Abstract
Neurexin 1 (NRXN1) is a large presynaptic transmembrane protein that has complex and variable patterns of expression in the brain. Sequence variants in NRXN1 are associated with differences in cognition, and with schizophrenia and autism. The murine Nrxn1 gene is also highly polymorphic and is associated with significant variation in expression that is under strong genetic control. Here, we use co-expression analysis, high coverage genomic sequence, and expression quantitative trait locus (eQTL) mapping to study the regulation of this gene in the brain. We profiled a family of 72 isogenic progeny strains of a cross between C57BL/6J and DBA/2J (the BXD family) using exon arrays and massively parallel RNA sequencing. Expression of most Nrxn1 exons have high genetic correlation (r>0.6) because of the segregation of a common trans eQTL on chromosome (Chr) 8 and a common cis eQTL on Chr 17. These two loci are also linked to murine phenotypes relevant to schizophrenia and to a novel human schizophrenia candidate gene with high neuronal expression (Pleckstrin and Sec7 domain containing 3). In both human and mice, NRXN1 is co-expressed with numerous synaptic and cell signaling genes, and known schizophrenia candidates. Cross-species co-expression and protein interaction network analyses identified glycogen synthase kinase 3 beta (GSK3B) as one of the most consistent and conserved covariates of NRXN1. By using the Molecular Genetics of Schizophrenia data set, we were able to test and confirm that markers in NRXN1 and GSK3B have epistatic interactions in human populations that can jointly modulate risk of schizophrenia.
Keywords: expression QTL, genetic epistasis, GSK3B, neurexin, schizophrenia
Introduction
Neurexins (NRXNs) are neuronal cell adhesion proteins inserted mainly into the presynaptic membrane. They serve as bridging molecules that directly bind the presynaptic neurotransmitter machinery with postsynaptic transmembrane proteins such as the neuroligins and neurotransmitter receptors.1, 2, 3, 4, 5, 6 The NRXNs have an important role in regulation of presynaptic Ca2+ channels and neurotransmitter release, and are also involved in the modulation of postsynaptic NMDA and γ-aminobutyric acid (A) receptor activity.6, 7, 8, 9
There are three members in the NRXN gene family—Nrxn1, Nrxn2 and Nrxn3. They are among the largest of mammalian genes, Nrxn1 being close to 1.1 Mb and Nrxn2 about 1.7 Mb, and all are highly conserved across vertebrates.10, 11 A fascinating aspect of the NRXNs is their enormously complex and extensive alternative splicing that can potentially produce about a thousand isoforms.12 Each NRXN gene has two promoters and transcribes a long version (α-NRXN) and a short version (β-NRXN). Multiple isoforms of both long and short variants are produced as a result of alternative splicing from five splicing regions within each gene.10, 11 Deletion of the larger α-NRXNs leads to marked deficits in synaptic function.13, 14
Among the NRXN genes, NRXN1 has been implicated in neurodevelopmental and psychiatric disorders. Genome-wide association studies have consistently found links between copy number variation (primarily deletions) in NRXN1 and autism and schizophrenia.15, 16, 17, 18, 19 Associations have also been found between sequence variants in NRXN1 and nicotine addiction and alcoholism.20, 21, 22 In mice, deletion of Nrxn1α causes a decrease in excitatory synaptic transmission in the hippocampus, and behavioral changes including enhancement of motor learning and decreased prepulse inhibition (PPI) to acoustic startle—an endophenotype that models sensory-gating deficits in schizophrenia.23
These studies in human and mice have established an important role for NRXN1 in the etiology of neuropsychiatric diseases. However, disorders such as schizophrenia and autism are inherently polygenic and emerge from the interplay of multiple sequence variants and environmental factors. A high-priority candidate such as NRXN1 is bound to operate within the context of a wider network of genes and is likely to involve non-linear and epistatic interactions that contribute to disease risk. Despite the importance of epistasis (gene–gene interactions) in disease process, current large-scale genetic studies rarely go beyond linear additive models.24 Those that do apply a network approach to genome-wide association studies25, 26, 27, 28 rely on known protein interactions and pathways, and are essentially constrained by prior knowledge.
Here, we ask whether we can use gene co-expression analysis in the mouse to define and refine genetic and molecular networks for Nrxn1 that provide insights into neuronal processes and possible disease mechanisms. To what extent can we translate from mouse to human at the level of genetic networks? Specifically, can data from a large family of mouse strains reveal novel partners for Nrxn1 that are relevant to human disease? The Nrxn1 locus is highly polymorphic in the BXD family and contains several single-nucleotide polymorphisms (SNPs) and insertions and deletions. Applying a reverse complex trait approach, we have treated these natural sequence variants and corresponding variations in transcript abundance as a starting point in the dissection of downstream phenotypes.29 We used expression QTL (eQTL) analysis to evaluate the genetic regulation of Nrxn1 and to build an interaction network around Nrxn1 that includes other genes and also key behavioral phenotypes. Finally, we used human data extracted from the Molecular Genetics of Schizophrenia (MGS) study30 to evaluate the translational relevance of the murine analysis and to test a specific hypothesis about an epistatic interaction between NRXN1 and glycogen synthase kinase 3 beta (GSK3B).
Materials and methods
Mouse strains and resources
The BXD inbred strains were derived by crossing the parental strains C57BL/6J (B6) and DBA/2J (D2) followed by >20 generations of inbreeding.31, 32, 33 The parental lines differ at approximately 4.8 million SNPs and at another 500 000 insertions and deletions and copy number variations and the family as a whole is highly polymorphic.34 The BXDs are a densely phenotyped and genotyped family and have been used as a genetic reference panel for over 30 years.35 A compendium of over 2000 phenotypes and over 45 microarray data sets for these strains is available from GeneNetwork (http://www.genenetwork.org).
For this study, transcript measurements were taken from the hippocampi of 70 BXD strains, the parental B6 and D2, and reciprocal F1 hybrids (B6D2F1 and D2B6F1). Details on the strain, sex and age of each animal can be accessed from http://www.genenetwork.org/dbdoc/UMUTAffyExon_0209_RMA.html. The mice were bred at the University of Tennessee Health Science Center or at the Jackson Laboratory, Bar Harbor, ME, USA. Hippocampal dissections were carried out at UTHSC or at the Beth Israel Deaconess Medical Center by our colleague, Dr Glenn D Rosen (details on dissection and tissue processing are in Supplementary methods). All animal procedures were approved by the Animal Care and Use Committees at UTHSC and BIDMC.
Microarray data processing
To provide a comprehensive analysis of natural variation in gene expression in the mouse hippocampus, we have previously generated transcriptome profiles using conventional Affymetrix arrays for ∼100 strains of mice.36 In this study, we used exon arrays (Affymetrix Exon 1.0 ST, Affymetrix, http://www.affymetrix.com) to provide a more detailed but still global analysis of the genetic control of alternative splicing in the hippocampus. The Affymetrix Exon 1.0 ST arrays contain a total of 1.2 million probe sets and uses at least four probes to interrogate an exon. Probe level intensity values were extracted from the CEL files using the GeneChip Operating Software provided by Affymetrix. Probe set level summarization and data normalization was done within GeneChip Operating Software using the Robust Multichip Averaging method. The expression values were then log-transformed and stabilized to a mean of 8 and an s.d. of 2 across the samples. The entire data set (BXD hippocampus mRNA UMUTAffy Hippocampus Exon (February 2009) RMA) can be downloaded from GeneNetwork data sharing site using the accession number GN206 (http://www.genenetwork.org/share/data/).
