Abstract
Bartonella spp. were detected in rats (Rattus norvegicus) trapped in downtown Los Angeles, California, USA. Of 200 rats tested, putative human pathogens, B. rochalimae and B. tribocorum were found in 37 (18.5%) and 115 (57.5%) rats, respectively. These bacteria among rodents in a densely populated urban area are a public health concern.
Keywords: Bartonella, bacteria, genetic diversity, gltA, rats, Rattus norvegicus, zoonoses, Los Angeles
Bartonella spp. are vector-borne bacteria associated with an increasing array of emerging zoonotic infections in humans and animals (1). Some Bartonella spp. are widely distributed among small mammals in the United States and potentially cause human health concerns because these bacteria may be associated with human diseases (2). However, limited surveys have been conducted to identify infectious agents involved in zoonotic infections in rodents within urban areas of the United States (3). Norway rats (Rattus norvegicus) have been shown to harbor several Bartonella species, including B. tribocorum, B. elizabethae, B. rattimassiliensis, B. phoceensis, B. queenslandensis, and a strain closely related to B. rochalimae (3–6), of which B. elizabethae, B. rochalimae, and B. tribocorum were implicated in human diseases (7–11).
Our study had 3 purposes. The first purpose was to investigate prevalence of Bartonella spp. infections in rodent populations in downtown Los Angeles, California, USA. The second purpose was to evaluate genetic diversity of Bartonella spp. in blood of urban rats by analyzing variations of the citrate synthase (gltA) gene. The third purpose was to compare rates of detection of Bartonella spp. infections in rats between culture and molecular assays.
The Study
Rats were sampled during 2003–2007 at 16 sites in downtown Los Angeles in a rodent management/disease surveillance program. The rats were captured by using Tomahawk live traps (Tomahawk Live Trap Co., Hazelhurst, WI, USA) and anesthetized with CO2. Blood samples were obtained by cardiac puncture, transferred to sterile cryovials, placed on dry ice, and preserved at −70°C until testing at the Bartonella Laboratory of the Centers for Disease Control and Prevention (Fort Collins, CO, USA).
Genomic DNA was extracted from animal blood according to the blood protocol of the QIAamp DNA Mini Kit (QIAGEN, Valencia, CA, USA). Primers CS140f and CS443f (12) and CS781f and CS1137n (13), including newly designed primers CS120f-5′-TTTCACTTATGATCCTGGCTT-3′ and CS1210r-5′-GATCYTCAATCATTTCTTTCCA-3′, which amplify DNA fragments ranging from 380 to 1,091 bp encompassing a 327-bp–specific zone (between bp positions 801 and 1127) of the gltA gene (14), were used in different combinations.
Eight PCRs were performed for each sample. PCR products were purified by using the QIAquick PCR Purification Kit (QIAGEN, Germantown, MD, USA). The same primers were used for DNA sequencing. The obtained DNA sequences were trimmed to 327 bp between positions 801 and 1127 because most Bartonella spp. gltA sequences available in GenBank are limited to this size.
Details of procedures used for isolation of Bartonella spp. from mammalian blood have been described (2). Rat blood was diluted 1:4 in brain–heart infusion medium containing 5% fungizone and pipetted onto heart infusion agar plates containing 10% rabbit blood. Plates were incubated aerobically at 35°C in an atmosphere of 5% CO2 for <4 weeks and monitored for bacterial growth at least 1× per week after initial plating. Colonies were subpassaged onto fresh agar plates until a pure culture, free from contamination, was obtained.
Analysis of DNA sequences and phylogenetic relationships was performed by using MEGA4 (www.megasoftware.net/) and the neighbor-joining method with the Kimura 2-parameter distance model. Stability of inferred phylogeny was assessed by using bootstrap analysis of 1,000 randomly generated trees. DNA sequences obtained in this study were deposited in GenBank under accession nos. JF429450–JF429625.
A total of 200 R. norvegicus rats were trapped in 16 sites in downtown Los Angeles. Bartonella DNA was detected in 135 (67.5%) of 200 rat blood samples. PCR-positive blood samples were subsequently cultured for viable Bartonella organisms. Fifty-nine (43.7%) of 135 blood samples were confirmed as bacteremic for bartonellae by culturing.
A total of 176 sequences were obtained either directly from blood samples (117 sequences) or from pure cultures (59 sequences). Among 176 gltA sequences, 23 genotypes with >1 nt difference were found, and sequence similarity between genotypes ranged from 85.3% to 99.7%. These 23 genotypes were closely related to B. tribocorum (n = 16), B. rochalimae (n = 2), B. queenslandensis (n = 1), or 4 potentially novel genotypes. A total of 130 DNA sequences (JF429450–JF429579) obtained from 115 rats were grouped into 16 genotypes (1–16). These genotypes showed 98.2%–99.7% sequence similarity with each other and were genetically related to B. tribocorum IBS506T (AJ005494) (97.9%–100% identity). All 16 genotypes clustered with B. tribocorum with a high bootstrap value, as shown in the phylogenetic tree (Figure A1).
