Abstract
To determine possible cosavirus association with clinical disease, we used real-time reverse transcription PCR to test children and HIV-positive adults in Brazil with and without gastroenteritis. Thirteen (3.6%) of 359 children with gastroenteritis tested positive, as did 69 (33.8%) of 204 controls. Low prevalence, frequent viral co-infections, and low fecal cosavirus RNA concentrations argue against human pathogenicity.
Keywords: Picornaviridae infections, human cosavirus, Brazil, communicable diseases, virus shedding, gastroenteritis, pathogenesis, viruses
The family Picornaviridae comprises 12 genera and includes several leading pathogens affecting human and animal health, e.g., foot-and-mouth-disease virus and polioviruses. With the advent of metagenomics, 3 novel human picornaviruses have been described since 2008: klassevirus, salivirus, and cosavirus (1–3).
Besides being detected in raw sewage (4), cosaviruses have been detected in human fecal specimens and were tentatively associated with gastroenteritis in 2 patients from Australia and Scotland (3,5). Cosavirus pathogenicity in humans has remained unknown, however, because detection rates in patients and healthy controls were similar in the only available cohort studies of patients with acute flaccid paralysis in Southeast Asia (3) and with gastroenteritis in China (6). We investigated cosavirus prevalence in 464 children and HIV-infected adults with gastroenteritis and 253 controls without gastroenteritis in Brazil.
The Study
Fecal specimens from 6 clinical cohorts in Salvador, northeastern Brazil, were analyzed (Table 1). Adult cohorts comprised HIV-infected patients with gastroenteritis (105 persons) and without (49 persons) gastroenteritis. Child cohorts included 359 children with gastroenteritis and 204 healthy children from child-care centers. Nasal swab specimens were obtained from controls attending child-care centers and from children with gastroenteritis and concomitant respiratory symptoms. All specimens were stored at −30° to −80°C until further processing.
Table 1. Clinical cohorts tested for cosavirus, Salvador, Brazil*.
| Cohort no. | Cohort description† | Sampling site‡ | Sampling time | Participant age, mo, mean (SD) | No. participants | No. (%) RT-PCR positive§ | Virus concentration, log10 RNA copies/g feces, mean (SD)¶ |
|---|---|---|---|---|---|---|---|
| 1 | HIV-infected adults with gastroenteritis | Infectious Diseases HIV Outpatient Department | 2007 Mar–2010 Mar | 487.6 (114.4) | 105 | 1 (1.0) | 4.43 |
| 2 | HIV-infected adults without gastroenteritis | 2007 Mar–2010 Mar | 533.8 (115.9) | 49 | 0 | – | |
| 3 | Children with gastroenteritis | Department of Pediatrics Metabolic Unit# | 2006 Feb–2007 Sep | 19.0 (15.6) | 359 | 13 (3.6) | 3.40 (0.93) |
| 4 | Control children without gastroenteritis | Community child-care center** | 2008 Dec | 29.6 (13.1) | 132 | 65 (49.2) | 2.97 (0.97) |
| 5 | Control children without gastroenteritis | Community child-care center** | 2011 Nov–2011 Dec | 14.3 (5.5) | 62 | 4 (6.5) | 3.41 (0.49) |
| 6 | Control children without gastroenteritis | University child-care center** | 2011 Oct–2011 Nov | 18.6 (4.2) | 10 | 0 | – |
| Total | 717 | 83 (11.6) |
*RT-PCR, reverse transcription PCR; –, no virus obtained. †Gastroenteritis was defined as acute diarrhea with >3 watery stools in the previous 24 h and within 13 d before hospital admission. ‡All hospital units were located within the Hospital Professor Edgard Santos, Federal University of Bahia. All sites were located in Salvador, Bahia, in northeastern Brazil. §Samples only considered if positive in nested RT-PCR as in (3) and in strain-specific real-time RT-PCR. ¶Measured by strain-specific RT-PCR. #Reports of diarrhea in the preceding 2 wk served as an exclusion factor. **Written consent was obtained from adult family members.