Array data analysis on GeneNetwork
QTL mapping was done for the probe set level data using QTL Reaper.37, 38 The results are presented as likelihood ratio statistic (LRS) scores (LOD score=LRS/4.61). We used 3795 informative microsatellite and SNP markers in the BXDs for interval mapping. The BXD genotypes can be downloaded from http://www.genenetwork.org/share/data/. Up to a million permutations were performed to compute the genome-wide suggestive and significant thresholds. Nrxn1 is located on chromosome (Chr) 17 between 90.432 and 91.492 Mb (mm9, build 37) on the minus strand and is targeted by 172 probe sets. For the probe sets that targeted exons in Nrxn1 and mapped as cis eQTL, we screened the probe sequences for overlapping SNPs that may confound the QTL analysis. Although Nrxn1 is SNP rich, most exons are devoid of SNPs. With the exception of exons 5 and 17 (each harbors a synonymous mutation), the exon targeting probe sets do not overlap SNPs and the consistent cis effect is reliable.
For the co-expression analysis in the mouse hippocampus, pair-wise Pearson product moment correlations were computed for all probe sets based on expression variation across the 74 lines of mice. The top 1000 probe sets correlated with Nrxn1 were filtered by expression level. Only probe sets with expression >8 on a log2 scale, and well-annotated probe sets that map to known genes were retained for subsequent analysis. Co-expression results are presented for a representative probe set (Affy 4930721) targeting a middle exon (exon 12) that has high expression, is associated with both eQTLs on Chrs 17 and 8 (see Results section), and is well correlated with other Nrxn1 exons. Analyses using other exons from the same correlated block of exons give similar results. We also used a publicly available human forebrain expression data (neocortex and hippocampus) generated using the Affymetrix U133 Plus 2.0 arrays (GEO accession number GSE5281).39, 40 This data set contains samples from Alzheimer's disease cohort and age-matched control subjects. We computed Pearson product correlations across 71 samples from normal subjects and retrieved the top 1000 covariates of NRXN1 from the pool of over 54 000 probe sets.
RNA sequencing
Total RNA was isolated from the hippocampus of adult female B6 and D2 mice. Ribosomal RNA was depleted using the RiboMinus eukaryotic kit (http://www.invitrogen.com). We prepared multiplexed bar-coded fragment libraries using the SOLiD Fragment Library Barcoding kits. Emulsion PCR was then done and libraries were amplified and sequenced on the ABI SOLiD 3 plus system (http://www.appliedbiosystems.com).
We generated a total of 77 million and 80 million 50 nucleotide RNA sequencing (RNA-seq) tags for B6 and D2, respectively. Of these, approximately 58 million and 63 million reads aligned to the mouse reference genome (NCBI37/mm9) using the ABI Bioscope software (http://www.solidsoftwaretools.com). The alignment results (BAM files) were uploaded to the Partek Genomic Suite (http://www.partek.com) and GeneNetwork's UCSC Genome Browser mirror (http://ucscbrowser.genenetwork.org/) for visualization. Number of sequence tags was counted for each exon feature within Nrxn1. The entire data can be downloaded from GeneNetwork's high-throughput sequence data sharing site at http://galaxy.genenetwork.org:8080/library. Additionally, read alignments can also be visualized on the GeneNetwork Genome Browser.
QTL analysis for behavioral traits
To identify behavioral phenotypes that may be downstream of Nrxn1 we used a point-wise P-value of 0.01. We selected phenotypes that correlated with marker rs6313030 (located on Chr 17 at 90.460122 Mb within the Nrxn1 gene) and then analyzed behavioral traits assayed in the BXDs using marker regression. The behavioral phenotypes with QTLs on distal Chr 17 are (1) prepulse inhibition, assayed by McCaughran et al.41 in a panel of 21 BXD strains (trait ID on Genenetwork is 10396), (2) anxiety trait measure by time spent in open quadrant of zero-maze, assayed in a larger panel of 57 BXD strains42 (trait ID 11696) and (3) handling induced convulsion as an index of ethanol withdrawal severity, measured in 25 BXD strains43 (trait ID 10065).
Gene–gene interaction analysis
To explore epistatic associations between NRXN1 and GSK3B, we first extracted all markers in NRXN1 and GSK3B and in the 20 kb flanking regions from the MGS data set.30 GAIN and non-GAIN data sets30 were combined and markers common to both were used. Association tests and minor allele frequency (MAF) were calculated using PLINK 2.05044 and markers with minor allele frequencies ⩾0.05 were selected (384 in NRXN1 and 17 in GSK3B). Sporadic missing genotypes in these markers were imputed by MDRdt Beta V0.4.3 (http://www.epistasis.org). Multifactor dimensionality reduction (MDR) method was used to conduct gene–gene interaction analyses (MDR 2.0 beta 8.1). The details of MDR are described elsewhere.24, 45, 46 We used two methods to further filter the markers. One is the odds ratio filter implemented in the MDR software, which uses the case control ratio in the data set to define high-risk and low-risk genotype combinations.46, 47, 48 Using this approach, the top five markers from each gene were selected. The other filter was based on single marker association P-values in which tagged SNPs49 with association P⩽0.15 in both genes were selected (a total of 13 markers in the two genes). SNPs selected by both methods were used for MDR analyses and best models were generated based on a 10-fold cross-validation consistency. In this study, we limited the search to a 2-loci and 3-loci interaction model using SNPs from the first and second filters, respectively. The significance of the best model was evaluated by 10 000 permutations (MDRpt 1.0 beta 2).50
Additional bioinformatics tools
Gene ontology and pathway enrichment analysis was done using DAVID 6.7 (http://david.abcc.ncifcrf.gov).51, 52 Protein–protein network analysis was done using the protein interaction database and network tool available on GeneMania (http://www.genemania.org).53 The protein network was based on known physical interactions curated from 114 sources, and the network was weighted by biological process.
Results
Genetic regulation of Nrxn1 exons
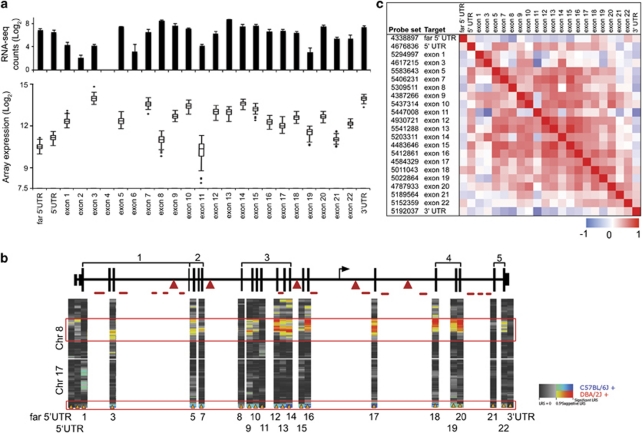
Nrxn1 has widespread expression in the brain. We used microarrays to measure its transcript at exon level resolution in the hippocampus of 74 members of the BXD family. Additionally, we measured expression in parental strains using RNA-seq. The very deep coverage provided by the arrays and RNA-seq uncovered significant inter-exon variability in Nrxn1 that may reflect alternative isoforms (Figure 1a). For example, low expressing exons (exons 2, 6, 11 and 19) are located within known splice sites10, 11 and the RNA-seq also showed a potentially skipped exon (exon 4 of Refseq gene NM_020252.3) (Figure 1b). To evaluate how tightly expression of exons is coupled with one another, we generated a correlation matrix of the exons and UTRs based on expression variation across the BXD family (Figure 1c). For the most part, exons form highly correlated expression blocks indicating that they are regulated as a single unit. The few exceptions are exons 1, 3, 11, 21, 22 and the UTRs, which show poor correlation with most of the other exons and may be alternatively spliced.
Figure 1.