Two genotypes (22 and 23) from 38 sequences (JF429588–JF429625) obtained from 37 rats had a similarity between 98.5% and 98.8% to B. rochalimae BMGH (DQ683195). These 2 genotypes joined with B. rochalimae with a high bootstrap branching pattern in phylogenetic analysis (Figure A1). The B. queenslandensis group consisted of a single genotype (genotype 17) from 4 sequences (JF429580–JF429583) from 4 rats. This genotype demonstrated a sequence similarity to B. queenslandensis AUST/NH12T (EU111800) of 99.0% for the gltA gene (Figure A1).
Conclusions
In this study, 3 novel Bartonella genogroups identified in 4 R. norvegicus rats were not genetically related to any known Bartonella spp. and might represent novel Bartonella spp. Genotypes 18 (JF429586) and 19 (JF429587) formed a Bartonella genogroup with 95% similarity to B. tribocorum as the closest species. Likewise, genotypes 20 (JF429584) and 21 (JF429585) have 91.4% and 92.6% sequence identities, respectively, with B. clarridgeiae, which is their closest related species (Figure A1).
Of 135 PCR-positive blood samples, 59 yielded cultures of 3 Bartonella spp.: B. tribocorum (n = 54), B. rochalimae (n = 4), and B. queenslandensis (n = 1). The number of CFU per 100 µL of blood varied: 800–1,400 CFU for B. tribocorum, 160–240 CFU for B. rochalimae, and 100 CFU for B. queenslandensis. Although 4 rats were positive by PCR for B. queenslandensis, this organism was cultured from only 1 of these animals.
Our study reports detection and identification of Bartonella spp. in urban rats in downtown Los Angeles, California. We demonstrated that R. norvegicus in downtown Los Angeles can serve as reservoirs of several Bartonella spp., such as B. rochalimae, B. tribocorum, B. queenslandensis, and possibly 3 additional novel species. Some genotypes identified in rats showed a high level of similarity (<98.8%) with a B. tribocorum isolate obtained from a febrile patient in Thailand (GenBank accession no. GQ225706) (11). Another species (B. rochalimae) was isolated from a patient with splenomegaly who had traveled to South America (14). This bacterium has also been isolated from dogs, foxes, rats, shrews, gerbils, and raccoons, suggesting that multiple reservoirs may be involved in maintenance of this species. One Bartonella genotype found in 4 rats in this study was 99.0% similar to B. queenslandensis (GenBank accession no. EU111800), which was originally isolated from R. fuscipes rats in Australia (15).
Because most identified Bartonella species have been reported as human infectious agents elsewhere, the finding that they were circulating among rodents in a densely populated urban area is of serious public health concern. In this context, further studies should be conducted on a larger collection of rodents and clinical human samples to determine evolutionary, genetic, and pathogenic relationships.
Acknowledgment
We thank Robert Flores for assistance with rat trapping and processing.
This study was supported in part by an appointment to the Emerging Infectious Diseases Fellowship Program administered by the Association of Public Health Laboratories and funded by the Centers for Disease and Control and Prevention.
Biography
Dr Gundi is an Emerging Infectious Diseases Fellow at the Bartonella Laboratory, Division of Vector-Borne Diseases, Centers for Disease Control and Prevention, Fort Collins, Colorado. His research interests include tropical and emerging infectious diseases.
Figure A1.
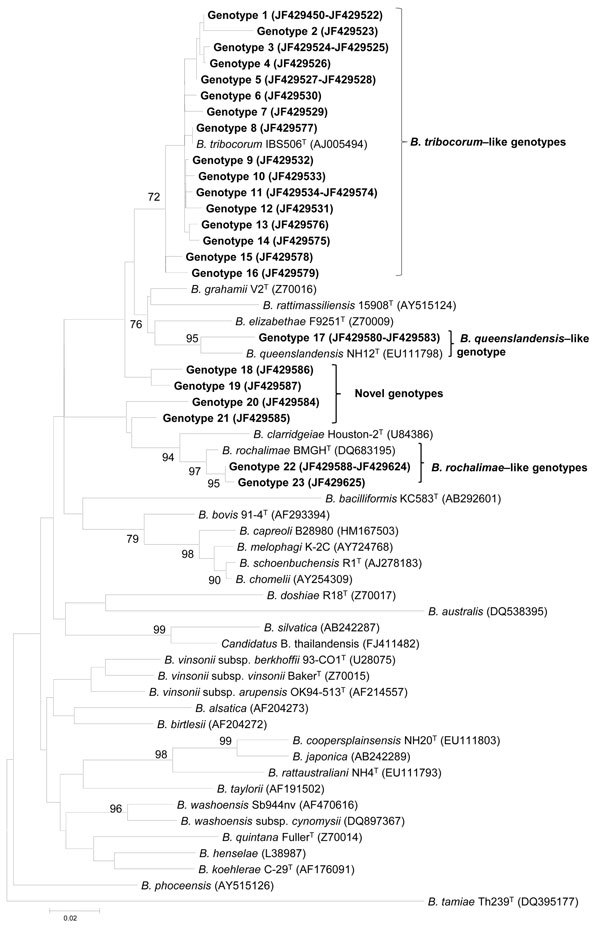
Phylogenetic classification of Bartonella spp. genotypes detected in Rattus norvegicus rats in downtown Los Angeles, California, USA, on the basis of citrate synthase (gltA) gene sequences. The phylogram was constructed by using the neighbor-joining method with the Kimura 2-parameter distance model. Only bootstrap values >70% obtained from 1,000 replicates are given. GenBank accession numbers for reference sequences are indicated in parentheses. Scale bar indicates nucleotide substitutions per site. Boldface indicates genotypes identified in rats.