Viral RNA was purified from ≈200 mg fecal specimen and 140 µL nasal swab specimen suspended in phosphate-buffered saline by using the Viral RNA Mini kit (QIAGEN, São Paulo, Brazil) as described (7). A nested reverse transcription PCR (3) detected cosavirus. After nucleotide sequencing of all PCR-positive specimens, we developed a specific real-time RT-PCR for quantifying viruses from Brazil (Table 2). Assay optimization and quantification relied on photometrically quantified cRNA in vitro transcripts, as described (8). After assay optimization, sensitivity was 6.8 copies per reaction.
Table 2. Oligonucleotides used for detection and quantification of cosaviruses*.
| Oligonucleotide identity | Sequence, 5′ → 3′ | Genomic target region, RT-PCR type | Use | Reference |
|---|---|---|---|---|
| DKV-N5U-F1 | CGTGCTTTACACGGTTTTTGA (+) | 5’-UTR, nested RT-PCR 1st round | Cosavirus detection† | (3) |
| DKV-N5U-R2 | GGTACCTTCAGGACATCTTTGG (–) | |||
| DKV-N5U-F2 | ACGGTTTTTGAACCCCACAC (+) | 5’-UTR, nested RT-PCR 2nd round | ||
| DKV-N5U-R3 | GTCCTTTCGGACAGGGCTTT (–) | |||
| HCosV-rtF735-1 | TTGTAGYGATGCTGTRTGTGTGTG (+) | 5’-UTR, real time RT-PCR | Brazilian cosavirus quantification† | This study |
| HCosV-rtP783 | FAM-AGCCTCACAGGCCRRAAGCCCTGTC-DDQ1 (+, Probe) | |||
| HCosV-rtR827-1 | CCAYTGTGTGGGTCCTTTCG (–) |
*RT-PCR, reverse transcription PCR; UTR, untranslated region; FAM, fluorescein; R, G/A; DDQ1, deep dark quencher 1; Y, C/T. †RT-PCR reactions were carried out using the QIAGEN One-step RT-PCR kit as described by the manufacturer (QIAGEN, São Paulo, Brazil), 300 nmol/L of each primer, 200 nmol/L of the probe (real time RT-PCR assay), 1 μg bovine serum albumin, and 5 μL RNA extract. Second-round reactions used the Platinum Taq DNA Polymerase Kit as described by the manufacturer (Invitrogen, São Paulo, Brazil) with 2.5 mol/L MgCl and 1 µL of first-round PCR product. Real time RT-PCR amplification involved 55°C for 15 min, 95°C for 15 min, and 45 cycles of 95°C for 15 s and 58°C for 30 s (fluorescence measured).Nested RT-PCR involved 30 min at 50°C; 15 min at 95°C; 10 cycles of 20 s at 94°C, 30 s starting at 60°C with a decrease of 1°C per cycle, and 50 s at 72°C; and 40 cycles of 20 s at 95°C, 30 s at 54°C, and 50 s at 72°C; and a final elongation step of 5 min at 72°C. Second-round reactions used 3 min at 94°C and thermal cycling as for the first round.
In the adult cohorts, 1 of 105 HIV-infected persons with gastroenteritis had positive results for cosavirus. In the control group of 49 HIV-infected adults without gastroenteritis, none had positive cosavirus results (χ2 0.5; p = 0.49).
Considerably higher prevalence was detected among child cohorts. Of children with gastroenteritis, 13 (3.6%) of 359 patients were cosavirus positive. The proportion of cosavirus-positive controls without gastroenteritis, sampled in 2008, was significantly higher: 65 (49.2%) of 132 controls were cosavirus positive (χ2 149.1; p<0.0001). On resampling in 2011, 4 (6.5%) of 62 controls were cosavirus positive. Although the difference was not statistically significant (χ2 1.1; p = 0.3), this result was almost double that for patients. In another child-care center, none of 10 healthy children were cosavirus positive.
