Abstract
Syndapin I (PACSIN 1) is a synaptically enriched membrane tubulating protein that plays important roles in activity-dependent bulk endocytosis and neuronal morphogenesis. While syndapin I is an in vitro phosphoprotein, it is not known to be phosphorylated in neurons. Here, we report the identification of two phosphorylation sites, S76 and T181, of syndapin I from nerve terminals. Both residues are located at the N-terminal helix-capping motifs (N-Cap) of different α-helices in the F-BAR domain, important for F-BAR homodimer curvature and dimer-dimer filament assembly, respectively. Phospho-mimetic mutations of these residues regulate lipid-binding and tubulation both in vitro and in cells. Neither phosphosite regulated syndapin I function in activity-dependent bulk endocytosis. Rather, T181 phosphorylation was developmentally regulated and inhibited syndapin I function in neuronal morphogenesis. This suggests a novel mechanism for phosphorylation control of an F-BAR function through the regulation of α-helix interactions and stability within the folded F-BAR domain.
Many cellular processes require dynamic changes in the plasma membrane to form vesicles, tubules, filopodia, and large vacuole-endosome-like structures. This is achieved by recruitment of proteins to assist in shaping and remodeling of the membrane, such as the Bin/amphiphysin/Rvs (BAR) superfamily (1). BAR domains are elongated homodimers of α-helical coiled coils, characterized by a curvature with a set of positive residues on one surface that interact with phospholipid membranes (2, 3). Structural studies have given insights into the molecular mechanisms that explain the membrane binding and curvature sensing properties of BAR domains, but no regulatory signals are known to directly control BAR domain function in cells.
Syndapin I (PACSIN 1) is a neuronal-specific member of the Fes-CIP4 homology BAR (F-BAR) subfamily (4). It has an N-terminal F-BAR and a C-terminal SH3 domain that interacts with endocytic proteins such as dynamin I and cytoskeletal proteins like N-WASP (5, 6). The two domains are connected by a linker containing multiple Asn-Pro-Phe motifs that bind Eps15 homology (EH) domain-containing proteins like EHD1 (7). As with other members of the family, the F-BAR domain of syndapin binds and tubulates phospholipid liposomes, which have a large and shallow curvature (8, 9). Its crystal structure reveals distinct features in addition to those of other F-BAR proteins. Notably, the tips of its homodimer are bent away from its central body, forming a uniquely twisted S shape. This adds a second dimensional curvature predicted to play a role in determining the overall preferred membrane curvature (9, 10).
Syndapin I is critical for synaptic vesicle (SV) endocytosis, where it primarily acts through phospho-regulated binding to dynamin I (5, 11, 12). Dynamin I dephosphorylation upon strong depolarization promotes complex formation with syndapin I in nerve terminals. Blocking their interaction or knockdown of syndapin I inhibits activity-dependent bulk endocytosis (ADBE) during strong stimulation without affecting clathrin mediated endocytosis (CME) of SVs (13). Syndapin I also regulates neuronal morphogenesis by linking membrane deformation to N-WASP-dependent actin polymerization (14) and actin nucleation mediated by Cobl (15). Syndapin isoforms are in vitro substrates for protein kinase C (PKC) and casein kinase II (CKII) (4). A signaling pathway regulated by inositol hexakisphosphate (InsP6) leads to the phosphorylation of syndapin I in cells (16). Despite this, there is no current understanding of the relevance of these signaling events to protein function.
A large group of endocytic proteins are regulated by phosphorylation in nerve terminals. The best characterized are the dephosphins, which includes dynamin I (17). Other synaptic proteins such as synapsin are controlled in a similar way (18). Our goal was to determine whether syndapin I might also be regulated by phosphorylation in the nerve terminal. We found it to be phosphorylated in synaptosomes and in whole brain tissue. We identified two in vivo phosphosites. These sites controlled lipid binding and tubulation and regulated the role of syndapin I in neurite development. We propose that phosphorylation can regulate the cellular function of an F-BAR protein by directly controlling the F-BAR structure.
Results
Purification of Brain Syndapin I and Phosphosite Identification.