Expression of neurexin 1 (Nrxn1) exons in mouse hippocampus. (a) Exon-level expression of Nrxn1 was measured by direct sequencing of mRNA (upper bar chart; error bars are s.d.) and by using exon arrays (lower box plots). Expression values are on a log2 scale (y axis). There is significant variation in expression of exons among the BXDs. On average, expression varies 2.5-fold within exons. Based on RNA sequencing, exons 2 and 6 have low expression and exon 4 is skipped (there are no array probe sets for these). (b) The gene model depicted is based on RefSeq gene NM_020252.3. Location of the 5′ UTR for β-Nrxn1 is shown in intron 17 (arrow) and known splice sites are indicated above (1–5). Approximate locations of insertion and deletion (indel) variants that are larger than 10 bps are shown below (lines for deletions and triangles for insertions in D2). Expression QTLs on chromosomes (Chrs) 8 (trans) and 17 (cis) are shown as heat maps for each exon and UTR. Allele from the DBA/2J parent has the positive effect on Chr 8 (red) while allele from the C57BL/6J parent has the positive effect on Chr 17 (blue). (c) The correlation matrix displays pair-wise correlations between the UTRs and exons in the BXDs. The lower cells are Pearson product moment and upper cells are Spearman ranked correlations. Exons 5–20 form a tightly correlated expression block (exon 11 being the exception). Correlation between the distal and proximal parts of the gene is generally low.
Nrxn1 expression is also highly variable across the strains with exons varying from 1.5-fold to 10-fold in abundance (Figure 1a). On average, the exons show a 2.5-fold variation among the BXDs. To track down genetic sources of this variation, we carried out eQTL mapping using conventional linkage analysis tools and genotypes embedded in GeneNetwork. The UTRs and canonical exons of Nrxn1 are partly controlled by polymorphisms near the gene itself (so-called cis eQTL) and partly by trans-acting polymorphisms (Figure 1b). A tightly correlated set of exons map as a cis eQTL to distal Chr 17 within 0.5 Mb of Nrxn1 itself. These exons are also strongly co-modulated by a trans regulatory QTL on mid-Chr 8. Several exons that are putatively subjected to splicing (for example, exons 1, 8, 11 and 21) are not modulated by either the cis or Chr 8 trans loci. Additionally, there are trans eQTLs on other Chrs associated with a minor set of exons (Supplementary Figure S1). For example, exons 12, 16 and 19 have trans eQTLs in mid-Chr 2 and exons 1, 3, 11 and 22 have trans eQTLs on Chr 12. These findings indicate a level of isoform specificity and joint eQTL control that corresponds to the complex splicing signatures in the exon co-expression matrix.
Sequence polymorphism in Nrxn1 between C57BL/6J and DBA/2J
The maternal parent of the BXD strains, B6, was the first mouse genome to be sequenced and is used as the mouse reference genome. The paternal strain, D2, has recently been resequenced at high density using next-generation systems (brief description of the 100x resequencing of the D2 genome is provided in Supplementary methods).34 This now makes it possible to carefully review all sequence variants in and around Nrxn1 to help identify variants that are candidates for the cis-effect on expression. We found numerous sequence differences between the B6 (B) and D2 (D) alleles. There are over 2900 SNPs in Nrxn1. Of these, only nine are in the 3′ UTR, two are synonymous mutations in exons 5 and 17, and the remaining SNPs are in introns. In all, 15 SNPs in introns have potentially high impact based on overlap with predicted functional elements in conserved regions of introns54 (Supplementary Table S1). The introns also contain a large number of insertions and deletions (defined as insertion or deletion in D relative to B). There are 436 deletions and 340 insertions ranging in length from 1 to 2418 bp (Figure 1b and Supplementary Table S1). Although none of these overlap canonical exons, such structural variations can have significant effects on gene expression. We selected a 74-bp insertion and a 107-bp deletion located in intron 17, which harbors the alternative promoter for the β-variant10, 11 and confirmed the variants by PCR amplification (Supplementary Figure S2).
Genetic analysis of Nrxn1 co-expression network
To elucidate a co-expression network associated with the Nrxn1 transcript, we ranked the top 1000 correlates of Nrxn1 expression from the pool of 1.2 million probe sets. These represent transcripts that have coordinated expression with Nrxn1 and are potential partners in cellular functions. These top covariates (Pearson product r>∣0.6∣, P<3.7 × 10−9) correspond to 572 known genes (Supplementary Material B). Consistent with the synaptic involvement of Nrxn1, this set of genes is enriched in synapse-related transcripts and genes known to directly interact with Nrxn1 (for example, Cask and Nlgn1) (Supplementary Material B). More interestingly, there is also enrichment in genes that are part of the WNT, axon guidance, mitogen-activated protein kinase and ErbB signaling pathways (for example, Gsk3b, Akt1 and Nrg3) and this suggests that there is close interaction between Nrxn1 and downstream cellular signaling. We also used the Genetic Association Database (http://geneticassociationdb.nih.gov) and were able to extract a set of 20 schizophrenia candidate genes (Figure 2a) in our top 572 Nrxn1 covariates (Benjamini corrected enrichment P<0.001).
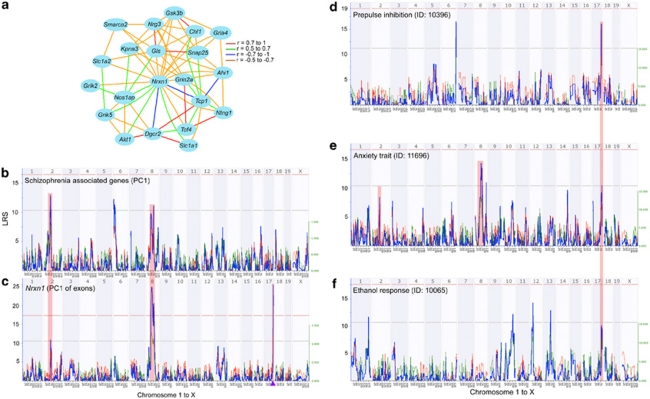
Figure 2.
Overlap of co-expression and behavioral QTLs. (a) An enriched set of twenty schizophrenia candidate genes was extracted from the covariates of neurexin 1 (Nrxn1) in the mouse hippocampus. These transcripts form a highly interconnected co-expression network with Nrxn1. (b) The co-expression network is modulated by expression QTLs (eQTLs) on chromosomes (Chrs) 2, 6 and 8. This is illustrated by the QTL map for the first principal component summarization (PC1) of the 20 schizophrenia candidates. The x axis represents Chrs 1 to X and the y axis shows the LRS score. The horizontal lines indicate the genome-wide suggestive and significant thresholds. (c) PC1 of the Nrxn1 exons has significant QTLs on Chrs 8 and 17, and suggestive QTL on Chr 2. Location of Nrxn1 in the cis eQTL is depicted by the triangle. Mouse behavioral traits that map close to the Nrxn1 locus at significant point-wise P-values (P<0.01) are prepulse inhibition (d), anxiety trait (e) and ethanol response (f). Overlapping QTL locations are highlighted.
eQTL mapping of the co-expression network members detected regulatory loci (that is, trans eQTL hotspots) on (1) mid-Chr 2 (67–77 Mb), (2) proximal Chr 6 (2–20 Mb) and (3) mid-Chr 8 (50–85 Mb). The first principal component (PC1) summarization of these highly correlated transcripts, also called an eigengene,55 retained this whole-genome eQTL pattern. This is illustrated by the eigengene for the tightly correlated schizophrenia network: the PC1 of the 20 transcripts accounts for ∼55% of the total variance and maps as suggestive QTLs to Chrs 2, 6 and 8 (Figure 2b). PC1 of the Nrxn1 exons (accounting for ∼50% of the total variance) has significant QTLs on Chrs 8 and 17, and suggestive QTL on Chr 2 (Figure 2c). Although exons 7, 8, 11 and 12 of Nrxn1 map as weak trans eQTL to proximal Chr 6 (Supplementary Figure S1), this association does not reach the suggestive threshold in the summarized map. Based on the comparison of eQTLs, Nrxn1 is not likely to be a QTL hotspot for the expression traits. Instead, the co-expression network is modulated mainly by eQTL hotspots on Chrs 2, 6 and 8. A portion of the variance in Nrxn1 comes from these trans-effects that connect Nrxn1 to the network.
