Abstract
The severity of attention-deficit/hyperactivity disorder (ADHD) symptoms is a major predictor of long-term ADHD outcome. To investigate if two-locus interactions might predict ADHD severity, we studied a sample of 1341 individuals from families clustering ADHD, using the Vanderbilt Assessment Scale for Parents. Latent class cluster analysis was used to construct symptom profiles and classify ADHD severity. Single nucleotide polymorphisms (SNPs) spanning ADHD-linked chromosomal regions on chromosomes 4, 5, 10, 11, 12 and 17 were genotyped. SNPs associated with ADHD severity were identified and potential two-locus genetic interactions were tested. We found that SNPs within the LPHN3 gene interact with SNPs spanning the 11q region that contains DRD2 and NCAM1 not only to increase the risk of developing ADHD but also to increase ADHD severity. All these genes are identified to have a major role in shaping both brain development and function. These findings demonstrate that genetic interactions may predict the severity of ADHD, which in turn may predict long-term ADHD outcome.
Keywords: ADHD, DRD2, epistasis, LPHN3, NCAM1, severity of symptoms
Introduction
Attention-deficit and hyperactivity disorder (ADHD) is one of the most frequent psychiatric disorders in childhood, affecting 10% of children and adolescents in the United States.1 ADHD is characterized by elevated levels of inattention and/or hyperactive or impulsive behaviors that cause significant impairment2 on a child's academic and social functioning.3
Several studies show that the severity of ADHD symptoms predicts long-term prognosis.4, 5, 6, 7 For example, Barry et al. reported that the strongest predictor of discrepancy between predicted achievement evaluated by measurements of intellectual function and actual academic performance is the severity of ADHD symptoms, defined as the index composed of averaged behavioral data obtained from the hyperactivity/impulsivity checklist of DSM-IV, attention problems and hyperactivity scores from the Behavior Assessment System for Children-Parent Rating Scale, and from the total score on the Home Situations Questionnaire.4 Similar results have been reported showing that the severity of inattention and hyperactivity negatively correlates with academic performance and has deleterious effects on other aspects related to quality of life, such as risk avoidance and satisfaction.5 Another study showed that ADHD severity was positively associated with the use of tobacco, alcohol and marijuana.6 In individuals without frank ADHD, higher levels of inattention have been found to have a negative effect on both academic performance and college adjustment.7
Due to this evidence, using a clinical approach that takes severity into account is relevant. Furthermore, the traditional approach using the diagnostic criteria for ADHD as defined in the DSM-IV discards those individuals who, despite the severity of some of the specific symptoms, do not fit the overall numeric criteria for the diagnosis of ADHD. However, the severity of their symptoms could have a marked functional impact and these individuals may benefit from interventions.8, 9, 10
In the etiology of ADHD, genetic factors are strongly implicated.11 Recently, we identified a common variant of the Latrophilin 3 gene (LPHN3) that increases the risk of developing ADHD by 1.2-fold. The ADHD population attributable risk (PAR%) attributed to this gene is ∼9%, suggesting that incidence of ADHD in the general population would be reduced by about 9% if it were possible to control the effect of the LPHN3 variant that confers susceptibility to ADHD.12 Functional studies revealed that LPHN3 is expressed in key brain regions related to attention and activity, and that its variants affect metabolism in neural circuits implicated in ADHD and are associated with response to stimulant medication. These findings were replicated in samples obtained from unrelated populations.12, 13 In addition to increasing knowledge of the genetic basis of ADHD, the discovery of the molecular substrates of ADHD opens up new avenues for the exploration of targeted therapies.
In related studies, we analyzed two-locus interactions models between LPHN3 variants on chromosome 4q and a susceptibility haplotype on 11q encompassing the NCAM1, TTC12, ANKK1 and DRD2 genes. The simultaneous presence of both genetic risk variants increases the risk of ADHD by ∼2.5 compared with the risk when none of these variants are present.14 In addition to previous evidence linking these two regions with human behavior,15, 16, 17, 18, 19, 20 we found that this interaction also explains differences in brain metabolism as observed through proton magnetic resonance spectroscopy (1H-MRS) data and pharmacogenetic response to stimulant medication better than the presence of either variant alone.
In the present study, we aimed to evaluate the hypothesis that the interaction between ADHD susceptibility factors on 4q and 11q, as well as other genomic regions, not only predicts ADHD susceptibility but also the severity of the disease, as ADHD severity is the best indicator of long-term outcome. In this way, we additionally expand upon our previous findings to place them in a context where they may have a greater impact in terms of defining interventions for affected and subsyndromal individuals.
