Abstract
The val66met polymorphism on the BDNF gene has been reported to explain individual differences in hippocampal volume and memory-related activity. These findings, however, have not been replicated consistently and no studies to date controlled for the potentially confounding impact of early life stress, such as childhood abuse, and psychiatric status. Using structural and functional MRI, we therefore investigated in 126 depressed and/or anxious patients and 31 healthy control subjects the effects of val66met on hippocampal volume and encoding activity of neutral, positive and negative words, while taking into account childhood abuse and psychiatric status. Our results show slightly lower hippocampal volumes in carriers of a met allele (n=54) relative to val/val homozygotes (n=103) (P=0.02, effect size (Cohen's d)=0.37), which appeared to be independent of childhood abuse and psychiatric status. For hippocampal encoding activity, we found a val66met–word valence interaction (P=0.02) such that carriers of a met allele showed increased levels of activation in response to negative words relative to activation in the neutral word condition and relative to val/val homozygotes. This, however, was only evident in the absence of childhood abuse, as abused val/val homozygotes showed hippocampal encoding activity for negative words that was comparable to that of carriers of a met allele. Neither psychiatric status nor memory accuracy did account for these associations. In conclusion, BDNF val66met has a significant impact on hippocampal volume independently of childhood abuse and psychiatric status. Furthermore, early adverse experiences such as childhood abuse account for individual differences in hippocampal encoding activity of negative stimuli but this effect manifests differently as a function of val66met.
Keywords: BDNF, childhood abuse, fMRI, hippocampus, human memory, val66met
Introduction
Brain-derived neurotrophic factor (BDNF) regulates the sprouting of axons and dendrites in the hippocampus, a key structure for emotion and memory processing.1, 2, 3, 4 Rodent studies, for example, have shown that BDNF modulates hippocampal neuronal differentiation5 and hippocampal dependent memory.6, 7, 8 Moreover, human studies have reported a positive relation between BDNF concentration, hippocampal volume and memory performance.9, 10
Studies focusing on a single nucleotide site in the DNA sequence of the BDNF gene; val66met (a valine (val) to methionine (met) insertion at codon 66) have partly confirmed the associations of BDNF protein expression with neurobiological and behavioral abnormalities. Egan et al.11 showed in vitro that the met allele is linked to a reduced activity-dependent expression of BDNF in hippocampal neurons of rats, a finding that was replicated by Chen et al.12 In addition, studies have shown that in the hippocampus the met allele is associated with diminished levels of N-acetyl-aspartate, a putative marker for neuronal integrity.11, 13 In line with these findings, some studies have shown that the met allele is associated with impaired episodic memory11 and executive functioning.14, 15 Structural and functional magnetic resonance imaging (MRI) studies further suggest that carriers of a met allele have smaller hippocampal volumes relative to val/val homozygotes16, 17, 18, 19 and altered hippocampal activity during the encoding of stimuli.11, 20 Nevertheless, these findings have not been consistently replicated,21, 22, 23, 24, 25, 26 which might be due to the inclusion of small samples and task characteristics such as the emotional valence of the stimuli. Furthermore, the occurrence of early trauma, such as childhood abuse and psychiatric status, represent sources of variation in hippocampal volume and function (reviewed in refs. 4,27) that have not been taken into account in previous studies. In addition, gene–environment interactions have been reported between BDNF val66met and abuse on brain structure and activity.28, 29 As a consequence, the earlier reported associations between BDNF val66met locus and hippocampal structure and function might be (partly) dependent on a history of childhood abuse or on psychiatric status.
The goal of this study, then, was to evaluate the effects of val66met on hippocampal volume and on encoding-related hippocampal activity while taking into account the potential influence of childhood abuse and diagnostic status. Given earlier conflicting findings,11, 20 we further aimed to extend previous findings by examining the effects of neutral, positive and negative emotional stimuli on hippocampal activity.
Materials and methods
Subjects
The data analyzed are from the imaging sample of the Netherlands Study of Depression and Anxiety (NESDA).30, 31 Included in the NESDA imaging sample were 301 subjects, of whom 233 were patients with a current depressive and/or anxiety disorder and 68 healthy control subjects. Genetic and high-quality functional and structural MRI data were available for 157 persons who participated in the NESDA MRI study of whom 126 were depressed and/or anxious patients and 31 were healthy controls. Subjects in the current study did not differ from subjects in the NESDA imaging sample (N=301) with regard to age (P=0.98), gender (P=0.22) and current diagnosis (P=0.07).
Subjects underwent imaging at three different locations in the Netherlands: Academic Medical Center (AMC), University of Amsterdam, University Medical Center Groningen (UMCG) and Leiden University Medical Center (LUMC). To be eligible, subjects had to be 18–57 years of age and fluent in Dutch. Exclusion criteria were having an Axis-I disorder other than a depressive and/or anxiety disorder (Diagnostic and Statistical Manual of Mental disorders fourth edition (DSM-IV),32 being on antidepressant treatment other than selective serotonin reuptake inhibitors (SSRIs) at a stable dose,33 a history of a major internal or neurological disorder, dependency on alcohol and/or drugs, smoking >5 cigarettes a day, or hypertension (>180/130 mm Hg). The protocol and procedures were approved by each of the ethical committees of participating institutes and all subjects signed an informed consent.
Diagnoses of depressive and anxiety disorders were established according to the criteria set forth in the DSM-IV32 on the basis of responses to the Composite International Diagnostic Interview 2.1 (CIDI) lifetime version,34 a reliable and validated diagnostic tool.35 The severity of depressive and anxiety symptoms was assessed with the Montgomery Åsberg Depression Rating Scale (MÅDRS)36 and the Beck Anxiety Inventory (BAI),37 both of which have been shown to have excellent psychometric characteristics.38, 39
Childhood abuse was assessed retrospectively using a semi-structured childhood trauma interview.40, 41 In this interview, participants were asked whether they had experienced emotional neglect or psychological abuse, physical abuse and/or sexual abuse before the age of 16 years. After an affirmative answer, subjects were asked for details on the frequency of the events. Based on the sum and the frequency of abusive events an index (range 0–8) was calculated for each subject (for details see Wiersma et al.42).