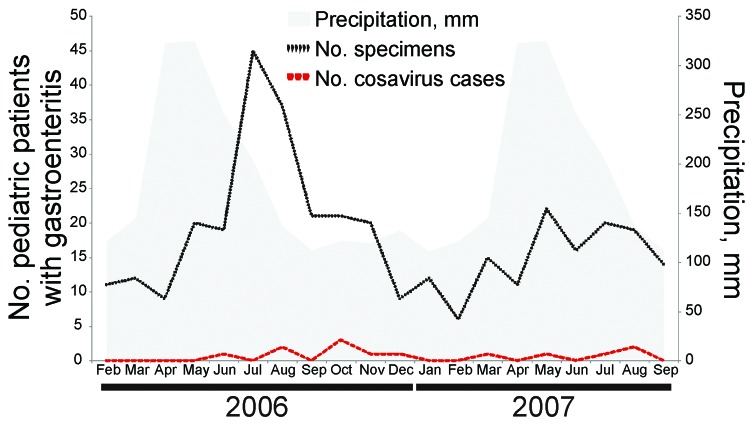
The higher prevalence detected in controls in 2008 could indicate a seasonal infection pattern because specimens were collected in a single weekend in a child-care center. The lower cosavirus-positive results in the same child-care center 3 years after the initial sampling support this idea. However, in sick children, cosavirus-positive specimens were obtained at similarly low rates throughout the year (Figure 1), which might argue against seasonal variation as a generic property of cosavirus infection.
Figure 1.
Detection pattern of cosavirus in children with gastroenteritis throughout different seasons during 2006–2007, Brazil. Temperature was not plotted because it varied little from mean 25.2°C through the year (range 23.6–26.7°C). Precipitation data were obtained from the German Weather Service and represent means throughout 1961–1990.
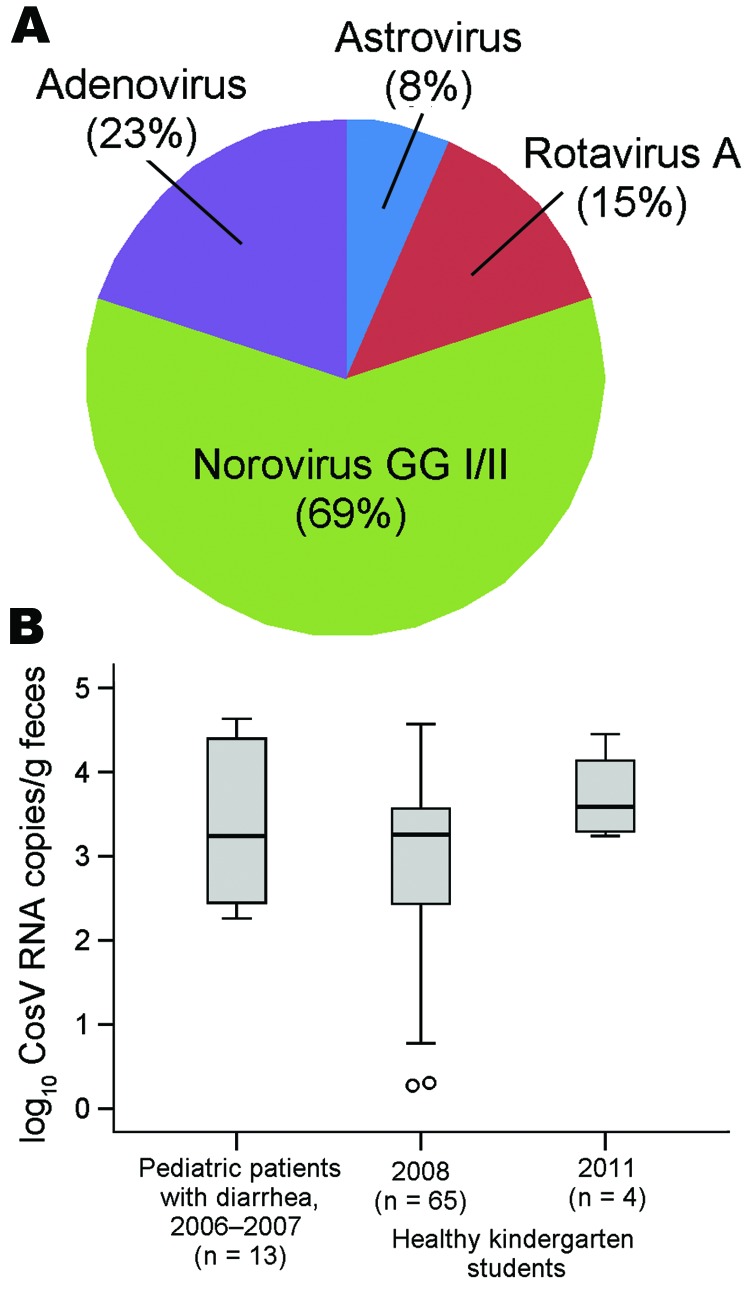
To evaluate whether cosavirus causes disease in ill children, we analyzed co-infections with common viral pathogens causing diarrhea (astrovirus, norovirus, rotavirus, adenovirus) (Figure 2, panel A). In 10 (76.9%) of 13 cosavirus-positive patients, >1 of these pathogens could be detected.
Figure 2.
Co-infections and fecal cosavirus (CosV) RNA concentrations. A) Co-infections with established viral causes of diarrhea in children with gastroenteritis who were positive for CosV. Viral RNA and DNA were detected by real-time PCR (methods available upon request) in the same eluates used for CosV detection. B) Boxplot generated with SPSS V19 (SPSS, Munich, Germany) of log10 CosV RNA concentrations per gram of feces in children with gastroenteritis and healthy control children from a child-care center in 2008 and 2011. Boxes show the medians and interquartile ranges (box length). The whiskers represent an extension of the 25th or 75th percentiles by 1.5× the interquartile range. Datum points beyond the whisker range are considered as outliers and marked as circles. GGI/II, genogroups I and II.
Acute infection with an enteric virus is usually associated with high viral RNA shedding. Cosavirus concentrations were low in outpatient and control children (≈103 copies/g) on 2 sampling occasions (Table 1) and did not differ significantly (analysis of variance F 2.0; p = 0.14; Figure 2, panel B).