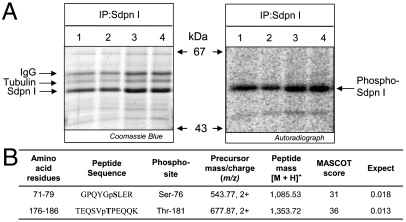
We immunoprecipitated syndapin I from unstimulated 32P-labeled rat brain synaptosome lysates, resolved the protein via SDS/PAGE, and detected its phosphorylation at 52 kDa by autoradiography (Fig. 1A). A number of phosphopeptides were identified by mass spectrometry; however, for this study we only describe the phosphorylation sites (phosphosites) located within the F-BAR domain. The peptides GPQYGSLER and TEQSVTPEQQK with phosphorylation matched to rat syndapin I at amino acids 71–79 and 176–186, respectively (Fig. 1B). The phosphopeptides and phosphosites were confirmed in both synaptosome and whole brain lysates to S76 and T181 (Fig. S1 A and B).
Fig. 1.
Purification and phosphosite analysis of syndapin I from 32P-labeled rat brain synaptosomes. (A) Syndapin I (Sdpn I) was immunoprecipitated (IP) from 32P-labeled rat brain synaptosome lysates. Proteins stained with Coomassie Blue, and autoradiography detected phosphorylated syndapin I (Phospho-Sdpn I). The quadruplicate samples are representative of three independent experiments. (B) Summary of two phosphosites identified in syndapin I by mass spectrometry (precursor mass/charge and peptide mass) followed by Mascot database searches (score and expect). The confirmed phosphosite in each peptide is bolded.
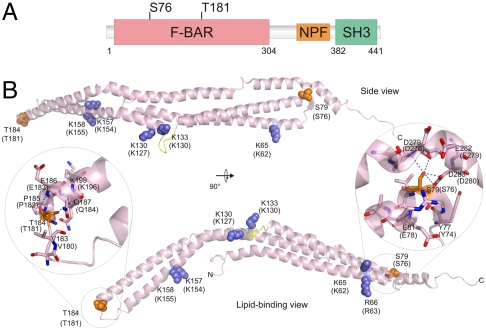
S76 and T181 Phosphosites Map to the Syndapin I F-BAR Domain.
S76 and T181 are located in the F-BAR domain (Fig. 2A). A sequence-based species alignment (Fig. S2A) of F-BAR domains from syndapin I, CIP4, FBP17, and FCHO2 showed that S76 and T181 are highly conserved between species and homologous F-BAR proteins. An alignment of crystal structures from human syndapin I (9), CIP4, (19), FBP17 (19), and FCHO2 (20) showed that the position of these residues at the beginning of an α-helix (N-cap position) was also conserved (Fig. S2B). In an N-capping motif, the side-chain hydroxyl of a Ser or Thr residue at the N-cap position forms a hydrogen bond with the backbone NH group of a residue at N-cap + 2 or N-cap + 3. The motif also includes a reciprocal hydrogen bond between the side chain of the residue at N-cap + 3 and the backbone NH of the N-cap residue thus further stabilizing the beginning of the α-helix (21, 22). S76 and T181 were mapped onto the crystal structure of human syndapin I along with residues (K62, R63, K127, K130, K154, and K155) (9) involved in binding the lipid bilayer (Fig. 2B) and with a modeled lipid bilayer (Fig. S2C). The S76 and T181 phosphosites are located in different orientations relative to the lipid-binding surface. There is > 98% identity between the rat and human sequences with the exception of a three residue insertion in a disordered N-terminal sequence. S76 is located at the beginning of helix α-2 (Fig. 2B) and is central to the six-helical bundle formed by the functionally relevant syndapin I F-BAR dimer (Fig. S2D). Although S76 is in an N-cap position at the boundary of helix α-2, the hydroxyl forms multiple interactions with residues in helix α-5 rather than with its own helix. For example, the side chain of S76 forms hydrogen bonds with the backbone carbonyl oxygen and NH of D276 and the side-chain acid of D280 in helix α-5 (Fig. 2B, Right Inset). We suspect that S76 forms N-capping interactions during protein folding; however, upon adopting a native fold this residue forms more favorable interactions with helix α-5 while helix α-2 is stabilized by the surrounding tertiary structure. In contrast, T181 is located at the distal tip of the dimer (Fig. 2B and Fig. S2B) where it is part of an N-capping box motif. Two hydrogen bonds form: one between the T181 side-chain hydroxyl and the backbone NH of E183 and the other between T181 backbone carbonyl oxygen and the side-chain amine of Q184 (Fig. 2B, Left Inset). The side chain of Q184 interacts with the backbone NH of T181 thus completing the N-capping box. The helix-capping motif including T181 is the site of helix–helix interactions that mediate BAR domain filament assembly (19). Therefore the unique positions of the phosphosites at two helix-capping motifs suggests they could regulate the F-BAR domain function through distinct mechanisms, possibly by altering the F-BAR domain curvature (S76) or filament assembly (T181).