Mouse behavioral phenotypes that map close to Nrxn1
We then asked whether mouse phenotypes that may be concordant with human traits are associated with the Nrxn1 locus. GeneNetwork maintains a database of over 2000 phenotypes collected from the BXDs. We specifically searched this database for behavioral traits that have linkage with markers near Nrxn1. We found three such traits that have significant point-wise QTLs on distal Chr 17. First, PPI to acoustic startle 41 mapped to distal Chr 17 with a point-wise P=0.0003 (LRS=16) (Figure 2d). Second, anxiety phenotype assayed on the zero-maze42 mapped to distal Chr 17 with point-wise P=0.005 (LRS=10.5). The anxiety trait also has striking match with the Nrxn1 exons in genome-wide QTL pattern with additional QTLs on mid-Chr 8 (LRS=14; point-wise P=0.0007) and Chr 2 (LRS of 8.4; point-wise P of 0.01) (Figure 2e). Finally, handling-induced convulsion as an index of alcohol withdrawal severity43 mapped to distal Chr 17 with a point-wise P of 0.007 (LRS=9.6) (Figure 2f).
We note that the behavioral traits also have QTLs that are distinct from the QTLs associated with Nrxn1 and its co-expression network. The associations between the QTLs and multi-level phenotypes presents a complex genetic structure and the effect of Nrxn1 on the behavior traits can either be directly mediated by the cis eQTL, or more likely, is transmitted via the co-expression network with major contributions by the set of trans eQTLs.
Cross-species analysis of the Nrxn1 covariates
We next examined if correlations at the mRNA level are preserved across species. Our aim was to identify strong and consistent correlations that are conserved in both mice and humans and that may be novel functional and genetic associates of Nrxn1. We used a human forebrain expression data set39, 40 and extracted the top 1000 covariates of NRXN1 (Pearson product r>∣0.55∣, P<3 × 10−7). This corresponds to 767 unique genes (Supplementary Material B). Of these 767 genes, only 42 overlap with the 572 co-expressed genes in mice and have conserved correlations with NRXN1 in human forebrain as well as in the mouse hippocampus (Supplementary Table S2). This includes GSK3B and NTNG1, which are both part of the Nrxn1 primed schizophrenia network derived from the mouse covariates.56, 57
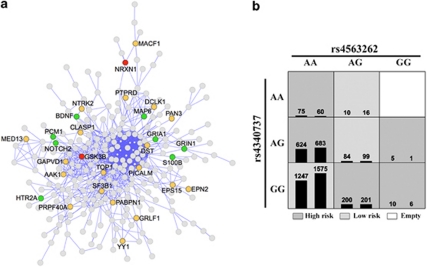
To explore protein interactions that may be encrypted in the mRNA co-expression set, we extracted a protein–protein interaction network from the list of 767 transcript covariates (Figure 3a). In all, 261 out of the 767 query genes (34%) formed a highly interconnected protein network that incorporated most of the conserved transcript correlates of NRXN1 including GSK3B, and also several other schizophrenia candidates. GSK3B has diverse roles in cellular signaling in the brain57, 58, 59 and we examined the expression covariance of GSK3B and NRXN1 in a number of different mouse and human brain regions and found generally strong correlations between the two (Supplementary Figure S3). The strong genetic correlation suggests interactions between NRXN1 and GSK3B and places NRXN1 as a synaptic gene that may also function as a gateway to cellular signaling cascades.
Figure 3.
Deriving protein–protein and genetic interactions from co-expressed genes. (a) The co-expression network for neurexin 1 (NRXN1) in the human brain was organized based on known protein–protein interactions. This resulted in a single large network that connected 261 out of 767 co-expressed transcripts. The network incorporates several known schizophrenia candidates (shaded green) and several conserved covariates of NRXN1, that is, are correlated with NRXN1 in both human and mouse (shaded orange). NRXN1 and glycogen synthase kinase 3 beta (GSK3B) are part of this protein network (shaded red). (b) Test for epistasis using the multifactor dimensionality reduction method found significant interaction between rs4563262 in NRXN1 and rs4340737 in GSK3B. Distribution of cases (left bar in cell) and controls (right bar in cell) stratified by multilocus genotype. Each multifactorial cell is labeled as ‘high risk' or ‘low risk'. Average balanced prediction accuracy for the model was 51.85%, permuted P=0.0123.
Test for epistasis between NRXN1 and GSK3B in human schizophrenia
We hypothesize that there is close functional relation between NRXN1 and GSK3B. We tested for gene–gene interactions in human using the MGS data set that has 2255 schizophrenia cases and 2641 controls.30 We found significant 2-loci interaction between rs4563262 (NRXN1) and rs4340737 (GSK3B) with a permuted P=0.012 (Figure 3b). Using a 3-loci model, there was significant interaction between rs6736816 and rs9309200 in NRXN1, and rs9826659 in GSK3B (permuted P=0.0373) (Table 1). No covariate sex effect was found. Among the top five SNPs in the two genes, we also found that rs4340737 in GSK3B and rs9309200 in NRXN1 are the only two SNPs that showed significant associations with schizophrenia (P=0.0171 for rs4340737 and P=0.0293 for rs9309200).
Table 1. Multilocus epistatic interaction models between NRXN1 and GSK3B.
| No. of loci | Best model | Testing balance accuracy (%) | Cross-validation consistency | 10 000 permutation P-value |
|---|---|---|---|---|
| 2 | rs4563262 (NRXN1) | 51.85 | 10/10 | 0.0123 |
| rs4340737 (GSK3B) | ||||
| 3 | rs6736816, rs9309200 (NRXN1) | 51.86 | 10/10 | 0.0373 |
| rs9826659 (GSK3B) |
Abbreviations: GSK3B, glycogen synthase kinase 3 beta; NRXN1, neurexin 1.
Evidence from mouse to prioritize potential candidates for schizophrenia
Our study demonstrates that gene co-expression analysis can effectively select related disease candidates and epistatic partners. We therefore used evidence from the co-expression analysis and mouse behavioral endophenotypes to further prioritize other potential candidates. For this, we considered genes in the MGS data set that were nominally associated with schizophrenia. The study by Shi et al.30 generated a list of 1202 SNPs in the European-American cohort and a list of 997 SNPs in the African-American cohort with association P<0.001. We mined the SNP lists for nearby genes with mouse homologues that are among the top covariates of Nrxn1 in the mouse hippocampus. Additional functional evidence was gathered from knockout studies and gene ontology. On the basis of this, we identified 30 genes that are nominally associated with schizophrenia and are co-expressed with Nrxn1. Several of these are already known candidates for neuropsychiatric disorders including schizophrenia, autism, attention deficit hyperactivity disorder (ADHD), and bipolar disorder (Table 2).60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73 Strikingly, 10 out of the 30 genes are also nominally associated with substance dependence and nicotine addiction.74, 75 In the mouse, deletion of six of these candidates (CTNND2, DMN, GSK3B, NRCAM, PLCB1 and RIMS1) lead to behavioral changes related to PPI, social and cognitive functions, hyperactivity, fear learning and anxiety-related traits.76, 77, 78, 79, 80, 81, 82, 83
Table 2. Covariates of Nrxn1 associated with schizophrenia in the MGS data set at P<0.001.