Low cosavirus concentrations in the enteric tract could result from swallowing viruses originating in the respiratory tract. Therefore, we tested for cosavirus in 96 nasal swab specimens from gastroenteritis patients, including 3 nasal swab specimens from persons with cosavirus-positive fecal specimens and 65 nasal swab specimens from cosavirus-positive controls sampled in 2008. None of the nasal specimens from controls with positive fecal specimens and only 1 nasal specimen from a patient unrelated to the 3 aforementioned persons was weakly positive (103 RNA copies/mL), which refutes this hypothesis.
To analyze whether a preceding point-source infection caused high cosavirus prevalence in the controls without gastroenteritis sampled in 2008, we determined the genomic sequence of the 5′ untranslated region PCR amplicons and phylogenetically analyzed the sequence (GenBank accession no. JN228118–JN228188). Cosaviruses from these controls were distributed across the phylogenetic tree (Technical Appendix). Maximum nucleotide distance within these cosaviruses was up to 22.5% in the analyzed 398-nt fragment, making a recent point-source infection unlikely.
Conclusions
Human cosavirus infections were reported previously from a limited number of persons and geographic areas (3–6). In Brazil, the 3.6% detection rate in children with gastroenteritis was comparable to the 1.8% rate in a cohort study of gastroenteritis patients in China (6). Although the 6.5% detection rate in 1 control cohort in Brazil was compatible with the 1.7% rate in 60 healthy controls in China, the combined 33.8% prevalence detected in controls from 3 different samplings in Brazil was much higher. Nonetheless, the prevalence was comparable to the 43.9% detected in 41 healthy Southeast Asian children in the only other cohort study (3). Detecting cosavirus in 1 of 154 adults in Brazil was compatible with finding a single cosavirus-positive patient among 1,000 adults with gastroenteritis in Scotland, confirming that cosaviruses are rare and probably neither pathogenic nor commensal in adults (3).