Subjects and methods
Subjects
We analyzed a total of 349 nuclear families consisting of a total of 1371 individuals. Participants were from a United States population, 4–65 years of age, ascertained from ADHD probands. Diagnosis of ADHD in children was established using the DSM-IV criteria;2 in adults, the Conners Adult ADHD Rating Scale (CAARS)21 was used. All participants were evaluated using the Vanderbilt Assessment Scale for Parents (VAS-P),22 which includes all 18 DSM-IV criteria for ADHD (questions 1–9 for inattention; questions 10–18 for hyperactivity/impulsivity), oppositional defiant disorder (ODD; questions 19–26), conduct disorder (CD; questions 27–40) and seven items from the Pediatric Behavior Scale23 screening for anxiety and depression (A/D; questions 41–47 in the VAS-P). Each question of the VAS-P is scored based on the frequency of presentation of the behavior on a 1 to 4 scale (1=never, 2=occasionally, 3=often and 4=very often).22 Complete information was available for 1341 individuals. More detailed information regarding clinical assessment and specific demographics of this sample is described elsewhere.8
Definition of severity
Latent class cluster analysis (LCCA) models containing 1–10 classes were fitted to the data using Latent GOLD 4.5 (Statistical Innovations, Belmont, MA, USA). Latent GOLD uses both EM and Newton–Raphson algorithms to find the maximum likelihood for each model after estimating model parameters.24 In the LCC models, separate analyses were performed for each of the five VAS-P domains (inattention, hyperactivity, ODD, CD and A/D). The number of clusters was selected using a likelihood ratio test evaluating whether increases in likelihood (L2) associated with increased latent classes justified their inclusion. Certainty of these clusters was assessed calculating P-values associated with L2 values after running 500 parametric bootstrap replicates. As covariates for all models, we used gender, ADHD diagnosis and age as categorical variables (children: 4–11 years; adolescents: 12–17 years; adults >17 years).12
As implemented in Latent GOLD, individuals are assigned posterior membership probabilities for belonging to each cluster based on their symptom profiles. Individuals are additionally assigned to the cluster for which the posterior probability is highest. Based on this assignment, VAS-P profiles are obtained for each cluster. Using these profiles, a severity scale was derived as follows: consider the LCC results for a particular VAS-P domain, for example, inattention, and let K be the number of clusters found, Q be the number of questions in that domain, ni be the number of individuals in that cluster (i=1,2,…,K) and NPi be the number of questions in cluster i with an average profile P. If NPi>Q/2, all ni individuals in cluster i will be classified as being severe for this domain, and not severe otherwise. In other words, individuals with severe symptoms will be those for whom the cluster they belong to, has at least Q/2+1 questions with a profile above ‘Ocassionally'. From now on, individuals with severe symptoms will be treated as ‘cases' and non-severely affected individuals as ‘controls'. For the analyses, this derivation allows more precise descriptions of each individual's characteristics (profiles), descriptions that would be difficult to define when relying solely on the categorical DSM-IV diagnosis.
Genotyping and genetic statistical analysis
DNA was prepared from peripheral-blood specimens and the Illumina genotyping service was used for genotyping, as previously described.12 Out of the 1341 individuals who completed the VAS-P questionnaire, blood samples were available for 1181; genetic data was available for 813 individuals. A total of 369 markers homogenously distributed on genomic regions of chromosomes 4 (156 markers), 5 (48 markers), 10 (7 markers), 11 (129 markers), 12 (15 markers) and 17 (14 markers), as described previously,12 were selected for analysis because of the presence of loci linked to ADHD in a genetic isolate from Colombia.25
Family-based association tests (FBATs) using a dominant model under the hypotheses of no linkage and no association were performed as implemented in Golden Helix's PBAT module (Golden Helix, Bozeman, MT, USA. Golden Helix PBAT Software http://www.goldenhelix.com). These hypotheses were selected because the linkage studies had been performed on a different population, and hence there was no previous knowledge about linkage or association in the United States population described here. As phenotypes, the VAS-P score, defined as the summation of the individual's responses in a particular domain, and the VAS-P severity scale, derived using LCCA, were used independently. The significance of markers was determined by using both the raw P-value and a permutation-based approach with B=10 000 permutations.
Interactions effects on the severity of symptoms
We used the generalized Cochran–Mantel–Haenszel test as implemented in R,26 to determine for all possible pairs of markers located on different chromosomes and significantly associated either with the VAS-P score or the severity of symptoms, whether the pair of single nucleotide polymorphisms (SNPs) are independent, conditionally on the levels of the severity phenotype (severe and not severe). This approach was stratified by domain, and using the definition of severity of symptoms previously described. A total of p pairs of markers were found to be of potential interest. By domain, the false discovery rate (FDR)27 was used to correct by multiple testing.
To perform the Cochran–Mantel–Haenszel test for a particular domain we used the severity scale as the stratum indicator, such that a series of p pairs of contingency tables, each of dimension r × c, can be obtained. In our context, p (p=587) is the number of possible pairs of markers located within different chromosomes, r (r=3) is the number of possible genotypes in marker 1 and c (c=3) the number of possible genotypes in marker 2. To avoid any bias inherent to family data, we selected only those families with (i) discordant parents and concordant children affected, (ii) two discordant siblings and unaffected parents, (iii) unaffected parents and several discordant children and (iv) discordant parents and discordant siblings. Families falling in the third case were further examined so only one affected and one unaffected child was selected. This selection was performed matching by gender and age range. Parents were excluded from families falling in the fourth case. At the end of the process, only one discordant pair was selected within those families that fulfilled the inclusion criteria.
Results
Severity of ADHD symptoms
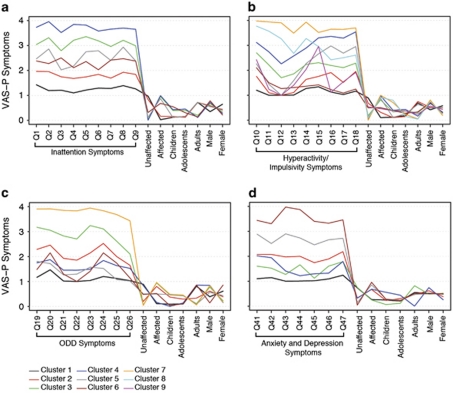
In our maximally expanded sample (n=1341), we identified six significant independent and mutually exclusive clusters for the VAS-P inattention domain, nine for hyperactivity/impulsivity, seven for ODD and six for the A/D (Table 1 and Figure 1), all of which were consistent with findings from previous studies.8, 12, 28, 29, 30, 31 Because of small variability in the predictors in the CD domain, LCCA models could not be fitted to this data. Individuals were considered as having severe symptoms when belonging to clusters 3, 4, 5 and 6 in the inattention domain (n=647, 49%); to clusters 3, 4, 5, 7 and 8 in the hyperactivity/impulsivity domain (n=532, 39.7%); to clusters 3 and 7 in the ODD domain (n=229, 17%) and to clusters 5 and 6 in the A/D domain (n=177, 13.2%) (Figure 1). Under this categorization, severe versus not severe status involves all of the independent variation conferred by the spectrum of symptoms that define ADHD and its comorbidities. Further, using this categorization for each domain, we performed association and interaction analyses for the severity phenotype.