Genotyping was performed by Perlegen Sciences (Mountain View, CA, USA). For a detailed description of the procedures according to which genotyping was performed, we refer to Boomsma et al.43 Val66met was extracted from whole-genome data using PLINK software version 1.07 (http://pngu.mgh.harvard.edu/~purcell/plink). In our sample, 103 subjects were val/val homozygotes (65.6%) and 54 subjects carried a met allele (34.4%). Two subjects (1.3%) with the met/met genotype were merged with heterozygous subjects into a group of met allele carriers. Genotype counts were 82 val66val, 42 val66met and 2 met66met in the patient group and 21 val66val, 10 val66met and 0 met66met in the healthy control group. Patient and healthy control samples did not differ with regard to genotype distribution (P=0.77). Allele frequencies were in Hardy–Weinberg equilibrium in the GAIN-MDD sample in which the genotyping was performed (N=3530, χ12=0.62, P=0.43) and in the sub-sample on which we present data (N=157, χ12=2.66, P=0.10).
Memory paradigm
In the scanner, subjects performed a subject-paced, event-related encoding task, similar to the paradigm described by Daselaar et al.44 and known to reliably activate the hippocampus. The task is described in detail elsewhere.45 Briefly, during the encoding phase of the task 120 words (40 of neutral valence, 40 of positive valence and 40 of negative valence) were presented in pseudo-randomized order. Subjects were instructed to classify these words according to valence. After a 10-minute retention interval, subjects were asked to complete a word recognition task. Subjects were instructed to indicate whether they had seen or probably had seen the word or whether the word was new. Discriminant accuracy was calculated as the proportion correctly recognized words minus the proportion false alarms.45
Image acquisition and data handling
Image acquisition and data handling are detailed elsewhere.31, 45 In sum, imaging data were collected using Philips 3-Tesla MRI scanners (Best, the Netherlands) using SENSE-6 and 8 channel head coils (AMC and UMCG/LUMC, respectively). Echo-planar images were obtained using a T2*-weighted gradient echo sequence with repetition time 2300 ms, a 30 ms echo time (UMCG 28 ms), a matrix size of 96 × 96 (UMCG 64 × 64), producing 35 axial slices of 3 mm thickness direction interleaved, 2.29 × 2.29 mm2 in-plain resolution (UMCG 3 × 3). Anatomical imaging included a sagittal 3-D gradient-echo T1-weighted sequence with a repetition time of 9 ms and a 3.5 ms echo time acquiring slices with a voxel size of 1 × 1 × 1 mm3. Imaging data were preprocessed with SPM5 (Statistic Parametric Mapping, http://www.fil.ion.ucl.ac.uk/spm/).
Preprocessing of the data included reorientation of the functional images to the anterior commissure, slice time correction, image realignment, registration of the T1-scan to the mean image, warping to Montreal Neurological Institute (MNI) space as defined by the SPM5 T1-template, reslicing to 3 × 3 × 3 mm3 voxels, and spatial smoothing using an 8-mm FWHM Gaussian kernel. Haemodynamic responses to each stimulus were modeled with a delta function convolved with a synthetic haemodynamic response function and modulated using response times.
Contrast images for ‘subsequent hits vs baseline' were calculated for the neutral, positive and negative word condition per subject on a voxel-by-voxel basis, based on subsequent recognition success and entered in a 2 (group: val/val homozygotes vs met carriers; independent factor) by 3 (condition: neutral, positive, negative (>baseline); dependent factor) MANCOVA with age, education and scan center as covariates. Mean BOLD signal change during successful encoding in the left and right hippocampus was extracted per condition (neutral/positive/neutral>baseline) using the MARSBAR toolbox.46 The hippocampal masks of the Automated Anatomical Labeling software package, implemented in the WFU Pick Atlas toolbox47 were used to define the left and right hippocampal region.
Anatomical images were processed using an optimized voxel-based morphometry approach, following the Diffeomorphic Anatomical Registration Through Exponentiated Lie algebra (DARTEL)48 using SPM5 software implemented in Matlab 7.1.0 (The MathWorks, Natick, MA, USA). For details see van Tol et al.45 To test for differences in regional brain volume, an independent samples t-test was set up for a voxel-wise comparison of the gray matter density images of the val/val homozygotes and met carriers, with age, scan center and total gray matter volume as covariates. Following a similar approach as for signal change extraction, the mean volume of the left and right hippocampus was additionally extracted, again using the binary masks of the hippocampus based on the Anatomical Automatic Labelling atlas. Data were exported to SPSS 18.0 (Chicago, IL, USA) for further analysis.
Statistical analyses
Computations were performed in SPSS 18.0. A P-value of <0.05 (2-tailed) was considered as the threshold for statistical significance. Demographical and clinical characteristics between val/val homozygotes and carriers of a met allele were compared using Student's t-tests for continuous- and χ2-tests for categorical data.
