Terracciano A. Internal consistency, retest reliability, and their implications for personality scale validity. Pers Soc Psychol Rev. 2011;15:28–50. doi: 10.1177/1088868310366253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang KL, McCrae RR, Angleitner A, Riemann R, Livesley WJ. Heritability of facet-level traits in a cross-cultural twin sample: support for a hierarchical model of personality. J Pers Soc Psychol. 1998;74:1556–1565. doi: 10.1037//0022-3514.74.6.1556. [DOI] [PubMed] [Google Scholar]
- Stoel RD, De Geus EJ, Boomsma DI. Genetic analysis of sensation seeking with an extended twin design. Behav Genet. 2006;36:229–237. doi: 10.1007/s10519-005-9028-5. [DOI] [PubMed] [Google Scholar]
- Bevilacqua L, Doly S, Kaprio J, Yuan Q, Tikkanen R, Paunio T, et al. A population-specific HTR2B stop codon predisposes to severe impulsivity. Nature. 2010;468:1061–1066. doi: 10.1038/nature09629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilia G, Chen WM, Scuteri A, Orrú M, Albai G, Dei M, et al. Heritability of cardiovascular and personality traits in 6148 sardinians. PloS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton University Press: Princeton, NJ; 1994. [Google Scholar]
- Allik J, Realo A, MÃμttus R, Esko T, Pullat J, Metspalu A. Variance determines self-observer agreement on the Big Five personality traits. J Res Person. 2010;44:421–426. [Google Scholar]
- De Fruyt F, De Bolle M, McCrae RR, Terracciano A, Costa PT, Jr, Collaborators of the Adolescent Personality Profiles of Cultures Project Assessing the universal structure of personality in early adolescence: the NEO-PI-R and NEO-PI-3 in 24 cultures. Assessment. 2009;16:301–311. doi: 10.1177/1073191109333760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelis M, Esko T, MÃgi R, Zimprich F, Toncheva D, Karachanak S, et al. Genetic structure of Europeans: a view from the north-east. PLoS One. 2009;4:e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Costa PT, Jr, McCrae RR. Personality plasticity after age 30. Person Soc Psychol Bull. 2006;32:999–1009. doi: 10.1177/0146167206288599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MJ, Martin NG. Brisbane adolescent twin study: outline of study methods and research projects. Aust J Psychol. 2004;56:65–78. [Google Scholar]
- Distel MA, Willemsen G, Ligthart L, Derom CA, Martin NG, Neale MC, et al. Genetic covariance structure of the four main features of borderline personality disorder. J Person Disord. 2010;24:427–444. doi: 10.1521/pedi.2010.24.4.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medland SE, Nyholt DR, Painter JN, McEvoy BP, McRae AF, Zhu G, et al. Common variants in the Trichohyalin gene are associated with straight hair in Europeans. Am J Hum Genet. 2009;85:750–755. doi: 10.1016/j.ajhg.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PT, Jr, McCrae RR. The NEO-PI/NEO-FFI Manual Supplement. Psychological Assessment Resources Inc.: Odessa, FL; 1989. [Google Scholar]
- Barker DJ, Osmond C, Forsen TJ, Kajantie E, Eriksson JG. Trajectories of growth among children who have coronary events as adults. N Engl J Med. 2005;353:1802–1809. doi: 10.1056/NEJMoa044160. [DOI] [PubMed] [Google Scholar]
- Eriksson JG, Osmond C, Kajantie E, Forsen TJ, Barker DJ. Patterns of growth among children who later develop type 2 diabetes or its risk factors. Diabetologia. 2006;49:2853–2858. doi: 10.1007/s00125-006-0459-1. [DOI] [PubMed] [Google Scholar]
- Rikkonen K, Pesonen AK, Heinonen K, Lahti J, Kajantie E, Forsen T, et al. Infant growth and hostility in adult life. Psychosom Med. 2008;70:306–313. doi: 10.1097/PSY.0b013e3181651638. [DOI] [PubMed] [Google Scholar]
- Boomsma DI, de Geus EJ, Vink JM, Stubbe JH, Distel MA, Hottenga JJ, et al. Netherlands Twin Register: from twins to twin families. Twin Res Hum Genet. 2006;9:849–857. doi: 10.1375/183242706779462426. [DOI] [PubMed] [Google Scholar]
- Willemsen G, de Geus EJ, Bartels M, van Beijsterveldt CE, Brooks AI, Estourgie-van Burk GF, et al. The Netherlands Twin Register biobank: a resource for genetic epidemiological studies. Twin Res Hum Genet. 2010;13:231–245. doi: 10.1375/twin.13.3.231. [DOI] [PubMed] [Google Scholar]
- Zuckerman M. Dimensions of sensation seeking. J Consult Clin Psychol. 1971;36:45–52. [Google Scholar]
- Aluja A, García Ó, García LF. Relationships among extraversion, openness to experience, and sensation seeking. Person Individ Differ. 2003;35:671–680. [Google Scholar]
- Zeggini E. Next-generation association studies for complex traits. Nat Genet. 2011;43:287–288. doi: 10.1038/ng0411-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perneger TV. What's wrong with Bonferroni adjustments. BMJ. 1998;316:1236–1238. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Tarone R, McLaughlin JK. The false-positive to false-negative ratio in epidemiologic studies. Epidemiology. 2011;22:450–456. doi: 10.1097/EDE.0b013e31821b506e. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci USA. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch KP, Timmesfeld N, Renner TJ, Halperin R, Roser C, Nguyen TT, et al. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm. 2008;115:1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- Chu TT, Liu Y. An integrated genomic analysis of gene–function correlation on schizophrenia susceptibility genes. J Hum Genet. 2010;55:285–292. doi: 10.1038/jhg.2010.24. [DOI] [PubMed] [Google Scholar]
- Francks C, Maegawa S, Lauren J, Abrahams BS, Velayos-Baeza A, Medland SE, et al. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia Mol Psychiatry 2007121129–1139.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe K, Chisaka O, Van Roy F, Takeichi M. Stability of dendritic spines and synaptic contacts is controlled by alpha N-catenin. Nat Neurosci. 2004;7:357–363. doi: 10.1038/nn1212. [DOI] [PubMed] [Google Scholar]
- Smith A, Bourdeau I, Wang J, Bondy CA. Expression of catenin family members CTNNA1, CTNNA2, CTNNB1 and JUP in the primate prefrontal cortex and hippocampus. Brain Res Mol Brain Res. 2005;135:225–231. doi: 10.1016/j.molbrainres.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Park C, Falls W, Finger JH, Longo-Guess CM, Ackerman SL. Deletion in Catna2, encoding alpha N-catenin, causes cerebellar and hippocampal lamination defects and impaired startle modulation. Nat Genet. 2002;31:279–284. doi: 10.1038/ng908. [DOI] [PubMed] [Google Scholar]
- Uemura M, Takeichi M. Alpha N-catenin deficiency causes defects in axon migration and nuclear organization in restricted regions of the mouse brain. Dev Dyn. 2006;235:2559–2566. doi: 10.1002/dvdy.20841. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon FJ, Akula N, Schulze TG, Muglia P, Tozzi F, Detera-Wadleigh SD, et al. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21.1. Nat Genet. 2010;42:128–131. doi: 10.1038/ng.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.