|
Association in human |
Mouse gene and syntenic region |
Mouse array probe ID |
Co-expression in mouse |
Mouse KO phenotype | Other human associations | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Locus | P-value | Gene | ID | Chr | Mb | r | P(r) | GO | |||
| European Americans | ||||||||||||
| rs10140896 | 14q31.3;Intr (intronic) | 9.4E–07 | EML5 | 319670 | 12 | 100.08 | 5399699 | 0.65 | 4.7E–11 | Microtubule cytoskeleton | ||
| rs7760476 | 6p22.3;Intr | 0.00002 | ATXN1 | 20238 | 13 | 45.66 | 4622958 | −0.64 | 1.4E–10 | Addiction; cognitive function; schizophrenia; | RNA processing | |
| rs2725045 | 8p23.2;Intr | 0.00004 | CSMD1 | 94109 | 8 | 17.50 | 5613572 | 0.61 | 2.0E–09 | Addiction | ||
| rs2192420 | 14q24.3;Upstr | 0.00006 | NRXN3 | 18191 | 12 | 90.90 | 5145873 | 0.68 | 1.7E–12 | Addiction; schizophrenia | Cell adhesion | |
| rs1386688 | 8p22;Intr | 0.00007 | PSD3 | 234353 | 8 | 70.22 | 5393349 | 0.65 | 4.6E–11 | Addiction | Cell signaling | |
| rs9341356 | 6q13;Intr | 0.00007 | RIMS1 | 116837 | 1 | 22.74 | 5163292 | 0.61 | 2.4E–09 | PPI; psychotomimetic drug response; social behavior | Cognitive performance | Neuro-transmission |
| rs231150 | 8q23.3;Dwnstr | 0.0001 | TRPS1 | 83925 | 15 | 50.61 | 4317283 | 0.61 | 2.1E–09 | Transcription factor | ||
| rs17171808 | 5q31.2;Intr | 0.0002 | JMJD1B | 277250 | 18 | 34.95 | 4508461 | 0.63 | 3.5E–10 | Nuclear oxidoreductase | ||
| rs4822743 | 22q12.1;Intr | 0.0002 | TPST2 | 22022 | 5 | 112.74 | 4312446 | −0.63 | 3.0E–10 | Sulfur metabolism | ||
| rs11097407 | 4q22.2;Intr | 0.0002 | SMARCAD1 | 13990 | 6 | 65.05 | 5186186 | 0.62 | 7.9E–10 | Chromatin modification | ||
| rs2530215 | 5p15.2;Intr | 0.0003 | CTNND2 | 18163 | 15 | 30.13 | 4694831 | 0.62 | 1.3E–09 | Fear learning; cognitive deficits | Addiction; schizophrenia | Cell adhesion |
| rs11223336 | 11q25;Intr | 0.0006 | OPCML | 330908 | 9 | 27.92 | 4747785 | 0.67 | 7.5E–12 | Addiction | Cell adhesion, opioid receptor | |
| rs12448737 | 16p13.2;Intr | 0.0007 | A2BP1 | 268859 | 16 | 6.81 | 4974807 | 0.62 | 1.5E–09 | Addiction; ADHD; bipolar schizoaffective; autism | RNA processing | |
| rs17286683 | 5q31.3;Upstr | 0.0008 | ANKHD1 | 108857 | 18 | 36.79 | 4412853 | 0.61 | 3.2E–09 | Translation regulation | ||
| rs4624596 | 3q13.33;Intr | 0.0009 | GSK3B | 56637 | 16 | 38.19 | 4314494 | 0.61 | 2.2E–09 | Impaired learning; behavioral despair | Bipolar; cognitive performance | ErbB, WNT signaling |
| rs4860425 | 4q13.1;Intr | 0.0009 | LPHN3 | 319387 | 5 | 82.12 | 5370748 | 0.66 | 1.8E–11 | ADHD | Neuropeptide signaling, cell adhesion | |
| rs1536142 | 1p32.2;Intr | 0.0009 | DAB1 | 13131 | 4 | 104.29 | 4375891 | −0.63 | 5.9E–10 | Addiction | Cell adhesion | |
| African Americans | ||||||||||||
| rs13065441 | 3q26.2;Intr | 0.00002 | TNIK | 665113 | 3 | 28.56 | 4843478 | 0.68 | 2.6E–12 | Addiction; schizophrenia | WNT, MAPK signaling | |
| rs7125438 | 11q25;Intr | 0.00002 | OPCML | 330908 | 9 | 27.92 | 4747785 | 0.67 | 7.5E–12 | Addiction | Cell adhesion, opioid receptor | |
| rs409797 | 7q31.1;Intr | 0.00008 | NRCAM | 319504 | 12 | 45.67 | 5569519 | 0.64 | 1.4E–10 | PPI; social, cognitive functions | Autism | Cell adhesion |
| rs6963574 | 7q32.3;Intr | 0.00008 | MKLN1 | 27418 | 6 | 31.44 | 4419118 | 0.71 | 8.4E–14 | Cytoskeletal response | ||
| rs596929 | 11q24.2;Intr | 0.0001 | KIRREL3 | 67703 | 9 | 34.32 | 5276457 | 0.61 | 2.9E–09 | ADHD; response to antipsychotics | Cell adhesion, synapse formation | |
| rs2826771 | 21q21.1;Intr | 0.0002 | NCAM2 | 17968 | 16 | 81.51 | 5268421 | 0.63 | 4.3E–10 | Psychiatric disorders | Cell adhesion | |
| rs1351569 | 8p23.2;Intr | 0.0002 | CSMD1 | 94109 | 8 | 17.50 | 5613572 | 0.61 | 2.0E–09 | Addiction | ||
| rs17446308 | 20p12.3;Intr | 0.0002 | PLCB1 | 18795 | 2 | 134.68 | 5117797 | 0.66 | 2.2E–11 | PPI; social behavior; hyperactivity; cognitive impairment | Cognitive performance | Phospholipid metabolism |
| rs11955233 | 5q14.1;Dwnstr | 0.0003 | JMY | 57748 | 13 | 94.22 | 5373932 | 0.64 | 1.1E–10 | Cytoskeleton, transcription regulation | ||
| rs2167566 | 2p15;Intr | 0.0004 | USP34 | 17847 | 11 | 23.27 | 5362770 | 0.62 | 1.5E–09 | Protein metabolism | ||
| rs1456729 | Xp21.1;Intr | 0.0005 | DMD | 13405 | X | 81.15 | 4416174 | 0.62 | 8.7E–10 | Anxiety related | Muscular dystrophy | |
| rs13250856 | 8q11.23;Intr | 0.0005 | RB1CC1 | 12421 | 1 | 6.24 | 4969100 | 0.64 | 1.2E–10 | |||
| rs12928607 | 16p13.2;Intr | 0.0006 | A2BP1 | 268859 | 16 | 6.81 | 4974807 | 0.62 | 1.5E–09 | Addiction; ADHD; bipolar schizoaffective; autism | RNA processing | |
| rs10249419 | 7q31.2;Intr | 0.0007 | CTTNBP2 | 30785 | 6 | 18.46 | 5198380 | −0.67 | 1.2E–11 | Cytoskeleton | ||
| rs2256900 | 10q23.1;Intr | 0.0009 | NRG3 | 18183 | 14 | 39.81 | 4390722 | 0.62 | 1.3E–09 | Addiction; schizophrenia | ErbB signaling | |
| rs6460537 | 7q11.22;Intr | 0.0009 | AUTS2 | 319974 | 5 | 132.93 | 5208797 | 0.65 | 5.6E–11 | Bipolar schizoaffective | ||
Abbreviations: GO, gene ontology; KO, knockout; MAPK, mitogen-activated protein kinase; NRXN1, neurexin 1; PPI, prepulse inhibition; SNP, single-nucleotide polymorphism.
Among the novel genes in Table 2, we highlight Pleckstrin and Sec7 domain containing 3 (PSD3) as both a potential new candidate for schizophrenia and a candidate for the trans-effect associated with the co-expressed transcripts in the mouse brain. This gene has high expression in the brain and is located in mid-Chr 8 within the trans eQTL for the Nrxn1 exons. Expression of Psd3 is associated with a significant cis eQTL in the BXDs and is also strongly correlated with Nrxn1 (Supplementary Figure S4).