Main effects of val66met on right, left and total hippocampal volume were calculated using a Repeated Measures (RM) ANCOVA with left vs right hippocampal volume as the within-subjects factor and age, gender, number of years of education, SSRI use (no vs yes), alcohol use (no vs yes), scan site and total gray matter volume as covariates. ANCOVAs were used to assess the effects of val66met on memory accuracy and hippocampal activity during the encoding of neutral, positive and negative words. To address val66met–valence interactions effects on memory accuracy and hippocampal encoding activity, we ran RM ANCOVAs with word valence (positive vs neutral and negative vs neutral) as within-subject factor and age, gender, number of years of education, SSRI use (no vs yes), alcohol use (no vs yes), scan site, hippocampal volume, memory accuracy and handedness as covariates. If indicated by between-group differences in memory accuracy, accuracy scores were included as covariates in the analyses on hippocampal encoding activity.
Possible interaction effects of val66met with abuse and diagnosis (dummy variables coding for healthy, depressed, depressed-anxious and anxious) on hippocampal volume, memory accuracy and hippocampal encoding activity were evaluated using hierarchical stepwise regression analyses, if indicated by statistically significant associations in the above described analyses. Regression analyses consisted of three steps: (1) covariates (see above), (2) val66met, childhood abuse and diagnosis and (3) the interaction terms val66met × abuse and val66met × diagnosis. Analyses were rerun with lifetime instead of current diagnosis (6-month recency). Tolerance of the predictors and normality of error variances were verified.
To assess regional specificity of val66met within the hippocampus and to explore effects of val66met on other brain regions, voxel-wise analyses were repeated on the whole brain gray matter density maps and contrast maps reflecting encoding related activity using SPM5, with the threshold set at P<0.001, uncorrected. Uncorrected whole brain results are reported in the supplement. For regions outside the hippocampus, a threshold of P<0.05, FWE corrected was set.
Results
The overall sample (N=157) had a mean age of 37.39±10.08 years and included 100 women (63.7%). Demographical and clinical characteristics of the sample are given in Table 1 by BDNF genotype. There were no statistically significant differences between the genotype groups in terms of demographical and clinical variables. Furthermore, val66met was not differentially associated with exposure to childhood abuse (dichotomous (yes vs no), nor with exposure to specific types of childhood abuse (all P's>0.75).
Table 1. Demographic and clinical characteristics (mean±s.d. or percentages) by BDNF genotype.
| val 66 val (n=103) | val 66 met (n=54) | P -value | |
|---|---|---|---|
| Females (%) | 64.1 (n=66) | 63.0 (n=34) | 0.89 |
| Age | 37.13±9.61 | 37.89±10.39 | 0.65 |
| Education (years) | 12.43±3.00 | 12.63±3.28 | 0.70 |
| Smoker (%) | 33.0 (n=34) | 23.2 (n=13) | 0.14 |
| Alcohol use (%) | 56.2 (n=58) | 60.0 (n=32) | 0.58 |
| SSRI use (%) | 30.1 (n=31) | 20.4 (n=11) | 0.19 |
| Right handed (%) | 91.3 (n=94) | 94.4 (n=51) | 0.48 |
| Childhood abuse indexa | 1.59±1.96 | 1.65±2.27 | 0.87 |
| Type of childhood abuse | |||
| Emotional abuse/neglect (%) | 52.7 (n=54) | 52.3 (n=28) | 0.95 |
| Sexual abuse (%) | 13.6 (n=14) | 15.9 (n=9) | 0.94 |
| Physical abuse (%) | 11.0 (n=11) | 11.4 (n=6) | 0.94 |
| Diagnostic status | 0.78b | ||
| Healthy controls (%) | 20.4 (n=21) | 18.5 (n=10) | 0.78 |
| Depression (%) | 26.2 (n=27) | 29.6 (n=16) | 0.65 |
| Anxiety (%)c | 19.4 (n=20) | 22.2 (n=12) | 0.68 |
| Depression and anxiety (%)c | 34.0 (n=35) | 29.6 (n=16) | 0.58 |
| Depression severity, MÅDRS | 11.67±8.83 | 13.62±11.65 | 0.23 |
| Anxiety severity, BAI | 11.79±9.19 | 13.34±11.23 | 0.84 |
Abbreviations: BAI, Becks Anxiety Inventory; MÅDRS, Montgomery Åsberg Depression Rating Scale; SSRI, selective serotonin reuptake inhibitor.
Range 0–8.
P-value for the omnibus χ2 (3 degrees of freedom) for differences in distribution of the met allele over diagnoses.
Included a diagnosis of social phobia, panic disorder, generalized anxiety disorder and/or agoraphobia.
BDNF val66met and hippocampal volume
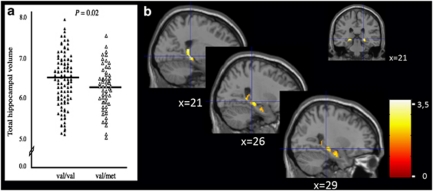
Total hippocampal volume was smaller in carriers of a met allele relative to val/val homozygotes (F1,180=5.33, P=0.02). The effect size of this difference (standardized Cohen's d, that is, the mean between-group difference divided by the pooled standard deviation)49 was 0.38 (see Figure 1 and Table 2 for covariate adjusted means on total, right and left hippocampal volume ±s.e.m. No interaction of val66met × right vs left hippocampus was observed (P=0.63). BDNF val66met had no effect on total gray matter volume (P=0.60). Voxelwise analyses in SPM5 confirmed these findings, with the peak voxel located in the posterior part of the hippocampus (MNI coordinate: Right hippocampus: (x=18, y=−33, z=8 and x=21, y=−30, z=−4), Z=3.61/3.42, k=29/17, PFWE_ROI=0.018; Left hippocampus: (x=−18, y=−36, z=8), Z=3.17, k=4, PFWE_ROI=0.062).
Figure 1.