Table 1. Summary of clusters derived by latent class analyses.
| Questions | Criteriaa | Cluster | Characteristic and affection status for broader phenotype | Predominant gender and age classes | Severity of symptoms | n (%) |
|---|---|---|---|---|---|---|
| Inattention | ||||||
| 1–9 | >5 | 1 | Minimal symptoms, unaffected for ADHD | Females-adults | Not severe | 370 (27.6) |
| 2 | Few symptoms, unaffected for ADHD | Males-adults | Not severe | 313 (23.4) | ||
| 3 | Mostly inattentive, high presence of symptoms, affected for ADHD | Males-children and adolescents (few adults) | Severe | 271 (20.2) | ||
| 4 | Mostly inattentive, higher presence of symptoms, affected for ADHD | Males-children and adolescents (few adults) | Severe | 182 (13.6) | ||
| 5 | Fewer symptoms, affected for ADHD | Males-adults | Severe | 151 (11.3) | ||
| 6 | Fewer symptoms, affected for ADHD | Males-children | Severe | 53 (4.0) | ||
| Hyperactivity/impulsivity | ||||||
| 10–18 | >5 | 1 | Minimal H/I symptoms, most individuals unaffected for ADHD | Females-adults | Not severe | 418 (31.2) |
| 2 | Minimal H/I symptoms, most individuals unaffected for ADHD | Males-adults | Not severe | 307 (22.9) | ||
| 3 | Few symptoms, affected for ADHD | Males-adolescents | Severe | 165 (12.3) | ||
| 4 | High presence of symptoms except for Q12, most individuals affected for ADHD | Males-children and adolescents (few Adults) | Severe | 165 (12.3) | ||
| 5 | Few symptoms for Q10–14 and fewer for Q15-Q18, individuals are mostly affected | Females-adults | Severe | 101 (7.5) | ||
| 6 | Few symptoms for Q10 and Q11, but minimal symptoms for the rest; ADHD affection status is equally present | Males-all ages | Not severe | 58 (4.3) | ||
| 7 | Mostly H/I individuals, all affected for ADHD; higher presence of symptoms | Males-children | Severe | 53 (4.0) | ||
| 8 | Mostly H/I individuals affected for ADHD; high presence of symptoms | Males-children | Severe | 48 (3.6) | ||
| 9 | Few-to-minimal symptoms except in Q15; ADHD affection status is equally present | Males-children | Not severe | 25 (1.9) | ||
| ODD | ||||||
| 19–26 | >3 | 1 | Minimal symptoms, most individuals unaffected for ADHD | Female-adults | Not severe | 365 (27.2) |
| 2 | Few symptoms, most individuals affected for ADHD | Male-all ages | Not severe | 293 (21.9) | ||
| 3 | High presence of symptoms in Q19–25 and few for Q26; most individuals affected for ADHD | Males-children | Severe | 182 (13.6) | ||
| 4 | Minimal symptoms in all questions; individuals are mostly unaffected for ADHD | Males-adults | Not severe | 161 (12) | ||
| 5 | Minimal symptoms in all questions; ADHD affection status is equally present | Males-children and adolescents (few adults) | Not severe | 149 (11.1) | ||
| 6 | Few symptoms in Q20 and Q24; ADHD affection status is equally present | Females-adults | Not severe | 143 (10.7) | ||
| 7 | Higher presence of symptoms in all questions; most individuals affected for ADHD | Males-children and adolescents (few adults) | Severe | 47 (3.5) | ||
| Anxiety and depression | ||||||
| 41–47 | >2 | 1 | Minimal symptoms in all questions; individuals are mostly unaffected for ADHD | Males-adults | Not severe | 375 (28.0) |
| 2 | Fewer symptoms; mostly affected individuals | Females-adults | Not severe | 376 (28.1) | ||
| 3 | Few symptoms; most individuals unaffected for ADHD | Males-children | Not severe | 255 (19) | ||
| 4 | Fewer symptoms in Q41, Q42 and Q47 with few on the rest; most individuals affected for ADHD | Males-children | Not severe | 157 (11.7) | ||
| 5 | High presence of symptoms; most individuals affected for ADHD | Females-adults | Severe | 132 (9.9) | ||
| 6 | Higher presence of symptoms in all questions; most individuals affected for ADHD | Females-adults | Severe | 45 (3.4) | ||
Abbreviations: ADHD, attention deficit hyperactivity disorder; adolescents, 12–17 years; adults, >17 years; children, 4–11 years; H/I, hyperactivity/impulsivity; ODD, oppositional defiant disorder; Q, question.
Number of questions being marked as ‘often' or ‘very often' in the Vanderbilt Assessment Scale for Parents (VAS-P) questionnaire.
Figure 1.