Discussion
Synopsis
Nrxn1 is highly polymorphic and is associated with significant variation in expression among mice. We have exploited this variation to trace co-expression networks and eQTLs that link Nrxn1 with other genes and also with higher order behavioral phenotypes. GSK3B is among the strongest and most consistent covariates of NRXN1 in both mice and humans and we detected a significant epistatic interaction between these genes that modulates schizophrenia risk. This cross-species analysis also highlights novel candidates for schizophrenia in human, especially PSD3.
Sub-transcript level analysis of gene expression regulation
For a fine-scale analysis of Nrxn1, we have treated each exon, intron and the UTRs as discrete expression units that can be individually subjected to genetic analysis. This revealed some level of complexity in transcript regulation. For instance, exons 8, 11 and 21 show low correlation with other exons and are also associated with distinct eQTLs. Aside from these exceptions, the majority of exons have tightly correlated expression, partly due to shared modulation by two prominent eQTLs—one a cis eQTL at Nrxn1, and the other, a trans eQTL on mid-Chr 8.
The mid-Chr 8 trans eQTL also modulates several key genes correlated with Nrxn1 including Psd3, Gsk3b, Snap25 and Kpna3. This interval corresponds to human 8p21, a region that contains many genes linked to brain developmental and psychiatric disorders.84 Polymorphisms in this region in humans may also have trans-regulatory effect on the expression of other schizophrenia susceptibility genes.85 In a striking concordance between mouse and human, this part of mouse Chr 8 is also a QTL hot spot for brain structural traits in the BXDs and is linked to variations in the volume of the amygdala, hippocampus, cerebellum and striatum.86, 87 Located in this mid-Chr 8 QTL is Psd3, a gene that is modulated by a cis eQTL and is also co-expressed with Nrxn1. In the human MGS data set, SNPs near PSD3 are nominally associated with schizophrenia (P=0.00007). Although little is currently known about the function of PSD3, the converging evidence from multiple sources makes this gene a high-priority candidate for the eQTL in mouse and a novel candidate for schizophrenia in human.
Variation in Nrxn1 expression may influence schizophrenia endophenotypes
In the BXDs, PPI to acoustic startle maps to distal Chr 17 close to the Nrxn1 locus. PPI is a classic endophenotype that models sensory gating deficits in schizophrenia patients,88 and polymorphisms in Nrxn1 are strong candidates with compelling face validity. Consistent with association to schizophrenia, deletion of Nrxn1α in mice results in decreased PPI.23 However, in this knockout, social behavior and anxiety traits are unaffected .23
Unlike studies using gene knockouts, we have evaluated naturally occurring variants that will generally have more subtle effects on phenotypes. Most of the SNPs and insertions and deletions in Nrxn1 are intronic and are unlikely to cause gene disruption. However, these more moderate variants model the type of variation seen in human populations. A detailed analysis of structural variations in the NRXN genes in human has found a range of deletions and duplications.89 The deletions in NRXN1, particularly those that span exons, are strongly associated with schizophrenia. Additionally, there are possible associations with other psychiatric conditions and alcoholism that indicate pleiotropic effects of NRXN1.89 This is consistent with our findings in mouse in which QTLs for ethanol response and anxiety map near Nrxn1.
Potential functional and epistatic partners: Nrxn1 and Gsk3b
Hundreds of susceptibility variants have been identified for schizophrenia in recent large-scale genomic studies. Associated genes fall into diverse functional categories and this supports the impressive genetic heterogeneity of complex neuropsychiatric diseases.90, 91 The approach we have taken here is to use a population model and exploit endogenous variations in a high-priority candidate as a starting point for a systematic gene-to-phenotype analysis. NRXN1 has been repeatedly linked to schizophrenia and autism15, 16, 17, 18, 19 and as we show, a co-expression network primed by this gene captures other genes that work closely with Nrxn1 and are involved in disease pathway.
Co-expression and eQTL analyses led us to genes already linked to Nrxn1 (for example, Cask, Nlgn1, γ-aminobutyric acid and glutamate receptors), and also led us to novel genes that have less obvious, but potentially interesting links to Nrxn1. Gsk3b is one such gene that we detected and then validated as a novel genetic partner of Nrxn1. GSK3B has diverse functions in the cell, including synaptic and intracellular signal transduction57, 58, 59 and could therefore be a conduit between the synaptic role of NRXN1 and cell signaling. Supporting this hypothesis, we found significant genetic interactions between SNPs in NRXN1 and GSK3B that indicate that susceptibility to schizophrenia can be modified to an extent by the combination of alleles in the two genes. For example, in the 2-loci model, the AA genotype at rs4563262 (NRXN1) and the AG and GG genotypes at rs4340737 (GSK3B) interact to increase or decrease vulnerability.
Despite the genetic interactions, to our knowledge, no study has yet demonstrated direct physical link between NRXN1 and GSK3B. However, the presynaptic region is a potential locus of cellular interaction between the two proteins. GSK3B, in addition to its many functions in cell signaling, also modulates presynaptic vesicle release by phosphorylating the voltage-dependent calcium channels and reducing intracellular Ca2+ levels.59 It also regulates the development of the active zone—the site of vesicle fusion and neurotransmitter release.92 NRXN1, a predominantly presynaptic protein, modulates release by selectively acting on voltage-dependent calcium channel (P/Q and N type), possibly by modifying the coupling of Ca2+ channels to release-ready vesicles.13 Variants of NRXN1 and GSK3B may therefore have a collective influence on the synaptic environment and consequently on disease susceptibility. Both the genetic interaction and a potential molecular interaction are consistent with a model in which neurotransmission and intracellular signaling are coupled in the etiology of schizophrenia and other psychiatric disorders.