(a) Scattergram of total hippocampal volume for subjects homozygous for the val allele (val/val, n=103) and carriers of a met allele (val/met, n=54). Horizontal lines indicate the mean for each group. Data are adjusted for age, gender, number of years of education, selective serotonin reuptake inhibitor use, alcohol use and scan site. (b) Coronal and sagittal views and a statistical map of t-transformed hippocampal volume differences by BDNF val66met genotype. Note: voxelwise analyses confirmed these findings with the peak voxel located in the posterior part of the hippocampus (MNI coordinate: right hippocampus: (x=18, y=−33, z=8 and x=21, y=−30, z=−4), Z=3.61/3.42, k=29/17; left hippocampus: (x=−18, y=−36, z=8), Z=3.17, k=4).
Table 2. Cerebral and hippocampal volumes and hippocampal-related encoding activity (mean±s.e.m.) by BDNF genotype and word valence (neutral, positive and negative).
| val66val (n=103) | val66met (n=54) | P-value | |
|---|---|---|---|
| Total gray matter volume (ml)a | 736.57±5.35 | 731.66±7.45 | 0.60 |
| Hippocampal volume (ml) a , b | |||
| Total | 6.45±0.03 | 6.31±0.04 | 0.02 |
| Right | 3.06±0.02 | 2.99±0.02 | 0.01 |
| Left | 3.39±0.02 | 3.31±0.02 | 0.05 |
| Hippocampal encoding activity a , c , d | |||
| Neutral words | 0.15±0.04 | 0.16±0.06 | 0.92 |
| Positive words | 0.15±0.05 | 0.23±0.07 | 0.32 |
| Negative words | 0.20±0.05 | 0.36±0.06 | 0.05 |
All mean values are corrected for gender, age, years of education, selective serotonin reuptake inhibitor and alcohol use and site of scanning.
Mean values are additionally corrected for total cerebral grey matter volume.
Values are expressed as change vs baseline and thus are in arbitrary units.
Mean values are additionally corrected for total hippocampal volume.
Regression analyses were used to evaluate whether the smaller hippocampal volume in met carriers as compared with val/val homozygotes were moderated by the effects of abuse or diagnostic status. Main effects of childhood abuse and psychiatric status, and interaction effects of val66met with childhood abuse and psychiatric status on hippocampal volume were not observed (all P's>0.10). The main effect of val66met remained statistically significant after the inclusion of childhood abuse and psychiatric status in the model (B=−0.13, 95% confidence interval (CI)=−0.24 to −0.02, P=0.02). Similar results were obtained in analyses with lifetime instead of current diagnosis and in analysis in which continuous measures for childhood abuse and depression severity (that is, total MÅDRS score) were included as predictors (data not shown). No effect of BDNF val66met was observed on other structures at the set threshold (see Supplementary Table 1).
BDNF val66met and task performance
There were no overall differences in discriminant accuracy as a function of genotype (covariate adjusted means±se: val/val homozygotes=0.57±0.01 vs met carriers=0.58±0.02; P=0.85). Interaction effects of val66met and word valence on memory accuracy were not observed, either (all P's>0.10). Furthermore, memory accuracy was unrelated to hippocampal volume (Pearson's r=0.13; P=0.10) and to hippocampal encoding activity (r=0.04; P=0.66).
BDNF val66met and hippocampal activity
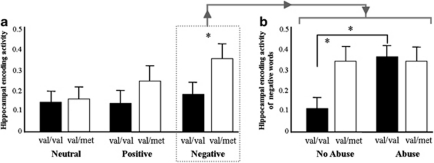
Main effects of val66met and word valence on hippocampal activity during the encoding of neutral and positive words were not observed (see Table 2). However, val66met interacted with neutral vs negative word valence (P=0.02) such that hippocampal activity was higher in carriers of a met allele in the negative word condition relative val/val homozygotes (P=0.05, Bonferroni corrected (P=NS) and to hippocampal activity in the neutral word condition (P=0.002, Bonferroni corrected (P=0.01). This was not observed in val/val homozygotes (see Figure 2a and Table 2 for covariate adjusted means±se by word valence). No val66met-neutral vs positive word valence interaction effect on encoding activity was found (P=0.17). Effects of lateralization were not observed. Voxel-wise analyses located the peak voxel of the interaction between negative vs neutral encoding × val66met cluster at the left posterior hippocampus ((x=−21, y=−27, z=−6), F1,461=14,11, Z=3.55, PFWE_ROI=0.024, K (number of voxels)=15). Exploratory voxel-wise whole brain analyses showed no statistical significant effects of val66met and val66met–word valence interactions in brain areas other than the hippocampus at the a priori set threshold of P<0.05, FWE corrected (see Supplementary Tables 2 and 3).
Figure 2.
(a) Mean total hippocampal activity during encoding by stimulus valence for subjects homozygous for the val allele (val/val, n=103) and carriers of a met allele (val/met, n=54). (b) Mean total hippocampal activity during the encoding of negative words by childhood abuse before the age of 16 years (yes vs no) for subjects homozygous for the val allele (55 abused, 48 non abused) and carriers of a met allele (25 abused, 29 non abused). The val66met–childhood abuse interaction effect is significant at P=0.01. Error bars reflect the s.e.m. Data are adjusted for age, gender, number of years of education, selective serotonin reuptake inhibitor use, alcohol use and scan site. *P<0.05.
Regression analyses were used to evaluate whether the higher hippocampal activity during the encoding of negative words were moderated by the effects of abuse or diagnostic status. Hippocampal encoding activity in response to words of negative valence was higher in abused subjects as compared with non-abused subjects (B=0.16, 95%CI=0.05–0.28, P=0.007). In addition, we found a val66met–childhood abuse interaction (B=−0.10, 95%CI=−0.17 to −0.02, P=0.01) showing that childhood abuse predicted increased hippocampal activation in response to negative words in val/val homozygotes (P=0.009) but not in carriers of a met allele (P=0.34) (see Figure 2b). Effects of psychiatric status (lifetime and current) and val66met by psychiatric status interaction effects were not observed (all P's>0.10). Adding memory accuracy as a predictor to the model did not change our results (data not shown) making it unlikely that these results are accounted for by genotype differences regarding attention or effort.