Fig. 2.
Location of S76 and T181 on the syndapin I F-BAR. (A) Schematic location of S76 and T181 phosphosites in the domain structure of rat syndapin I. (B) Cartoon of the human syndapin I F-BAR (PDB ID code 3HAI). The side view (Top) and lipid-binding view (Bottom) of monomer are shown. The homologous human residues for S76 and T181 phosphosites are shown in orange spheres, and the basic residues involved in lipid binding are shown in blue spheres. Numbering for the rat sequence residues are in brackets. The close-up of S76 and T181 show potential hydrogen bonds (dashed line) with D276 and D280, and E183 and Q184 residues, respectively. Note that the sequences of rat and human syndapin I differ such that rat S76 is S79 in humans, and rat T181 is T184 in humans.
S76 and T181 Phosphosites Regulate Lipid Binding and Tubulation.
F-BAR domains can bind phospholipid-containing liposomes and induce membrane tubulation in cells and in vitro (3, 8, 19). We generated point mutations of each phosphosite to Ala which prevents phosphorylation, or to a Glu that mimics phosphorylation in the GST- and GFP-tagged syndapin I F-BAR domain (1–304). Circular dichroism analysis of the recombinant mutant proteins showed that the mutations did not affect helical folding of the F-BAR (Fig. S3). To determine whether the phosphosites affect syndapin self-oligomerization (23), the mutants were coexpressed in COS7 cells with myc-tagged syndapin I full-length (FL) wild type (WT) followed by immunoprecipitation with an anti-GFP antibody. They all coimmunoprecipitated with the myc-syndapin I, indicating that the phosphosites did not detectably affect self-oligomerization via the F-BAR domain (Fig. S4).
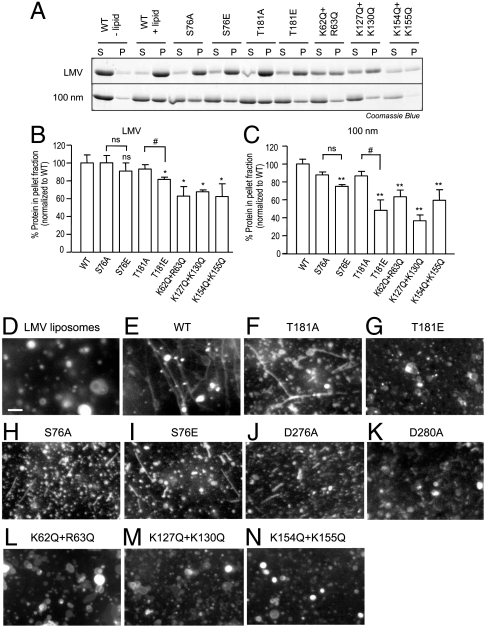
Next, we analyzed the ability of these mutants to bind and tubulate phospholipid liposomes. Purified syndapin I F-BAR-WT preferentially binds large multilamellar vesicles (LMV), which are a mixture of nonuniform-sized liposomes, compared to homogenous 400-nm- and 100-nm-sized liposomes (Fig. 3A and Fig. S5A), consistent with previous observations (9). Neither S76A nor S76E affected binding to LMV or 400-nm liposomes; however, S76E showed reduced binding to 100-nm liposomes (Fig. 3 A–C and Table S1 and Fig. S5 A and B). Similarly, T181A did not affect binding to liposomes of any size, whereas T181E showed reduced binding to liposomes of all sizes. The effect of T181E was greater than for S76E and was similar in magnitude to that observed for the K62Q+R63Q, K127Q+K130Q, K154Q+K155Q mutants (Fig. 3 A–C and Fig. S5 A and B and Table S1). The different liposome-binding characteristics of the two phosphosites correlates with our F-BAR domain-lipid binding model where S76 is orientated away from the lipid interacting surface and T181 toward it (Fig. S2C).