Acknowledgments
We thank Drs David Kulp and Manjunatha Jagalur for providing analytical support with the Affymetrix Exon ST array data. We thank the UTHSC Center for Integrative and Translational Genomics, and NIH Grants from NIAAA (NIAAA U01AA017590, U01AA13499, U24AA13513 and U01AA014425), and a Human Brain Project from NIDA, NIMH and NIAAA (P20-DA 21131).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Biederer T, Sudhof TC. Mints as adaptors. Direct binding to neurexins and recruitment of munc18. J Biol Chem. 2000;275:39803–39806. doi: 10.1074/jbc.C000656200. [DOI] [PubMed] [Google Scholar]
- Hata Y, Butz S, Sudhof TC. CASK: a novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J Neurosci. 1996;16:2488–2494. doi: 10.1523/JNEUROSCI.16-08-02488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichtchenko K, Hata Y, Nguyen T, Ullrich B, Missler M, Moomaw C, et al. Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell. 1995;81:435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- Petrenko AG, Perin MS, Davletov BA, Ushkaryov YA, Geppert M, Sudhof TC. Binding of synaptotagmin to the alpha-latrotoxin receptor implicates both in synaptic vesicle exocytosis. Nature. 1991;353:65–68. doi: 10.1038/353065a0. [DOI] [PubMed] [Google Scholar]
- Sugita S, Saito F, Tang J, Satz J, Campbell K, Sudhof TC. A stoichiometric complex of neurexins and dystroglycan in brain. J Cell Biol. 2001;154:435–445. doi: 10.1083/jcb.200105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Atasoy D, Arac D, Yang X, Fucillo MV, Robison AJ, et al. Neurexins physically and functionally interact with GABA(A) receptors. Neuron. 2010;66:403–416. doi: 10.1016/j.neuron.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattenstroth G, Tantalaki E, Sudhof TC, Gottmann K, Missler M. Postsynaptic N-methyl-D-aspartate receptor function requires alpha-neurexins. Proc Natl Acad Sci USA. 2004;101:2607–2612. doi: 10.1073/pnas.0308626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, et al. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Ushkaryov YA, Petrenko AG, Geppert M, Sudhof TC. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257:50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- Rowen L, Young J, Birditt B, Kaur A, Madan A, Philipps DL, et al. Analysis of the human neurexin genes: alternative splicing and the generation of protein diversity. Genomics. 2002;79:587–597. doi: 10.1006/geno.2002.6734. [DOI] [PubMed] [Google Scholar]
- Tabuchi K, Sudhof TC. Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics. 2002;79:849–859. doi: 10.1006/geno.2002.6780. [DOI] [PubMed] [Google Scholar]
- Missler M, Sudhof TC. Neurexins: three genes and 1001 products. Trends Genet. 1998;14:20–26. doi: 10.1016/S0168-9525(97)01324-3. [DOI] [PubMed] [Google Scholar]
- Dudanova I, Sedej S, Ahmad M, Masius H, Sargsyan V, Zhang W, et al. Important contribution of alpha-neurexins to Ca2+-triggered exocytosis of secretory granules. J Neurosci. 2006;26:10599–10613. doi: 10.1523/JNEUROSCI.1913-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Rohlmann A, Sargsyan V, Aramuni G, Hammer RE, Sudhof TC, et al. Extracellular domains of alpha-neurexins participate in regulating synaptic transmission by selectively affecting N- and P/Q-type Ca2+ channels. J Neurosci. 2005;25:4330–4342. doi: 10.1523/JNEUROSCI.0497-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISC Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum J, Xu Q, Payne TJ, Ma JZ, Huang W, Gelernter J, et al. Significant association of the neurexin-1 gene (NRXN1) with nicotine dependence in European- and African-American smokers. Hum Mol Genet. 2008;17:1569–1577. doi: 10.1093/hmg/ddn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HC, Chang CC, Lin CY, Chen CL, Fann CS. A genome-wide scanning and fine mapping study of COGA data. BMC Genet. 2005;6 (Suppl 1 doi: 10.1186/1471-2156-6-S1-S30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci USA. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH, Williams SM. Epistasis and its implications for personal genetics. Am J Hum Genet. 2009;85:309–320. doi: 10.1016/j.ajhg.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dushlaine C, Kenny E, Heron E, Donohoe G, Gill M, Morris D, et al. Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry. 2010;16:286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- Jia P, Wang L, Meltzer HY, Zhao Z. Common variants conferring risk of schizophrenia: a pathway analysis of GWAS data. Schizophrenia Res. 2010;122:38–42. doi: 10.1016/j.schres.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Human Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattin KA, Moore JH. Exploiting the proteome to improve the genome-wide genetic analysis of epistasis in common human diseases. Hum Genet. 2008;124:19–29. doi: 10.1007/s00439-008-0522-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW. Headlong into a genomic singularity. Front Neurosci. 2010;4:25. doi: 10.3389/neuro.15.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe'er I, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BA, Wnek C, Kotlus BS, Roemer N, MacTaggart T, Phillips SJ. Genotyping new BXD recombinant inbred mouse strains and comparison of BXD and consensus maps. Mamm Genome. 1999;10:335–348. doi: 10.1007/s003359900998. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Gu J, Qi S, Lu L. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol. 2001;2:RESEARCH0046. doi: 10.1186/gb-2001-2-11-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Agarwala R, Capra JA, Chen Z, Church DM, Ciobanu DC, et al. High-throughput sequencing of the DBA/2J mouse genome from UT-ORNL-KBRIN Bioinformatics Summit 2010. BMC Bioinform. 2010;11 (Suppl 4:O7. [Google Scholar]
- Morse HC, III, Chused TM, Hartley JW, Mathieson BJ, Sharrow SO, Taylor BA. Expression of xenotropic murine leukemia viruses as cell-surface gp70 in genetic crosses between strains DBA/2 and C57BL/6. J Exp Med. 1979;149:1183–1196. doi: 10.1084/jem.149.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall RW, Kempermann G, Peirce J, Lu L, Goldowitz D, Gage FH, et al. Genetics of the hippocampal transcriptome in mouse: a systematic survey and online neurogenomics resource. Front Neurosci. 2009;3:55. doi: 10.3389/neuro.15.003.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozhui K, Ciobanu DC, Schikorski T, Wang X, Lu L, Williams RW. Dissection of a QTL hotspot on mouse distal chromosome 1 that modulates neurobehavioral phenotypes and gene expression. PLoS Genet. 2008;4:e1000260. doi: 10.1371/journal.pgen.1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JL, Li H, Wang J, Manly KF, Hitzemann RJ, Belknap JK, et al. How replicable are mRNA expression QTL. Mamm Genome. 2006;17:643–656. doi: 10.1007/s00335-005-0187-8. [DOI] [PubMed] [Google Scholar]
- Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Walker DG, et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics. 2007;28:311–322. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, et al. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci USA. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughran J, Jr, Bell J, Hitzemann R. On the relationships of high-frequency hearing loss and cochlear pathology to the acoustic startle response (ASR) and prepulse inhibition of the ASR in the BXD recombinant inbred series. Behav Genet. 1999;29:21–30. doi: 10.1023/a:1021433705004. [DOI] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, et al. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav. 2010;9:129–159. doi: 10.1111/j.1601-183X.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC. Provisional mapping of quantitative trait loci for chronic ethanol withdrawal severity in BXD recombinant inbred mice. J Pharmacol Exp Ther. 1998;286:263–271. [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24:150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez DR, White BC, Motsinger AA, Bush WS, Ritchie MD, Williams SM, et al. A balanced accuracy function for epistasis modeling in imbalanced datasets using multifactor dimensionality reduction. Genet Epidemiol. 2007;31:306–315. doi: 10.1002/gepi.20211. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Coffey CS, Hebert PR, Ritchie MD, Krumholz HM, Gaziano JM, Ridker PM, et al. An application of conditional logistic regression and multifactor dimensionality reduction for detecting gene-gene interactions on risk of myocardial infarction: the importance of model validation. BMC Bioinform. 2004;5:49. doi: 10.1186/1471-2105-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38 (Web Server issue:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa M, Aoki M, Yamada K, Iwayama-Shigeno Y, Takao H, Meerabux J, et al. Case-control association study of human netrin G1 gene in Japanese schizophrenia. J Med Dent Sci. 2004;51:121–128. [PubMed] [Google Scholar]