Discussion
We addressed the effects of val66met on hippocampal volume and function while taking into account the possible confounding effects of childhood abuse and psychiatric status.
In line with some previous studies,17, 18, 19 but not all (for example, ref. 26) we find smaller hippocampal volumes in carriers of a met allele relative to val/val homozygotes. This effect has generally been explained by abnormal intracellular trafficking and impaired activity secretion of BDNF and by extension aberrant trophic support in carriers of a met allele relative to the val/val homozygotes that have been shown in in vitro experiments.11, 12 As atrophy of the hippocampus has also been associated with (early) stress and/or a current or remitted depressive episode,4 it is crucial to exclude the possible confounding effects of these variables. Our data suggest that the association between the met allele and hippocampal volume is independent of childhood abuse. This finding is at odds with those of Gatt et al.,28 who modeled the interaction of early life stress and val66met in the prediction of hippocampal volume and found that the combination of carrying a met allele and being exposed to early life stress was associated with smaller hippocampal volumes in healthy adults. It could be that the observed discrepancy between the results of Gatt et al.28 and ours might be explained by a broader definition of early life stress by Gatt et al.28 who included, for example, also illness and exposure to natural disasters as stressful events, whereas we focused on childhood abuse including physical, sexual and emotional abuse. Furthermore, Gatt et al.28 studied healthy control subjects, whereas we studied mostly patients. However, exactly how these differences between the studies could have led to a different pattern of results is unclear. In line with Frodl et al.,19 we show that lifetime and current psychiatric status does not thrive the val66met genotype effect on hippocampal volume, providing evidence for a direct association between the met allele and small hippocampal volume that further appears to be specific to the hippocampus.
In addition to reduced hippocampal volume, we show that val66met interacts with word valence such that encoding activity is increased in carriers of a met allele during the negative word condition and not in the neutral or positive word condition. This effect was not observed in other brain areas than the hippocampus and is consistent with studies in which emotional stimuli were used,24, 25 but not with studies in which neutral stimuli were used.50 We could not replicate the finding of higher hippocampal activation in carriers of a met allele in response to neutral stimuli as is reported in the seminal study by Egan et al.12 On the basis of a recent study that showed that negative affectivity increased more in response to social stress in met carriers as compared with val/val homozygotes,51 one may speculate that carriers of a met allele are more sensitive or reactive to negative stimuli. Owing to a possible relation between higher hippocampal activity and psychopathology,52, 53 this finding might concur with studies that show a link between the met allele and depression (reviewed in ref. 53). We further found, in line with some studies that childhood abuse predicts higher levels of hippocampal encoding activity.52, 53, 54 However, from our data it appears that childhood abuse is associated with a relative increase in hippocampal activity in val/val homozygotes only and not in carriers of a met allele. Although speculative, an interpretation may be that higher levels of hippocampal activity after exposure to childhood abuse in val/val homozygotes reflect a higher sensitivity for emotionally negative stimuli in that in carriers of a met allele is present regardless of exposure to childhood abuse. This idea is in line with studies that report hippocampal dysfunction in various severe psychiatric illnesses, particularly if exposure to childhood abuse is documented.4, 52, 53, 54, 55
Despite differences in hippocampal volume and activity between val/val homozygotes and carriers of a met allele we did not find differences in memory accuracy and clinical variables (for example, depression severity) as a function of BDNF genotype. This may suggest on the one hand that our findings are relevant for both healthy individuals and patients. Moreover, it is pertinent to the debate on the relationship between hippocampal volume and function with behavioral performance. In line with the absence of associations between hippocampal volume, hippocampal function and memory performance in our study, a recent review on 80 studies showed that the model: ‘a bigger brain structure → greater brain response → better performance' may not reflect reality.56
A notable strength of our study is that the findings are derived from a genetically homogeneous sample and it thus is unlikely that our results are devoid by population stratification.57 Furthermore, we studied the effects of val66met in the context of childhood abuse and emotional valence of stimuli, and our results clearly highlight the importance of including such variables. A few weaknesses of our study also merit attention. Obviously, we cannot exclude the possibility that other polymorphisms on the BDNF gene or on other genes, notably those that constitute the neurotrophic pathway (for example, CREB1 and NTRK2)29 might have contributed to the effects that we observed. With regard to our self-reported measurement of childhood abuse, it should be noted that the validity and reliability of recall might vary by diagnosis and time since abuse took place. Furthermore, in the face of negative findings, statistical power is important to take into account. Overall we had a comparatively large sample size, but our analysis on psychiatric status might have been underpowered particularly because the size of the control samples may have been too small (for example, only 31 healthy control subjects) to detect effect sizes that are reported to be moderate at best.58, 59 Finally, although we speculate that carriers of a met allele are more reactive to emotionally negative laden stimuli as compared with val/val homozygotes we are not able to confirm this because we have no subjective ratings of the stimuli by our participants.
In sum, our results suggest that BDNF val66met has a small effect on hippocampal volume and this effect appears to be independent of childhood abuse and psychiatric status. Furthermore, gene–environment interactions between val66met and childhood abuse account for individual differences in hippocampal encoding activity of negative stimuli. Important venues for future research are to delineate the exact mechanisms, in vivo, through which the met allele produces its effect on hippocampal volume and function. In addition, it remains to be investigated, in longitudinal designs, whether or not the effects of val66met on hippocampal volume and activity are predictive for individual cognitive functioning and psychological well-being.