Profile plots derived using latent class cluster analysis applied to attention-deficit and hyperactivity disorder symptoms as measured by the Vanderbilt Assessment Scale for Parents (VAS-P) questionnaire. For each domain, VAS-P symptoms profiles within clusters are shown in a scale from 1 to 4 (1=never, 2=occasionally, 3=often and 4=very often). Demographic characteristics for each cluster are shown in a scale from 0 to 1, representing the proportion of individuals from the population with such characteristics. (a) Inattention (Q1–Q9): Q1: careless, inattentive; Q2: sustains attention poorly; Q3: appears to not listen; Q4: poor follow through; Q5: disorganized; Q6: avoids/dislikes sustained mental effort; Q7: loses needed objects; Q8: easily distracted; and Q9: often forgetful. (b) Hyperactivity/impulsivity items ( Q10–Q18): Q10: fidgets or squirms; Q11: cannot stay seated; Q12 restless; Q13: loud; noisy; Q14: always ‘on the go' Q15: talks excessively; Q16: blurts out; Q17: impatient; and Q18: intrusive. (c) Oppositional defiant disorder (ODD; Q19–Q26): Q19: loses temper; Q20: argues with adults; Q21: defies adults' rules; Q22: annoys others; Q23: shifts blames to others; Q24: touchy; Q25: angry/resentful; and Q26: vindictive. (d) Anxiety and depression (Q41–Q47): Q41: fearful, worried; Q42: fear of making mistakes; Q43: feels useless; Q44: blames self; Q45; feels unloved; Q46: sad; and Q47: embarrassed.
FBAT
Once the severity of symptoms for each of the four VAS-P domains was identified, an FBAT was performed, taking into account the complete family structure and the severity status (severe and not severe) as disease indicator (outcome). In complementary analyses, the overall VAS-P score for each dimension was also used as the outcome for the FBAT as a general indicator of severity.
Table 2 presents the FBAT results for both the LCCA-derived severity status and the overall VAS-P score for each domain. After permutation, a total of 38 genetic markers were found to be significantly associated either with the VAS-P score or the severity of symptoms by dimension.
Table 2. Raw and permuted P-values from family-based association tests using the VAS-P score and the severity of symptoms derived using latent class analyses, as qualitative and quantitative traits, respectively.
| Marker | Gene | Allele | Frequency | Effect |
Score |
Severity |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inattention | H/I | ODD | CD | A/D | Inattention | H/I | ODD | A/D | |||||
| Chromosome 4 | |||||||||||||
| rs1947275 | LPHN3 | C | 0.814 | + | 0.00819a | ||||||||
| rs2132074 | LPHN3 | G | 0.618 | + | 0.0017a | ||||||||
| rs4552500 | LPHN3 | G | 0.608 | + | 0.0031b | ||||||||
| rs13124636 | LPHN3 | G | 0.046 | − | 0.0414b | ||||||||
| rs4860091 | LPHN3 | T | 0.386 | + | 0.0054b | ||||||||
| rs335322 | LPHN3 | G | 0.575 | + | 0.007b | ||||||||
| rs10015239 | LPHN3 | A | 0.577 | + | 0.0098b | ||||||||
| rs35106420 | LPHN3 | G | 0.985 | + | 0.0154b | ||||||||
| rs12646895 | LPHN3 | G | 0.508 | + | 0.0019b | ||||||||
| rs1510920 | LPHN3 | C | 0.061 | + | 0.0368b | ||||||||
| rs186750 | LPHN3 | A | 0.243 | + | 0.0216b | ||||||||
| rs734644 | LPHN3 | T | 0.271 | + | 0.0117b | ||||||||
| rs6813183 | LPHN3 | G | 0.300 | + | 0.0114b | ||||||||
| rs6551670 | LPHN3 | A | 0.300 | + | 0.0105b | ||||||||
| rs6551669 | LPHN3 | C | 0.300 | + | 0.0105b | ||||||||
| Chromosome 5 | |||||||||||||
| rs249637 | NDFIP1 | G | 0.074 | − | 0.0216b | ||||||||
| rs37022 | SLC6A3 | T | 0.831 | + | 0.0052a | ||||||||
| rs250682 | SLC6A3 | C | 0.798 | + | 0.0082b | ||||||||
| rs6596271 | SLC25A48 | G | 0.042 | − | 0.0052b | ||||||||
| rs164080 | GNPDA1 | C | 0.526 | + | 0.0015b | ||||||||
| Chromosome 11 | |||||||||||||
| rs10891551 | DRD2 | A | 0.122 | + | 0.0253b | ||||||||
| rs719804 | TTC12 | G | 0.223 | − | 0.0117b | ||||||||
| rs4938006 | NCAM1/TTC12 | G | 0.109 | + | 0.034b | 0.0106a | 0.0359a,c | ||||||
| rs12799083 | DRD2 | C | 0.031 | − | 0.0107b | ||||||||
| rs4245148 | DRD2 | T | 0.122 | + | 0.0251b | ||||||||
| rs17596017 | NCAM1 | T | 0.024 | − | 0.0191b | ||||||||
| rs1381246 | BSX | C | 0.451 | − | 0.0057b | ||||||||
| rs652285 | NCAM1 | T | 0.057 | + | 0.0097b | 0.0082b | 0.0214b | ||||||
| rs675646 | NCAM1 | C | 0.071 | + | 0.0107b | 0.0807b | 0.0105a | ||||||
| rs649568 | NCAM1 | T | 0.044 | + | 0.0171b | 0.0162b | |||||||
| rs702966 | PHRF1 | C | 0.276 | − | 0.0121b | ||||||||
| rs2574829 | NCAM1 | G | 0.394 | + | 0.0084b | ||||||||
| rs11214521 | NCAM1 | G | 0.035 | − | 0.0226 | ||||||||
| rs11214505 | NCAM1 | G | 0.082 | − | 0.0292 | ||||||||
| rs1055076 | TTC12 | A | 0.175 | + | 0.0343 | ||||||||
| rs12222469 | NCAM1 | A | 0.035 | + | 0.0183b | ||||||||
| Chromosome 12 | |||||||||||||
| rs17722134 | TPH2 | G | 0.029 | − | 0.0085b | ||||||||
| Chromosome 17 | |||||||||||||
| rs7208257 | ARRB2 | C | 0.055 | + | 0.0164b | ||||||||
Abbreviations: A/D, anxiety and depression; CD, conduct disorder; H/I, hyperactivity/impulsivity; ODD, oppositional defiant disorder; VAS-P, Vanderbilt Assessment Scale for Parents.