- Hur EM, Zhou FQ. GSK3 signalling in neural development. Nat Rev Neurosci. 2010;11:539–551. doi: 10.1038/nrn2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyberg Z, Ferrando SJ, Javitch JA. Roles of the Akt/GSK-3 and Wnt signaling pathways in schizophrenia and antipsychotic drug action. Am J Psychiatry. 2010;167:388–396. doi: 10.1176/appi.ajp.2009.08121873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LQ, Liu D, Hu J, Cheng J, Wang SH, Wang Q, et al. GSK-3 beta inhibits presynaptic vesicle exocytosis by phosphorylating P/Q-type calcium channel and interrupting SNARE complex formation. J Neurosci. 2010;30:3624–3633. doi: 10.1523/JNEUROSCI.5223-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Sabatti C, Geurts van Kessel A, et al. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Human Genet. 2008;83:504–510. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Ramoz N, Reichert JG, Corwin TE, Kryzak L, Smith CJ, et al. Association analysis of the NrCAM gene in autism and in subsets of families with severe obsessive-compulsive or self-stimulatory behaviors. Psychiatr Genet. 2006;16:251–257. doi: 10.1097/01.ypg.0000242196.81891.c9. [DOI] [PubMed] [Google Scholar]
- Potkin SG, Turner JA, Guffanti G, Lakatos A, Fallon JH, Nguyen DD, et al. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophrenia Bull. 2009;35:96–108. doi: 10.1093/schbul/sbn155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak G, Boukhadra J, Shaikh SA, Kennedy JL, Le Foll B. Association of a polymorphism in the NRXN3 gene with the degree of smoking in schizophrenia: a preliminary study. World J Biol Psychiatry. 2009;10 (4 Part 3:929–935. doi: 10.1080/15622970903079499. [DOI] [PubMed] [Google Scholar]
- Need AC, Keefe RS, Ge D, Grossman I, Dickson S, McEvoy JP, et al. Pharmacogenetics of antipsychotic response in the CATIE trial: a candidate gene analysis. Eur J Human Genet. 2009;17:946–957. doi: 10.1038/ejhg.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M, Hine E, Wright MJ, Duffy DL, MacMillan J, Martin NG. Effects of SCA1, MJD, and DPRLA triplet repeat polymorphisms on cognitive phenotypes in a normal population of adolescent twins. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:95–100. doi: 10.1002/ajmg.b.30413. [DOI] [PubMed] [Google Scholar]
- Hamshere ML, Green EK, Jones IR, Jones L, Moskvina V, Kirov G, et al. Genetic utility of broadly defined bipolar schizoaffective disorder as a diagnostic concept. Br J Psychiatry. 2009;195:23–29. doi: 10.1192/bjp.bp.108.061424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacos M, Costas J, de Cid R, Bayes M, Gonzalez JR, Baca-Garcia E, et al. Identification of new putative susceptibility genes for several psychiatric disorders by association analysis of regulatory and non-synonymous SNPs of 306 genes involved in neurotransmission and neurodevelopment. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:808–816. doi: 10.1002/ajmg.b.30902. [DOI] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA, et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Human Genet. 2005;77:918–936. doi: 10.1086/497703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domene S, Stanescu H, Wallis D, Tinloy B, Pineda DE, Kleta R, et al. Screening of human LPHN3 for variants with a potential impact on ADHD susceptibility. Am J Med Genet B Neuropsychiatr Genet. 2011;156:11–18. doi: 10.1002/ajmg.b.31141. [DOI] [PubMed] [Google Scholar]
- Cirulli ET, Kasperaviciute D, Attix DK, Need AC, Ge D, Gibson G, et al. Common genetic variation and performance on standardized cognitive tests. Eur J Human Genet. 2010;18:815–820. doi: 10.1038/ejhg.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla K, Luo Y, Buchan T, Beachem MA, Guzauskas GF, Ladd S, et al. Alterations in CDH15 and KIRREL3 in patients with mild to severe intellectual disability. Am J Human Genet. 2008;83:703–713. doi: 10.1016/j.ajhg.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anney RJ, Lasky-Su J, O'Dushlaine C, Kenny E, Neale BM, Mulligan A, et al. Conduct disorder and ADHD: evaluation of conduct problems as a categorical and quantitative trait in the international multicentre ADHD genetics study. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1369–1378. doi: 10.1002/ajmg.b.30871. [DOI] [PubMed] [Google Scholar]
- Adkins DE, Aberg K, McClay JL, Bukszar J, Zhao Z, Jia P, et al. Genomewide pharmacogenomic study of metabolic side effects to antipsychotic drugs. Mol Psychiatry. 2011;16:321–332. doi: 10.1038/mp.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, et al. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry. 2008;65:683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Liu QR, Zhang PW, Walther D, Li CY, et al. Genome wide association for substance dependence: convergent results from epidemiologic and research volunteer samples. BMC Med Genet. 2008;9:113. doi: 10.1186/1471-2350-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillend C, Ungerer A. Behavioral characterization of mdx3cv mice deficient in C-terminal dystrophins. Neuromuscul Disord. 1999;9:296–304. doi: 10.1016/s0960-8966(99)00029-2. [DOI] [PubMed] [Google Scholar]
- O'Brien WT, Harper AD, Jove F, Woodgett JR, Maretto S, Piccolo S, et al. Glycogen synthase kinase-3beta haploinsufficiency mimics the behavioral and molecular effects of lithium. J Neurosci Official J Soc Neurosci. 2004;24:6791–6798. doi: 10.1523/JNEUROSCI.4753-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy SS, Nonneman RJ, Young NB, Demyanenko GP, Maness PF. Impaired sociability and cognitive function in Nrcam-null mice. Behav Brain Res. 2009;205:123–131. doi: 10.1016/j.bbr.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McOmish CE, Burrows E, Howard M, Scarr E, Kim D, Shin HS, et al. Phospholipase C-beta1 knockout mice exhibit endophenotypes modeling schizophrenia which are rescued by environmental enrichment and clozapine administration. Mol Psychiatry. 2008;13:661–672. doi: 10.1038/sj.mp.4002046. [DOI] [PubMed] [Google Scholar]
- Koh HY, Kim D, Lee J, Lee S, Shin HS. Deficits in social behavior and sensorimotor gating in mice lacking phospholipase Cbeta1. Genes Brain Behavior. 2008;7:120–128. doi: 10.1111/j.1601-183X.2007.00351.x. [DOI] [PubMed] [Google Scholar]
- Kimura T, Yamashita S, Nakao S, Park JM, Murayama M, Mizoroki T, et al. GSK-3beta is required for memory reconsolidation in adult brain. PLoS One. 2008;3:e3540. doi: 10.1371/journal.pone.0003540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israely I, Costa RM, Xie CW, Silva AJ, Kosik KS, Liu X. Deletion of the neuron-specific protein delta-catenin leads to severe cognitive and synaptic dysfunction. Curr Biol. 2004;14:1657–1663. doi: 10.1016/j.cub.2004.08.065. [DOI] [PubMed] [Google Scholar]
- Blundell J, Kaeser PS, Sudhof TC, Powell CM. RIM1alpha and interacting proteins involved in presynaptic plasticity mediate prepulse inhibition and additional behaviors linked to schizophrenia. J Neurosci Official J Soc Neurosci. 2010;30:5326–5333. doi: 10.1523/JNEUROSCI.0328-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabares-Seisdedos R, Rubenstein JL. Chromosome 8p as a potential hub for developmental neuropsychiatric disorders: implications for schizophrenia, autism and cancer. Mol Psychiatry. 2009;14:563–589. doi: 10.1038/mp.2009.2. [DOI] [PubMed] [Google Scholar]
- Bray NJ, Holmans PA, van den Bree MB, Jones L, Elliston LA, Hughes G, et al. Cis- and trans- loci influence expression of the schizophrenia susceptibility gene DTNBP1. Hum Mol Genet. 2008;17:1169–1174. doi: 10.1093/hmg/ddn006. [DOI] [PubMed] [Google Scholar]
- Airey DC, Lu L, Williams RW. Genetic control of the mouse cerebellum: identification of quantitative trait loci modulating size and architecture. J Neurosci. 2001;21:5099–5109. doi: 10.1523/JNEUROSCI.21-14-05099.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozhui K, Hamre KM, Holmes A, Lu L, Williams RW. Genetic and structural analysis of the basolateral amygdala complex in BXD recombinant inbred mice. Behav Genet. 2007;37:223–243. doi: 10.1007/s10519-006-9122-3. [DOI] [PubMed] [Google Scholar]
- Kellendonk C, Simpson EH, Kandel ER. Modeling cognitive endophenotypes of schizophrenia in mice. Trends Neurosci. 2009;32:347–358. doi: 10.1016/j.tins.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujescu D, Ingason A, Cichon S, Pietilainen OP, Barnes MR, Toulopoulou T, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- Burmeister M, McInnis MG, Zollner S. Psychiatric genetics: progress amid controversy. Nat Rev Genet. 2008;9:527–540. doi: 10.1038/nrg2381. [DOI] [PubMed] [Google Scholar]
- Viquez NM, Fuger P, Valakh V, Daniels RW, Rasse TM, DiAntonio A. PP2A and GSK-3beta act antagonistically to regulate active zone development. J Neurosci. 2009;29:11484–11494. doi: 10.1523/JNEUROSCI.5584-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.