Fig. 3.
Syndapin I phosphosites affect lipid binding and tubulation. (A) Protein–lipid binding of syndapin I F-BAR mutants were examined. Purified syndapin I F-BAR wild-type (WT) and point mutant proteins (5 μg) were incubated with synthetic large multilamellar vesicles (LMV) or uniform 100-nm liposomes (50 μg) made of 60% L-α-phosphatidyl-L-serine (PS), 20% L-α-phosphatidyl-L-choline (PC), and 20% L-α-phosphatidylethanolamine (PE). Samples were centrifuged and the supernatant (S) and pellet (P) fractions were analyzed using SDS/PAGE followed by Coomassie Blue staining. (B and C) Quantitative representation of A. Three independent experiments were performed and the protein band intensity was measured. The error bars indicate standard error of the mean (± SEM, n = 3). A one-way analysis of variance (ANOVA) was applied: ∗ P < 0.05 against WT, #P < 0.05 against A mutants, ns = non-significant. (D–N) In vitro protein–lipid tubulation of syndapin I F-BAR mutants. LMV (50 μg) containing 60% PS, 20% PC, 10% PE, and 10% fluorescein-conjugated PE were incubated with purified syndapin I F-BAR-WT, and point mutant proteins. Lipid tubulation was analyzed via fluorescence microscopy. Scale bar, 10 μm. Images are representative of at least two independent experiments.
We next investigated the ability of the phosphosite mutants to tubulate lipid. Purified syndapin I F-BAR-WT incubated with fluorescent LMV induced long tubules (Fig. 3E), but shorter tubules were induced with 400-nm liposomes (Fig. S5D), consistent with previous findings (9). T181A induced tubules with similar morphology to WT albeit with lower frequency (Fig. 3F). T181E, K62Q+R63Q, K127Q+K130Q, and K154Q+K155Q all showed reduced lipid binding and induced no tubulation (Fig. 3 G and L–N). In contrast, S76A and S76E induced short tubules with LMV (Fig. 3 H and I and Table S1) and no tubulation with 400-nm liposomes (Fig. S5 G and H). This, along with the selective liposome size binding effects, suggests that S76 mutants only tubulated larger curvature liposomes (> 400 nm), which were present in the nonuniform-sized population of LMV.
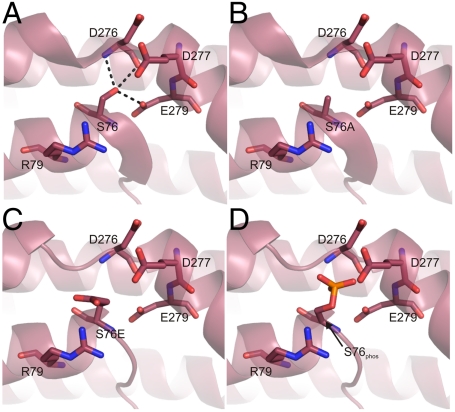
A potential molecular explanation for why both S76A and S76E induced similar phenotypes in lipid tubulation can be seen from the structure of the syndapin I F-BAR dimer (Fig. S2D). The two S76 residues from the dimer are located in a hinge between the central six-helical bundle and the tip. Modifications to S76 would be expected to alter the curvature of the lipid interacting surface and therefore its effect on the length of lipid tubulation and selective binding and tubulation of liposomes (100 nm) with higher curvature. To test this, we prepared structural models of the S76A, S76E, and the phosphorylated Ser modifications using the crystal structure and performed an energy minimization of surrounding residues using Sybyl-X (Fig. 4). As expected, the hydrogen bonds between the Ser and residues in helix α-5 in the crystal structure were disrupted in all models. This would cause a destabilization of this region, resulting in a change of the bend angle of the F-BAR domain. The placement of the phospho-mimetic S76E in the model initially placed a negatively charged group in the vicinity of D276, E279, and D280 residues; however, after energy minimization the Glu side chain flipped away from the acidic residues to interact with the positively charged R79 (Fig. 4C). Phosphorylation of S76 is therefore expected to have a strong effect on the hinge angle. Consistent with this, the structural model showed that the phosphate was unable to rotate out of the electronegative pocket provided by D276, E279, and D280 residues (Fig. 4D). This would lead to repulsion between groups and result in a change in bend angle. To test this, we examined liposome tubulation with either D276A or D280A mutations. Both mutants induced short tubules with LMV, similar to the S76A and S76E mutants (Fig. 3 J and K). Therefore S76 and T181 phosphosites regulate the F-BAR function through altered lipid-binding and tubulation properties using a distinct mechanism for each phosphosite.