The higher prevalence of cosavirus found in controls than in patients, the frequent co-infections with established pathogens, and the unusually low RNA virus concentrations give evidence against cosavirus involvement in human gastroenteritis. Viruses that replicate in the human gut generally reach concentrations 1,000- to 100,000-fold higher than those of cosavirus. This finding is exemplified by genetically related picornaviruses (Aichi viruses, parechoviruses, and cardioviruses) and established enteric pathogens (e.g., noroviruses and rotaviruses) (8–12). Notably, the aforementioned study on cardioviruses included the same specimens from Brazil, which indicates that poor sample quality was not a factor.
These low concentrations would be compatible with absence of replication in the enteric tract and passive virus ingestion, e.g., from nutritional sources, drinking water, or the respiratory tract. However, nutritional patterns of the tropical countries in which cosavirus have been detected certainly differ. Furthermore, in Brazil, adults are unlikely to have a completely different diet from infants and children. Moreover, the unprecedented detection of cosavirus in a respiratory tract specimen makes ingestion of viruses from nutritional sources alone unlikely, although a link to fluid droplets from drinking water in the respiratory tract is hypothetically possible.
Another explanation for low cosavirus RNA levels in fecal samples is that a cosavirus infection occurred early in the person’s life and produced partial mucosal immunity and limited subsequent cosavirus replication in the gut. This is exemplified for viruses transmitted by the fecal–oral route by up to 100-fold higher fecal shedding of vaccine rotavirus and poliovirus among seronegative persons than among seropositive or previously vaccinated persons (13,14). However, this explanation would be incompatible with the high prevalence of cosavirus in many control children, who were generally older than patients.
Prolonged low concentrations of picornavirus shedding has been demonstrated, e.g., by detectable hepatitis A virus RNA up to 3 months after acute infection (15). Nonetheless, this circumstance is unlikely to explain the low cosavirus concentrations, given the overall high number of persons with positive results.
Although our study extends the known geographic occurrence of cosavirus, whether it is a human pathogen remains to be determined. Future studies would be enhanced by serologic analyses and investigations focusing on nutrition and drinking water in tropical countries.
Supplementary Material
Cosavirus (CosV) partial 5′ untranslated region phylogeny.
Acknowledgments
We thank Tobias Bleicker, Sebastian Brünink, Monika Eschbach-Bludau, Célia Pedroso, Carlos Brites, Vanusa dos Santos Estrela, Maria Goreth Barberino, Ana-Rute Santos Oliveira, and Milena Carvalho Bastos for outstanding support and Alexander N. Lukashev for helpful suggestions. We are also grateful to the child-care center staff, children, and parents involved in this study.
The study was funded by the Foundation for Research Support of the State of Bahia (Fundação de Amparo à Pesquisa do Estado da Bahia), project codes APR 125/2006/ethics committee protocol 120/2005 and SUS0004/2007/ethics committee protocol 06/2007, and the European Union FP7 project European Management Platform for Emerging and Re-emerging Infectious Disease Entities (grant agreement no. 223498).
Biography
Dr Stöcker is a physician affiliated with the Federal University of Bahia, Salvador, Brazil, and the University of Bonn, Germany. His primary research interest is the implementation of molecular diagnostics in resource-limited settings.
Footnotes
Suggested citation for this article: Stöcker A, Souza BFCD, Ribeiro TCM, Netto EM, Araujo LC, Corrêa JI, et al. Cosavirus infection in persons with and without gastroenteritis, Brazil. Emerg Infect Dis [serial on the Internet]. 2012 Apr [date cited]. http://dx.doi.org/10.3201/eid1804.111415
These authors contributed equally to this article.