Acknowledgments
The NESDA study infrastructure is financed by the Geestkracht program of ZonMW, the Dutch Scientific Organization-Medical Sciences (Grant No. 10.000.1002) and by complementary funding from participating mental healthcare institutions (GGZ Buitenamstel, GGZ Drenthe, GGZ Friesland, GGZ Geestgronden, GGZ Rivierduinen and Lentis) and Universities (Leiden University Medical Center, University Medical Center Groningen, and VU University Medical Center) and the Center for Molecular and Systems Biology (CMSB). Genotyping was funded by the Genetic Association Information Network (GAIN) for the US National Institutes of Health (NIH). This particular project was further financed with NWO (Dutch Scientific Organization) VIDI-grant (Grant no. 016.085.353) awarded to Dr Bernet Elzinga and a collaborative grant from the Young Academy (Royal Netherlands Academy of Sciences) awarded to Dr Elzinga, Dr Penninx and Dr Aleman.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Murer MG, Yan Q, Raisman-Vosari R. Brain-derived neurotrophic factor in the control human brain, and in Alzheimer's disease and Parkinson's disease. Prog Neurobiol. 2001;63:71–124. doi: 10.1016/s0301-0082(00)00014-9. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Messaoudi E. BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Prog Neurobiol. 2005;76:99–125. doi: 10.1016/j.pneurobio.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Komulainen P, Pedersen M, Hanninen T, Bruunsgaard H, Lakka TA, Kivipelto M, et al. BDNF is a novel marker of cognitive function in ageing women: the DR's EXTRA study. Neurobiol Learning Mem. 2008;4:596–603. doi: 10.1016/j.nlm.2008.07.014. [DOI] [PubMed] [Google Scholar]
- MacQueen G, Frodl T. The hippocampus in major depression: evidence for the convergence of the bench and bedside in psychiatric research. Mol Psychiatry. 2011;16:252–264. doi: 10.1038/mp.2010.80. [DOI] [PubMed] [Google Scholar]
- Taliaz D, Stall N, Dar DE, Zangen A. Knockdown of brain-derived neurotrophic factor in specific brain sites precipitates behaviors associated with depression and reduces neurogenesis. Mol Psychiatry. 2010;15:80–92. doi: 10.1038/mp.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Gottschalk W. Modulation of hippocampal synaptic transmission and plasticity by neurotrophins. Prog Brain Res. 2000;128:231–241. doi: 10.1016/S0079-6123(00)28020-5. [DOI] [PubMed] [Google Scholar]
- Poo MM. Neurotrophins as synaptic modulators. Nat Rev Neurosci. 2001;2:24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- Choi DC, Maguschak KA, Ye K, Jang S-W, Myers KM, Ressler KJ. Prelimbic cortical BDNF is required for memory of learned fear but not extinction or innate fear. Proc Natl Acad Sci USA. 2010;107:2675–2680. doi: 10.1073/pnas.0909359107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunstad J, Benitez A, Smith J, Glickman E, Spitznagel MB, Alexander T, et al. Serum brain-derived neurotrophic factor is associated with cognitive function in healthy older adults. J Geriatr Psychiatry Neurol. 2008;21:166–170. doi: 10.1177/0891988708316860. [DOI] [PubMed] [Google Scholar]
- Erickson KI, Prakash RS, Voss MW, Chaddock L, Heo S, McLaren M. Brain-derived neurotrophic factor is associated with age-related decline in hippocampal volume. J Neurosci. 2010;30:5368–5375. doi: 10.1523/JNEUROSCI.6251-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, et al. The BDNF Val66Met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, et al. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24:4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern AJ, Savostyanova AA, Goldman A, Barnett AS, van der Veen JWC, Caillicot JH, et al. Impact of the brain-derived neurotrophic factor val66met polymorphism on levels of N-Acetyl-Aspartate assessed by magnetic resonance spectroscopic imaging at 3 Tesla. Biol Psychiatry. 2008;64:856–862. doi: 10.1016/j.biopsych.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakowski A, Czerski PM, Skibinska M, Hauser J. Polymorphisms on the brain-derived neurotrophic factor gene and performance on a cognitive prefrontal test in bipolar patients. Bipolar Disord. 2003;5:468–472. doi: 10.1046/j.1399-5618.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- Erickson KI, Kim JS, Suever BL, Voss MW, Francis BM, Kramer AF. Genetic contributions to age-related decline in executive function: a 10-year longitudinal study of COMT and BDNF polymorphisms. Front Hum Neurosci. 2008;2:1–9. doi: 10.3389/neuro.09.011.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezawas L, Verchinski BA, Mattay VS, Callicot JH, Kolochana BS, Straub RE, et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeszko PR, Lipsky R, Mentschel C, Robinson D, Gunduz-Bruce H, Sevy S, et al. Brain-derived neurotrophic factor val66met polymorphism and volume of hippocampal formation. Mol Psychiatry. 2005;10:631–636. doi: 10.1038/sj.mp.4001656. [DOI] [PubMed] [Google Scholar]