Fig. 4.
Model of S76 modifications on syndapin I F-BAR domain. The syndapin I F-BAR crystal structure (PDB ID code 3HAI), was modeled with an energy minimization of residues surrounding S76 using Sybyl-X 1.2 based on (A) WT, (B) S76A, (C) S76E, or a (D) phosphorylated S76. Protein is shown as cartoon with residues in stick. Hydrogen bonds are shown as dashed lines. Residues are labeled according to the rat syndapin I sequence.
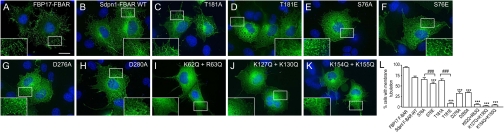
We next asked whether the in vitro effects of the phosphosites on lipid tubulation occurred in intact cells. COS7 cells were transfected with GFP-syndapin I F-BAR constructs, and the percentage of cells with membrane tubulation was scored. Expression of WT produced numerous tubules stemming from the perinuclear region toward the cell periphery (Fig. 5B). Tubulation was less extensive than with GFP-FBP17 F-BAR (8) (Fig. 5A). T181A overexpressing cells have similar morphology to WT cells, while the T181E was unable to induce any tubulation (Fig. 5 C and D) similar to that observed for the basic amino acid mutants of K62Q+R63Q, K127Q+K130Q, and K154Q+K155Q (Fig. 5 I–K). S76A and S76E induced short tubulation (Fig. 5 E and F) similar to that induced by D276A and D280A (Fig. 5 G and H). However, D276A and D280A had significantly stronger effects than S76A or S76E (Fig. 5L). Overall these experiments agree with the in vitro lipid tubulation assay and suggest that the two phosphosites have distinct roles in membrane tubulation in cells.
Fig. 5.
In-cell membrane tubulation analysis of syndapin I F-BAR phosphosite mutants. (A–K) Membrane tubulation was analyzed by overexpressing GFP-syndapin I F-BAR-WT (SdpnI F-BAR-WT), the phospho-mutants and lipid-binding mutants in COS7 cells. Nuclei were stained blue with DAPI to show the position of the cells. GFP-FBP17 F-BAR was a positive control for tubulation (A). The images are representative of at least four independent experiments. Scale bar, 20 μm. (L) Quantitative representation of data from experiments in A–K. One hundred cells per experiment overexpressing each GFP- construct were scored for tubulation and results presented as a % of total cell count with tubulation, n = 4 ± SEM. One-way ANOVA was applied: ∗∗ ∗ P < 0.0001 against WT, ###P < 0.0001 against A mutants.
S76 and T181 Phosphosites Regulate Neuronal Morphogenesis.