Nonsignificant P-values after B=10,000 permutations are shown in italic.
No correction for the number of questions interrogated was performed in genetic association analyses.
Permutated P-value <0.01.
Permutated P-value <0.05.
Effect on the severity towards opposite direction to the one described in the ‘effect' column.
In the inattention domain, a total of eight markers were found to be associated with either the LCCA-derived severity status or the VAS-P score. When considering the VAS-P score, significant associated markers were identified within LPHN3 (4q), NDFIP1 (5q), and DRD2 (11q) and TTC12 (11q); for the severity scale, all significantly associated markers are within NCAM1 (11q).
For hyperactivity/impulsivity, two markers within SLC6A3 (5p) were found to be associated with the VAS-P score and three markers within NCAM1 were associated with the severity of symptoms.
For ODD, four markers were found to be associated with the VAS-P score and four with the severity of symptoms. For the former, three of the four markers are within LPHN3 and one is within NCAM1. For the severity of symptoms, all significant markers are within LPHN3.
For CD, six markers were found to be associated with the VAS-P score, three within LPHN3 and three within NCAM1.
In the A/D domain, a total of eight markers were found to be associated with the VAS-P score and six with the severity of symptoms. Among the markers found to be associated with the VAS-P score, four are within LPHN3, two are within NCAM1, one is within GNPDA1 and one is within TPH2 (12q). Regarding severity of symptoms, a total of six markers were found to be associated, four within NCAM1, one within GNPDA1, and one at an intergenic region between NCAM1 and TTC12 on chromosome 11.
LPHN3 × 11q interaction
By domain, a total of 567 unique pairs of markers located at different chromosomes were tested for SNP × SNP interactions. After breaking up the family data using the strategy previously mentioned, the number of severe (cases)/not severe (controls) pairs of individuals available for analysis was 184 for the inattention domain, 169 for hyperactivity/impulsivity, 104 for ODD and 76 for A/D. Figure 2 presents the results of the Cochran–Mantel–Haenszel tests after correcting by multiple testing using the FDR. Regardless of the VAS-P domain being considered, statistically significant SNP × SNP interactions include markers within LPHN3 and within the region containing NCAM1, DRD2 and TTC12 genes (Table 2 and Figure 2).
Figure 2.
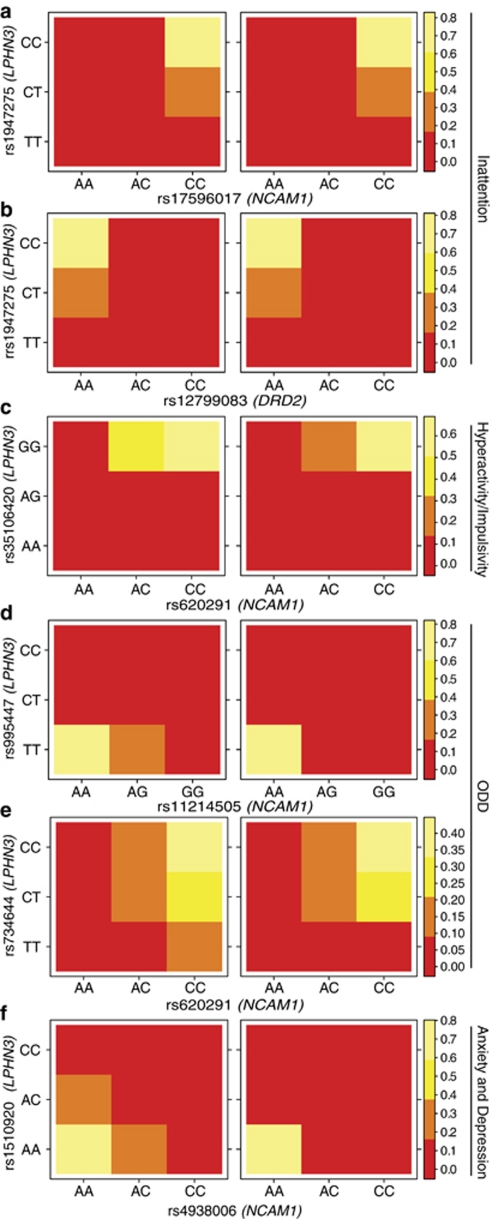
Genotypic frequency distribution for pairs of single nucleotide polymorphisms (SNPs) contributing to interaction effects and assessed by the Cochran–Mantel–Haenszel test. Significant interaction effects involve markers in the LPHN3 gene and a region in 11q that harbors the NCAM1 and DRD2 genes. Epistatic effects are depicted by changes in color that represent significant differences in genotypic distribution among severe (cases) vs not severe (controls) individuals. In there, the genotypes for one marker are held fixed whereas genotypes on the other marker vary. Inattention: (a) markers rs1947275 harbored in LPHN3 and rs17596017, in NCAM1, contribute to the severity of symptoms in this domain (M2=33.163, FDR-corrected P-value <0.001); (b) markers rs1947275 harbored in LPHN3 and rs12799083, in DRD2, produce a significant interacting effect contributing to the severity of symptoms (M2=28.456, FDR-corrected P-value <0.005). H/I: (c) markers rs35106420, in LPHN3, and rs620291, in NCAM1, produce an interacting effect contributing to the severity of symptoms (M2=20.497, FDR-corrected* P-value <0.05). ODD: (d) markers rs995447, in LPHN3, and rs11214505, in NCAM1, interact to modify the severity of symptoms (M2=41.379, FDR-corrected P-value <0.0001); (e) rs734644, in LPHN3, and rs4938006 produce an epistatic effect contributing to the severity of symptoms (M2=26.795, FDR-corrected P-value <0.01). A/D: (f) markers rs1510920, in LPHN3, and rs4938006 localized closest to NCAM1 in an intragenic region of chromosome 11q, interact to modify the severity of symptoms (M2=41.379, FDR-corrected P-value <0.0001). *FDR-corrected P-values calculated on the basis of 567 independent tests, corresponding to the maximum number of SNP pairs for each of the four domains of the VAS-P questionnaire from which the severity of symptoms was derived. Abbreviations as in Figure 1.