- Bueller JA, Aftab M, Sen S, Gomez-Hassan D, Burmeister M, Zubieta JK. BDNF val66met allele is associated with reduced hippocampal volume in healthy subjects. Biol Psychiatry. 2006;59:812–815. doi: 10.1016/j.biopsych.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Frodl T, Schule C, Schmitt G, Born C, Baghai T, Zill P, et al. Association of brain-derived neurotrophic factor val66met polymorphism with reduced hippocampal volumes in major depression. Arch Gen Psychiatry. 2007;64:410–416. doi: 10.1001/archpsyc.64.4.410. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolchana BS, Callicot JH, Egan MF, et al. Brain-derived neurotrophic factor val66met polymorphism affects human-memory related hippocampal activity and predicts memory performance. J Neurosci. 2003;23:6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin S, McQuoid DR, Potter G, Payne ME, MacFall JR, Steffens DC, et al. The brain-derived neurotrophic factor val66met polymorphism, hippocampal volume, and cognitive function in geriatric depression. Am J Ger Psychiatry. 2010;18:323–331. doi: 10.1097/JGP.0b013e3181cabd2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik MS, Wang L, Barch DM, Morris JC, Csernansky JG. BDNF polymorphism rs6265 and hippocampal structure and memory performance in healthy control subjects. Psychiatry Res. 2010;178:425–429. doi: 10.1016/j.psychres.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield PR, Williams LM, Paul RH, Gatt JM, Brown K, Luty A, et al. Disturbances in selective information processing associated with the val66met Polymorphism: evidence from cognition, the P300 and fronto-hippocampal systems. Biol Psychology. 2009;80:176–188. doi: 10.1016/j.biopsycho.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Lau JYF, Goldman D, Buzas B, Hodgkinson C, Leibenluft E, Nelson E, et al. BDNF gene polymorphism (val66met) predicts amygdala and anterior hippocampus responses to emotional faces in anxious and depressed adolescents. Neuroimage. 2010;53:952–961. doi: 10.1016/j.neuroimage.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis NA, Cabeza R, Need AC, Waters-Metenier S, Goldstein DB, LaBar KS.Brain-derived neurotrophic factor val66met polymorphism and hippocampal activation during episodic memory encoding and retrieval tasks Hippocampus 2010. e-pub ahead of print, 20 May; doi 10:1002/hipo.20809 . [DOI] [PMC free article] [PubMed]
- Gerritsen L, Tendolkar I, Franke B, Vasquez AA, Buitelaar J, Fernandez G, et al. BDNF val66met genotype modulates the effect of childhood adversity on subgenual anterior cingulate cortex volume in healthy subjects Mol Psychiatry 2011. e-pub ahead of print, 17 May; doi 10:10.1038/mp.2011.51. [DOI] [PubMed]
- Bremner JD, Elzinga BM, Schmahl C, Vermetten E. Structural and functional plasticity of the human brain in post-traumatic stress disorder. Prog Brain Res. 2007;167:171–186. doi: 10.1016/S0079-6123(07)67012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatt JM, Nemeroff CB, Dobson-Stone C, Paul RH, Bryant RH, Schofield R, et al. Interactions between BDNF Val66Met polymorphism predict brain and arousal pathways to syndromal depression and anxiety. Mol Psychiatry. 2009;14:681–695. doi: 10.1038/mp.2008.143. [DOI] [PubMed] [Google Scholar]
- Juhasz G, Dunham JS, McKie S, Thomas E, Downey D, Chase D, et al. The CREB1-BDNF-NTRK2 pathway in depression: Multiple gene-cognition-environment interactions Biol Psychiatry 201069762–771.e-pub ahead of print, 18 November; doi: 10.1016/j.biopsych. 2010.11.019 [DOI] [PubMed] [Google Scholar]
- Penninx BJW, Beekman ATH, Smit JH, Zitman FG, Nolen WA, Spinhoven P, et al. For the NESDA Research Consortium. The Netherlands Study of Depression and Anxiety (NESDA): rationale, objectives and methods. Int J Methods Psychiatr Res. 2008;17:121–140. doi: 10.1002/mpr.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tol MJ, van der Wee NJ, van den Heuvel OA, Demenescu LR, Nielen MM, Renken R, et al. Regional brain volume in depression and anxiety disorders Arch Gen Psychiatry 2010671002–1011.e-pub ahead of print October 2010; doi: 10.1001/archgen psychiatry. 2010.121.e [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association 1994Diagnostic and Statistical Manual of Mental Disorders4th ednAmerican Psychiatric Association: Washington, DC [Google Scholar]
- World Health Organization (WHO) Collaborating Centre for Drug Statistics Methodology. Anatomical Therapeutic Chemical (ATC) Classification, 2008. . http://www.whocc.no/atcddd/ (accessed January 2011).