Syndapin I is the phosphorylation-dependent dynamin I binding partner in nerve terminals (11). However, the two phosphosites did not conversely regulate dynamin I binding (Fig. S6 A and B). The dynamin I-syndapin I interaction triggers ADBE during intense stimulation in primary neurons (13). A potential function for syndapin I in ADBE is aiding dynamin I recruitment via the membrane sensing and tubulating properties of the F-BAR domain. We next asked whether the two phosphosites regulate ADBE. Knockdown of syndapin I expression in cultured cerebellar granular neurons (CGN) using an shRNA vector caused > 50% reduction of dextran uptake, indicating its importance for ADBE (13). This ADBE block was rescued by expression of WT full-length mouse syndapin I (Fig. S6C). The phospho-deficient or phospho-mimetic S76 and T181 mutants also rescued the block (Fig. S6D), indicating they do not regulate syndapin I function in ADBE. Their lack of effect suggests that the dynamin–syndapin interaction is more important for ADBE than the lipid-binding and tubulating ability of its F-BAR domain alone. Because our ADBE assay is an end-point assay, this does not rule out potential effects on ADBE kinetics, bulk endosome morphology, bulk endosome fate, or a role for other non-F-BAR phosphosites in syndapin I.
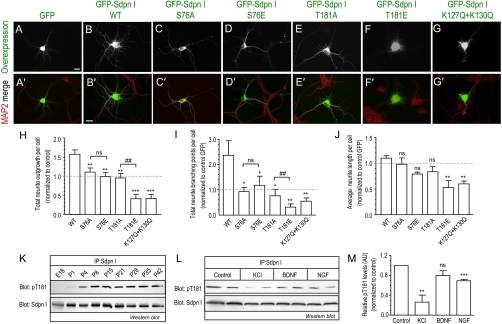
Syndapin I mediates neuronal morphogenesis via the membrane targeting, binding, and deformation properties of its F-BAR domain (14). We transfected hippocampal neurons at DIV4 with either GFP-syndapin I FL-WT, the phosphosite mutants (S76A, S76E, T181A, T181E), or the lipid-binding deficient mutant, K127Q + K130Q, alone and examined the morphology of the developing neuron in culture (Fig. 6). Neurons transfected with WT have increased total neurite outgrowth and branching (Fig. 6 A and B) consistent with a previous report (14). Both total neurite outgrowth (Fig. 6H) and branching (Fig. 6I) increased > 50% in WT compared to control neurons, but the average length of neurites was unaffected (Fig. 6J). Overexpression of the phosphosite mutants produced two clear phenotypes. Firstly, the helix-capping mutants, S76A or T181A, or the phospho-mimetic mutant S76E, did not increase total neurite outgrowth and branching elicited by overexpressing WT and had no effect on neurite length (Fig. 6 C–E and H–J). Secondly, the phospho-mimetic mutant T181E showed a significant reduction in total neurite outgrowth, branching, and neurite length compared to control GFP (Fig. 6 F–G and H–J). This was strongly mimicked by the lipid-binding deficient mutant, K127Q+K130Q. The different neuronal morphological phenotypes of the S76 and T181 correlated with the lipid-binding and tubulation phenotypes from the earlier in vitro assays (Table S1).
Fig. 6.
Syndapin I F-BAR phosphosites mutants alter neurite development. (A–G) Hippocampal neurons were transfected at DIV4 with GFP- syndapin I FL-WT and mutants and processed for immunofluorescence at DIV6. Anti-MAP2 staining shows neuronal morphology and the merged images with overexpression are shown in A′–G′. Images are representative of at least three independent experiments. Scale bar, 50 μm. (H–J) Quantitative analysis of the morphological effects per neuron transfected with GFP alone or GFP syndapin I phosphomutants for (H) total neurite outgrowth from each cell body, (I) total neurite branching points, and (J) average length of neurite outgrowth from the cell body. Relative mean values (± SEM) for H–J are normalized and expressed as a ratio to control GFP (dashed line) transfected neurons from at least three independent experiments. Total of 45–80 neurons for each condition were analyzed. One-way ANOVA was applied: ∗∗ ∗ P < 0.0001, ∗∗ P < 0.001, ∗ P < 0.05, ns = non-significant against WT, ##P < 0.001 against A mutants. (K–M) Western blot analyses of physiological changes to phospho-T181 (pT181) levels with total syndapin I as protein level control (Sdpn I). (K) pT181 in syndapin I immunoprecipitated (IP) from total rat brain lysates of different aged rats. The amount of total brain lysates from the different aged rats used for syndapin I IP were normalized to the relative increasing total syndapin I protein expression (Fig. S6B) so that total syndapin I protein was equal. (L) pT181 changes in syndapin I IP from CGN at DIV10 stimulated for 2 h with either 80 mM KCl, 100 ng/mL of brain-derived neurotrophic factor (BDNF) or nerve growth factor (NGF). (M) Quantitation of stimulation-dependent changes to pT181 phosphorylation from M using densitometry analysis. pT181 levels were normalized to total syndapin I protein and expressed as a relative value to control (nonstimulated) samples. Duplicate samples from n = 3 ± SEM, Student’s t test applied between control and each stimulation ∗∗ P < 0.01, ∗∗ ∗ P < 0.001. All blots are representative of three independent experiments.