For the inattention domain, the interaction between rs1947275 and rs17596017 was found to be statistically significant (M2=33.163, PFDR−corrected=0.00067). In this case, comparison of the genotypes distribution for these markers between cases and controls showed that decreasing the number of copies of the T allele in LPHN3 (from CT to CC in rs1947275) yields a reduction of the number double homozygous individuals for the C allele (marker rs17596017) (Figure 2).
For the hyperactivity/impulsivity domain, in which the interaction between rs35106420 and rs620291 was significant (M2=20.497, PFDR−corrected=0.02766), similar results were obtained. For instance, when the G allele is present in LPHN3 and the allele C in NCAM1 (while keeping the other fixed), the number of cases with a particular genotype decreases when compared with the controls (Figure 2).
For ODD, two interactions were found to be statistically significant, one defined by the markers rs995447 and rs11214505 (M2=41.379, PFDR−corrected=0.00008), and one by the markers rs734644 and rs620291 (M2=26.795, PFDR−corrected=0.00196). In the former, the presence of one copy of the G allele in NCAM1 yields a reduction in the number of cases with two copies of the C allele in LPHN3 and an increment of those with only one copy of it. In the latter, regardless the number of copies of the T allele in LPHN3, the number of cases with one or more copies of the C allele in NCAM1 is greater than the number of controls (Figure 2).
In the A/D domain, only the interaction between rs1510920 and rs4938006 was statistically significant (M2=23.973, PFDR−corrected=0.00655). For instance, when the one copy of the C allele is present in NCAM1, it yields a reduction in the number of cases with one or more copies of the A allele in LPHN3 (Figure 2).
Discussion
We found that several genetic markers interact to increase the severity of ADHD symptoms in our population. These markers, distributed along five chromosomal regions that have previously been linked to ADHD in a genetic isolate from Colombia,25 are harbored in genes that have a potential role in the neurobiology of ADHD. In our sample, two genetic interactions, LPHN3 × NCAM1 and LPHN3 × DRD2, modified the severity of symptoms in ADHD. The former modifies the inattention, hyperactivity/impulsivity, ODD and A/D symptoms in ADHD, the latter acts on the inattention domain only.
Severity was estimated in our sample by two methods, one using the overall VAS-P score, and the second quantifying the magnitude of inattention, hyperactivity/impulsivity, CD, ODD and A/D symptoms using LCC. By using this methodology, we could include in the analysis all the clinical information from the individuals regardless of their categorically diagnosed status. This approach may have a more direct translation to the clinical context, where severity of symptoms is a major predictor of functional outcome.
Of special interest in our findings is the association and interactions found in regions that harbor genes previously described as being associated with ADHD, such as LPHN312 and DRD2,20 as well as the novel interaction between LPHN3 and NCAM1 recently described by our group.14 These genes are expressed in areas of the brain that have highly related roles, which makes them relevant candidates to explain the type and severity of ADHD symptoms.
LPHN3 is a novel finding in association with ADHD. Our group has recently described the association between a specific LPHN3 haplotype and the increased risk of ADHD, particularly related to a higher risk of behavioral problems such as conduct disorder and substance use disorder.12 In this study, we found that the interaction of three genetic areas highly related to neurodevelopmental aspects as well as function of the reward system predicts severity of ADHD symptoms. This is a very interesting observation that deserves further replication in a bigger sample.
The region located in chromosome 11q23 includes the NCAM1, TTC12, ANKK1 and DRD2 genes, all of which are functionally linked to dopamine in the brain. Many association studies of DRD2 and substance dependence, including alcohol dependence and drug dependence have been reported.32, 33
NCAM1 is expressed in all cell types in the nervous system, where it promotes neuron–neuron and neuron–glia adhesion. It is also involved in signal transduction and critical developmental events such as migration and proliferation, neurite outgrowth and fasciculation, synaptogenesis and synaptic plasticity.34, 35 It has important roles in the development of the pre-frontal cortex19 as well as in the regulation of dopaminergic pathways15 and response to stress.18
Our previous results show that the association of the marker rs65511665 (harbored in the LPHN3 gene) is stronger in families from the Paisa population, a genetic isolate in Colombia that has a high prevalence of ADHD and a subtype highly co-morbid with disruptive behaviors.12, 36 Furthermore, the same marker is associated with response to stimulant medication12, 36 and, more recently, an interaction between this marker and an haplotype in 11q increases the risk of developing ADHD ∼2.5-fold.14 The present study expands previous work by adding an additional clinically relevant distinction. The role of NCAM1 in ADHD has not been extensively evaluated. However, according to our current findings, this gene and the genetic markers in its vicinity deserve further evaluation in the context of the implications that it may have in the clinical expression of ADHD symptoms.