Footnotes
Suggested citation for this article: Gundi VAKB, Billeter SA, Rood MP, Kosoy MY. Bartonella spp. in rats and zoonoses, Los Angeles, California, USA. Emerg Infect Dis [serial on the Internet]. 2012 Apr [date cited]. http://dx.doi.org/10.3201/eid1804.110816
References
- 1.Breitschwerdt EB, Maggi RG, Chomel BB, Lappin MR. Bartonellosis: an emerging infectious disease of zoonotic importance to animals and human beings. J Vet Emerg Crit Care (San Antonio). 2010;20:8–30. 10.1111/j.1476-4431.2009.00496.x [DOI] [PubMed] [Google Scholar]
- 2.Kosoy MY, Regnery RL, Tzianabos T, Marston EL, Jones DC, Green D, et al. Distribution, diversity, and host specificity of Bartonella in rodents from the southeastern United States. Am J Trop Med Hyg. 1997;57:578–88. [DOI] [PubMed] [Google Scholar]
- 3.Ellis BA, Regnery RL, Beati L, Bacellar F, Rood M, Glass GG, et al. Rats of the genus Rattus are reservoir hosts for pathogenic Bartonella species: an Old World origin for a New World disease? J Infect Dis. 1999;180:220–4. 10.1086/314824 [DOI] [PubMed] [Google Scholar]
- 4.Heller R, Riegel P, Hansmann Y, Delacour G, Bermond D, Dehio C, et al. Bartonella tribocorum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int J Syst Bacteriol. 1998;48:1333–9. 10.1099/00207713-48-4-1333 [DOI] [PubMed] [Google Scholar]
- 5.Gundi VA, Davoust B, Khamis A, Boni M, Raoult D, La Scola B. Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J Clin Microbiol. 2004;42:3816–8. 10.1128/JCM.42.8.3816-3818.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin JW, Chen CY, Chen WC, Chomel BB, Chang CC. Isolation of Bartonella species from rodents in Taiwan including a strain closely related to ‘Bartonella rochalimae’ from Rattus norvegicus. J Med Microbiol. 2008;57:1496–501. 10.1099/jmm.0.2008/004671-0 [DOI] [PubMed] [Google Scholar]
- 7.Daly JS, Worthington MG, Brenner DJ, Moss CW, Hollis DG, Weyant RS, et al. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comer JA, Diaz T, Vlahov D, Monterroso E, Childs JE. Evidence of rodent-associated Bartonella and Rickettsia infections among intravenous drug users from central and east Harlem, New York City. Am J Trop Med Hyg. 2001;65:855–60. [DOI] [PubMed] [Google Scholar]
- 9.Smith HM, Reporter R, Rood MP, Linscott AJ, Mascola LM, Hogrefe W, et al. Prevalence study of antibody to ratborne pathogens and other agents among patients using a free clinic in downtown Los Angeles. J Infect Dis. 2002;186:1673–6. 10.1086/345377 [DOI] [PubMed] [Google Scholar]
- 10.Eremeeva ME, Gerns HL, Lydy SL, Goo JS, Ryan ET, Mathew SS, et al. Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N Engl J Med. 2007;356:2381–7. 10.1056/NEJMoa065987 [DOI] [PubMed] [Google Scholar]
- 11.Kosoy M, Bai Y, Sheff K, Morway C, Baggett H, Maloney SA, et al. Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. Am J Trop Med Hyg. 2010;82:1140–5. 10.4269/ajtmh.2010.09-0778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birtles RJ, Raoult D. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int J Syst Bacteriol. 1996;46:891–7. 10.1099/00207713-46-4-891 [DOI] [PubMed] [Google Scholar]
- 13.Norman AF, Regnery R, Jameson P, Greene C, Krause DC. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.La Scola B, Zeaiter Z, Khamis A, Raoult D. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 2003;11:318–21. 10.1016/S0966-842X(03)00143-4 [DOI] [PubMed] [Google Scholar]
- 15.Gundi VA, Taylor C, Raoult D, La Scola B. Bartonella rattaustraliani sp. nov., Bartonella queenslandensis sp. nov. and Bartonella coopersplainsensis sp. nov., identified in Australian rats. Int J Syst Evol Microbiol. 2009;59:2956–61. 10.1099/ijs.0.002865-0 [DOI] [PubMed] [Google Scholar]