References
- 1.Li L, Victoria J, Kapoor A, Blinkova O, Wang C, Babrzadeh F, et al. A novel picornavirus associated with gastroenteritis. J Virol. 2009;83:12002–6. 10.1128/JVI.01241-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greninger AL, Runckel C, Chiu CY, Haggerty T, Parsonnet J, Ganem D, et al. The complete genome of klassevirus—a novel picornavirus in pediatric stool. Virol J. 2009;6:82. 10.1186/1743-422X-6-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapoor A, Victoria J, Simmonds P, Slikas E, Chieochansin T, Naeem A, et al. A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc Natl Acad Sci U S A. 2008;105:20482–7. 10.1073/pnas.0807979105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blinkova O, Rosario K, Li L, Kapoor A, Slikas B, Bernardin F, et al. Frequent detection of highly diverse variants of cardiovirus, cosavirus, bocavirus, and circovirus in sewage samples collected in the United States. J Clin Microbiol. 2009;47:3507–13. 10.1128/JCM.01062-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holtz LR, Finkbeiner SR, Kirkwood CD, Wang D. Identification of a novel picornavirus related to cosaviruses in a child with acute diarrhea. Virol J. 2008;5:159. 10.1186/1743-422X-5-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai XQ, Hua XG, Shan TL, Delwart E, Zhao W. Human cosavirus infections in children in China. J Clin Virol. 2010;48:228–9. 10.1016/j.jcv.2010.03.024 [DOI] [PubMed] [Google Scholar]
- 7.Drexler JF, Baumgarte S, Luna LK, Stöcker A, Almeida PS, Ribeiro TC, et al. Genomic features and evolutionary constraints in Saffold-like cardioviruses. J Gen Virol. 2010;91:1418–27. 10.1099/vir.0.018887-0 [DOI] [PubMed] [Google Scholar]
- 8.Baumgarte S, de Souza Luna LK, Grywna K, Panning M, Drexler JF, Karsten C, et al. Prevalence, types, and RNA concentrations of human parechoviruses, including a sixth parechovirus type, in stool samples from patients with acute enteritis. J Clin Microbiol. 2008;46:242–8. 10.1128/JCM.01468-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop RF. Natural history of human rotavirus infection. Arch Virol Suppl. 1996;12:119–28. [DOI] [PubMed] [Google Scholar]
- 10.Drexler JF, Luna LK, Stocker A, Almeida PS, Ribeiro TC, Petersen N, et al. Circulation of 3 lineages of a novel Saffold cardiovirus in humans. Emerg Infect Dis. 2008;14:1398–405. 10.3201/eid1409.080570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drexler JF, Baumgarte S, de Souza Luna LK, Eschbach-Bludau M, Lukashev AN, Drosten C. Aichi virus shedding in high concentrations in patients with acute diarrhea. Emerg Infect Dis. 2011;17:1544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henke-Gendo C, Harste G, Juergens-Saathoff B, Mattner F, Deppe H, Heim A. New real-time PCR detects prolonged norovirus excretion in highly immunosuppressed patients and children. J Clin Microbiol. 2009;47:2855–62. 10.1128/JCM.00448-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson EJ. Rotavirus vaccines: viral shedding and risk of transmission. Lancet Infect Dis. 2008;8:642–9. 10.1016/S1473-3099(08)70231-7 [DOI] [PubMed] [Google Scholar]
- 14.Laassri M, Lottenbach K, Belshe R, Wolff M, Rennels M, Plotkin S, et al. Effect of different vaccination schedules on excretion of oral poliovirus vaccine strains. J Infect Dis. 2005;192:2092–8. 10.1086/498172 [DOI] [PubMed] [Google Scholar]
- 15.Yotsuyanagi H, Koike K, Yasuda K, Moriya K, Shintani Y, Fujie H, et al. Prolonged fecal excretion of hepatitis A virus in adult patients with hepatitis A as determined by polymerase chain reaction. Hepatology. 1996;24:10–3. 10.1002/hep.510240103 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cosavirus (CosV) partial 5′ untranslated region phylogeny.