Acknowledgments
We would like to extend our deepest gratitude to all patients and families who took part in our research on ADHD. We thank Dr Benjamin D Solomon for critical reading and input to this manuscript. This research was supported by the Division of Intramural Research, National Human Genome Research Institute, National Institutes of Health, and Department of Health and Human Services, USA. MLB was supported by Ministerio de Educación, Chile, Grant MECESUP 0608. Julia Fekecs provided graphical assistance with the figures.
The authors declare no conflict of interest.
References
- Visser S, Bitsko R, Danielson M, Perou R. Increasing prevalence of parent-reported attention-deficit/hyperactivity disorder among children --- United States, 2003 and 2007. Mortal Morb Wkly Rep. 2010;59:1439–1443. [PubMed] [Google Scholar]
- American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders4th edn, text revision ed.American psychiatric Association: Washington, DC; 2000 [Google Scholar]
- Danckaerts M, Sonuga-Barke EJ, Banaschewski T, Buitelaar J, Dopfner M, Hollis C, et al. The quality of life of children with attention deficit/hyperactivity disorder: a systematic review. Eur Child Adolesc Psychiatry. 2010;19:83–105. doi: 10.1007/s00787-009-0046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry T, Lyman R, Klinger L. Academic underachievement and attention-deficit/hyperactivity disorder: the negative impact of symptom severity on school performance. J Sch Psychol. 2002;40:259–283. [Google Scholar]
- Riley AW, Spiel G, Coghill D, Dopfner M, Falissard B, Lorenzo MJ, et al. Factors related to health-related quality of life (HRQoL) among children with ADHD in Europe at entry into treatment. Eur Child Adolesc Psychiatry. 2006;15 (Suppl 1:I38–I45. doi: 10.1007/s00787-006-1006-9. [DOI] [PubMed] [Google Scholar]
- Upadhyaya HP, Carpenter MJ. Is attention deficit hyperactivity disorder (ADHD) symptom severity associated with tobacco use. Am J Addict. 2008;17:195–198. doi: 10.1080/10550490802021937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norvilitis JM, Sun L, Zhang J. ADHD symptomatology and adjustment to college in China and the United States. J Learn Disabil. 2010;43:86–94. doi: 10.1177/0022219409345012. [DOI] [PubMed] [Google Scholar]
- Acosta MT, Castellanos FX, Bolton KL, Balog JZ, Eagen P, Nee L, et al. Latent class subtyping of attention-deficit/hyperactivity disorder and comorbid conditions. J Am Acad Child Adolesc Psychiatry. 2008;47:797–807. doi: 10.1097/CHI.0b013e318173f70b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd RD, Sitdhiraksa N, Reich W, Ji TH, Joyner CA, Heath AC, et al. Discrimination of DSM-IV and latent class attention-deficit/hyperactivity disorder subtypes by educational and cognitive performance in a population-based sample of child and adolescent twins. J Am Acad Child Adolesc Psychiatry. 2002;41:820–828. doi: 10.1097/00004583-200207000-00014. [DOI] [PubMed] [Google Scholar]
- Volk HE, Henderson C, Neuman RJ, Todd RD. Validation of population-based ADHD subtypes and identification of three clinically impaired subtypes. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:312–318. doi: 10.1002/ajmg.b.30299. [DOI] [PubMed] [Google Scholar]
- Acosta MT, Arcos-Burgos M, Muenke M. Attention deficit/hyperactivity disorder (ADHD): complex phenotype, simple genotype. Genet Med. 2004;6:1–15. doi: 10.1097/01.gim.0000110413.07490.0b. [DOI] [PubMed] [Google Scholar]
- Arcos-Burgos M, Jain M, Acosta MT, Shively S, Stanescu H, Wallis D, et al. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol Psychiatry. 2010;15:1053–1066. doi: 10.1038/mp.2010.6. [DOI] [PubMed] [Google Scholar]
- Ribases M, Ramos-Quiroga JA, Sanchez-Mora C, Bosch R, Richarte V, Palomar G, et al. Contribution of LPHN3 to the genetic susceptibility to ADHD in adulthood: a replication study. Genes Brain Behav. 2010;10:149–157. doi: 10.1111/j.1601-183X.2010.00649.x. [DOI] [PubMed] [Google Scholar]
- Jain M, Vélez J, Acosta M, Balog J, Roessler E, Palacio L, et al. A cooperative interaction between LPHN3 and 11q doubles the risk for ADHD Mol Psychiatry 2011. May 24 (e-pub ahead of print). [DOI] [PMC free article] [PubMed]
- Xiao MF, Xu JC, Tereshchenko Y, Novak D, Schachner M, Kleene R. Neural cell adhesion molecule modulates dopaminergic signaling and behavior by regulating dopamine D2 receptor internalization. J Neurosci. 2009;29:14752–14763. doi: 10.1523/JNEUROSCI.4860-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucht M, Samochowiec A, Samochowiec J, Jasiewicz A, Grabe HJ, Geissler I, et al. Influence of DRD2 and ANKK1 genotypes on apomorphine-induced growth hormone (GH) response in alcohol-dependent patients. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:45–49. doi: 10.1016/j.pnpbp.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Schiff M, Weinhold B, Grothe C, Hildebrandt H. NCAM and polysialyltransferase profiles match dopaminergic marker gene expression but polysialic acid is dispensable for development of the midbrain dopamine system. J Neurochem. 2009;110:1661–1673. doi: 10.1111/j.1471-4159.2009.06267.x. [DOI] [PubMed] [Google Scholar]