- Wittchen HU, Robins LN, Cottler LB, Sartorius N, Burke JD, Regier D. Feasibility, reliability and sources of variance of the Composite International Diagnostic Interview (CIDI). The multicentre WHO/ADAMHA field trials. Br J Psychiatry. 1991;159:645–653. doi: 10.1192/bjp.159.5.645. [DOI] [PubMed] [Google Scholar]
- Wacker HR, Battegay R, Mullejans R, Schlosser C.2006Using the CIDI-C in the general populationIn: Stefanis CN, Rabavilas AD, Soldatos CR (eds).Psychiatry: A World Perspective Elsevier Science Publishers: Amsterdam [Google Scholar]
- Montgomery SA, Asberg MA. A new depression scale designed to be sensitive to change. Br J Psychiatry. 1979;134:382–389. doi: 10.1192/bjp.134.4.382. [DOI] [PubMed] [Google Scholar]
- Beck AT, Epstein N, Brown G, Steer RA. An inventory for measuring clinical anxiety: psychometric properties. J Consult Clin Psychol. 1988;56:893–897. doi: 10.1037//0022-006x.56.6.893. [DOI] [PubMed] [Google Scholar]
- Davidson J, Turnbull CD, Strickland R, Miller R, Graves K. The Montgomery-Asberg depression scale: reliability and validity. Acta Psychiatr Scand. 1986;73:544–548. doi: 10.1111/j.1600-0447.1986.tb02723.x. [DOI] [PubMed] [Google Scholar]
- Kabacoff RI, Segal DL, Hersen M, van Hasselt VB. Psychometric properties and diagnostic utility of the Beck anxiety inventory and the state-trait anxiety inventory with older adult psychiatric outpatients. J Anx Dis. 1997;11:33–47. doi: 10.1016/s0887-6185(96)00033-3. [DOI] [PubMed] [Google Scholar]
- de Graaf R, Bijl RV, ten Have M, Beekman ATF, Vollebergh WAM. Pathways to comorbidity: the transition of pure mood, anxiety and substance abuse disorder into comorbid conditions in a longitudinal population based study. J Affect Dis. 2004a;82:461–467. doi: 10.1016/j.jad.2004.03.001. [DOI] [PubMed] [Google Scholar]
- de Graaf R, Bijl RV, ten Have M, Beekman ATF, Vollebergh WAM. Rapid onset of comorbidity of common mental disorders: findings from the Netherlands Mental Health Survey and Incidence Study (NEMESIS) Acta Psychiatr Scand. 2004b;109:55–63. doi: 10.1046/j.0001-690x.2003.00222.x. [DOI] [PubMed] [Google Scholar]
- Wiersma JE, Hovens JGFM, van Oppen P, Giltay EJ, van Schaik JF, Beekman ATF, et al. The importance of childhood trauma and childhood life events for chronicity of depression in adults. J Clin Psychiatry. 2009;70:983–989. doi: 10.4088/jcp.08m04521. [DOI] [PubMed] [Google Scholar]
- Boomsma DI, Willemsen G, Sullivan PF, Heutink P, Meijer P, Sondervan D. Genome-wide association of major depression: description of samples for the GAIN major depressive disorder study: NTR and NESDA biobank projects. Eur J Hum Genet. 2008;16:335–342. doi: 10.1038/sj.ejhg.5201979. [DOI] [PubMed] [Google Scholar]
- Daselaar SM, Veltman DJ, Rombouts SA, Raaijmakers JG, Jonker C. Neuroanatomical correlates of episodic encoding and retrieval in young and elderly subjects. Brain. 2003;126:43–56. doi: 10.1093/brain/awg005. [DOI] [PubMed] [Google Scholar]
- van Tol M-J, Demenescu LR, van der Wee NJA, Kortekaas R, Nielen MMA, Den Boer JA, et al. Emotional word encoding and recognition in depression and anxiety disorders Biol Psychiatry 2011; e-pub ahead of print, 28 December; doi: 10.1016/j.biopsych.2011.11.016 [DOI] [PubMed]
- Brett M, Anton J-L, Valabregue R, Poline JB.Region of interest analysis using an SPM toolbox [abstract]. Presented at the 8th International Conference on Functional Mapping of the Human Brain; 2–6 June 2002; Sendai, Japan. Available on CD-ROM in NeuroImage 2002; 16.
- Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. Neuroimage. 2003;19:1233–1239. doi: 10.1016/s1053-8119(03)00169-1. [DOI] [PubMed] [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Cohen J. Statistical Power Analysis for the Behavioral Sciences. Lawrence Erlbaum Associates: Hillsdale, NJ; 1988. [Google Scholar]
- Banner H, Bhat V, Etchamendy N, Joober R, Bohbot VD. The brain-derived neurotrophic factor val66met polymorphism is associated with reduced functional magnetic resonance imaging activity in the hippocampus and increase use of caudate nucleus-dependent strategies in a human virtual navigation task. Eur J Neurosci. 2011;33:968–977. doi: 10.1111/j.1460-9568.2010.07550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichers M, Kenis G, Jacobs N, Myin-Germeys I, Schruers K, Mengelers R, et al. The psychology of psychiatric genetics: evidence that positive emotions in females moderate genetic sensitivity to social stress associated with the BDNF val66met polymorphism. J Ab Psychology. 2008;117:699–704. doi: 10.1037/a0012909. [DOI] [PubMed] [Google Scholar]
- Werner NS, Meindl T, Engel RR, Rosner R, Riedel M, Reiser M, et al. Hippocampal function during associative learning in patients with posttraumatic stress disorder. J Psychiatric Res. 2009;43:309–318. doi: 10.1016/j.jpsychires.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Thomaes K, Dorrepaal E, Draijer NPJ, de Ruiter MB, Elzinga BM, van Balkom AJ, et al. Increased activation of the left hippocampus region in complex PTSD during encoding and recognition of emotional words: a pilot study. Psychiatric Res: Neuroimaging. 2009;171:44–53. doi: 10.1016/j.pscychresns.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Heim C, Shugart M, Craighead WE, Nemeroff CB. Neurobiological and psychiatric consequences of childhood abuse and neglect. Dev Psychobiology. 2010;52:671–690. doi: 10.1002/dev.20494. [DOI] [PubMed] [Google Scholar]
- Verhagen M, van der Meij A, van Deurzen PAM, Janzing JGE, Arias-Vasquez A, et al. Meta-analysis of the BDNF val66met polymorphism in major depressive disorder: effects of gender and ethnicity. Mol Psychiatry. 2010;15:260–271. doi: 10.1038/mp.2008.109. [DOI] [PubMed] [Google Scholar]
- Eyler LT, Sherzai A, Kaup AR, Jeste DV.A review of functional brain imaging correlates of successful cognitive aging Biol Psychiatry 201170115–122.e-pub ahead of print, 13 December 2010; doi: 10.1016/j.biopsych.2010.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Campbell S, Marriott M, Nahmias C, Macqueen GM. Lower hippocampal volumes in patients suffering from depression: a meta-analysis. Am J Psychiatry. 2004;161:598–607. doi: 10.1176/appi.ajp.161.4.598. [DOI] [PubMed] [Google Scholar]
- Videbech P, Ravnkilde B. Hippocampal volume and depression: a meta-analysis of MRI studies. Am J Psychiatry. 2004;161:1957–1966. doi: 10.1176/appi.ajp.161.11.1957. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.