- Bisaz R, Schachner M, Sandi C. Causal evidence for the involvement of the neural cell adhesion molecule, NCAM, in chronic stress-induced cognitive impairments. Hippocampus. 2011;21:56–71. doi: 10.1002/hipo.20723. [DOI] [PubMed] [Google Scholar]
- Cox ET, Brennaman LH, Gable KL, Hamer RM, Glantz LA, Lamantia AS, et al. Developmental regulation of neural cell adhesion molecule in human prefrontal cortex. Neuroscience. 2009;162:96–105. doi: 10.1016/j.neuroscience.2009.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollins SH, Anastopoulos AD, Lachiewicz AM, FitzGerald D, Morrissey-Kane E, Garrett ME, et al. SNPs in dopamine D2 receptor gene (DRD2) and norepinephrine transporter gene (NET) are associated with continuous performance task (CPT) phenotypes in ADHD children and their families. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1580–1588. doi: 10.1002/ajmg.b.30876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conners C, Erhardt D, Sparrow E. The Conners Adult ADHD Rating Scale (CAARS) Multi-Health Systems Inc.: Toronto; 1998. [Google Scholar]
- Wolraich ML, Lambert W, Doffing MA, Bickman L, Simmons T, Worley K. Psychometric properties of the Vanderbilt ADHD diagnostic parent rating scale in a referred population. J Pediatr Psychol. 2003;28:559–567. doi: 10.1093/jpepsy/jsg046. [DOI] [PubMed] [Google Scholar]
- Lindgren S, Koeppl G.Assessing child behavior problems in a medical setting: development of the Pediatric Behavior ScaleIn: Prinz R, (ed).Advances in Behavioral Assessment of Children and Families JAI Press: Greenwich, CT; 198757–90. [Google Scholar]
- Vermunt J, Magidson J.Latent class cluster analysisIn: Haagenars J, AL M, (eds).Applied Latent Class Analysis Cambridge University Press: Cambridge, UK; 2002 [Google Scholar]
- Arcos-Burgos M, Castellanos FX, Pineda D, Lopera F, Palacio JD, Palacio LG, et al. Attention-deficit/hyperactivity disorder in a population isolate: linkage to loci at 4q13.2, 5q33.3, 11q22, and 17p11. Am J Hum Genet. 2004;75:998–1014. doi: 10.1086/426154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna; 2010. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Hudziak JJ, Heath AC, Madden PF, Reich W, Bucholz KK, Slutske W, et al. Latent class and factor analysis of DSM-IV ADHD: a twin study of female adolescents. J Am Acad Child Adolesc Psychiatry. 1998;37:848–857. doi: 10.1097/00004583-199808000-00015. [DOI] [PubMed] [Google Scholar]
- Rohde LA, Barbosa G, Polanczyk G, Eizirik M, Rasmussen ER, Neuman RJ, et al. Factor and latent class analysis of DSM-IVADHD symptoms in a school sample of Brazilian adolescents. J Am Acad Child Adolesc Psychiatry. 2001;40:711–718. doi: 10.1097/00004583-200106000-00017. [DOI] [PubMed] [Google Scholar]
- Jain M, Palacio LG, Castellanos FX, Palacio JD, Pineda D, Restrepo MI, et al. Attention-deficit/hyperactivity disorder and comorbid disruptive behavior disorders: evidence of pleiotropy and new susceptibility loci. Biol Psychiatry. 2007;61:1329–1339. doi: 10.1016/j.biopsych.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Elia J, Arcos-Burgos M, Bolton KL, Ambrosini PJ, Berrettini W, Muenke M. ADHD latent class clusters: DSM-IV subtypes and comorbidity. Psychiatry Res. 2009;170:192–198. doi: 10.1016/j.psychres.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito-Smythers C, Spirito A, Rizzo C, McGeary JE, Knopik VS. Associations of the DRD2 TaqIA polymorphism with impulsivity and substance use: preliminary results from a clinical sample of adolescents. Pharmacol Biochem Behav. 2009;93:306–312. doi: 10.1016/j.pbb.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes EE, Brown SM, Kimak M, Ferrell RE, Manuck SB, Hariri AR. Genetic variation in components of dopamine neurotransmission impacts ventral striatal reactivity associated with impulsivity. Mol Psychiatry. 2009;14:60–70. doi: 10.1038/sj.mp.4002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochlamazashvili G, Senkov O, Grebenyuk S, Robinson C, Xiao MF, Stummeyer K, et al. Neural cell adhesion molecule-associated polysialic acid regulates synaptic plasticity and learning by restraining the signaling through GluN2B-containing NMDA receptors. J Neurosci. 2010;30:4171–4183. doi: 10.1523/JNEUROSCI.5806-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci. 2007;10:19–26. doi: 10.1038/nn1827. [DOI] [PubMed] [Google Scholar]
- Palacio JD, Castellanos FX, Pineda DA, Lopera F, Arcos-Burgos M, Quiroz YT, et al. Attention-deficit/hyperactivity disorder and comorbidities in 18 Paisa Colombian multigenerational families. J Am Acad Child Adolesc Psychiatry. 2004;43:1506–1515. doi: 10.1097/01.chi.0000142279.79805.dc. [DOI] [PubMed] [Google Scholar]