Because WT overexpression had a strong effect on neuronal phenotype on its own [Fig. 6B, as previously reported (14)] we extended these experiments by using syndapin I knockdown followed by functional rescue to confirm our observations by a different strategy. Hippocampal neurons were cotransfected with an shRNA vector targeting endogenous rat syndapin I together with the RNAi-resistant mouse GFP-syndapin I FL-WT and phosphomutant constructs and examined the rescued neuronal morphology (Fig. S7). Quantitative analysis indicated that syndapin knockdown increased total neurite outgrowth, branching and neurite length (Fig. S7A) and overexpression of WT protein restored the neuron morphology, consistent with a previous report (14). Strikingly, phospho-deficient and phospho-mimetic S76 and T181 mutants coexpressed in this rescue experiment produced qualitatively the same pattern of neuron morphology (Fig. S7 B–D) as above when neurons were overexpressed (Fig. 6 H–J). Therefore S76 or T181 phospho-regulation changes F-BAR domain function, differentially affecting the morphology of developing neurons.
Effects on neuronal morphology such as these have been partly ascribed to N-WASP mediated actin dynamics in addition to the F-BAR (14). However, the phosphosite mutants did not affect syndapin I binding to N-WASP, using pulldowns with GST-syndapin I FL-WT and the phosphosite mutants (Fig. S6 A and B). Therefore, the effects of the phosphosite mutants on neuronal morphology are more likely due to direct regulation of the F-BAR domain than an indirect effect on actin.
Phosphorylation at T181 Dynamically Changes During Brain Development and in Response to Stimulation.
We raised an anti-syndapin I T181 phosphosite-specific (pT181) antibody (Fig. S8A) to directly assess whether the F-BAR phosphorylation is regulated in vivo during development. It has previously been shown that syndapin I expression increases during brain development (14) (Fig. S8B). Therefore we measured T181 phosphorylation after normalization of immunoprecipitated syndapin I protein levels in whole brain lysates from different aged rats (E18 to P42) (Fig. 6K). The relative proportion of syndapin I phosphorylated at T181 increased dramatically with brain development suggesting that T181 phosphorylation is developmentally regulated. We next investigated three stimulus conditions known to affect neuronal morphogenesis in culture (24). Stimulation of CGN at DIV10 with elevated KCl dramatically reduced T181 phosphorylation (Fig. 6 L and M). Stimulation of these neurons with a growth factor (NGF) also reduced phosphorylation at T181 but not with a neurotrophic factor (BDNF). Therefore two classes of stimuli cause dephosphorylation of syndapin I at T181 in neurons, suggesting that a depolarization-dependent and/or growth factor signaling pathway regulates syndapin I function in neuronal morphogenesis. The two F-BAR phosphosites were not specifically phosphorylated by the two known syndapin I protein kinases, PKC and CKII in vitro (Fig. S8C), indicating that other pathways are involved in regulating the two F-BAR phosphosites.
Discussion
We demonstrate that syndapin I is phosphorylated at two sites, S76 and T181, on its F-BAR domain in neurons which regulate its ability to bind phospholipids and tubulate membranes. T181 phosphorylation was developmentally regulated and found to inhibit syndapin I function in neuronal morphogenesis. Both phosphosites are part of two different helix-capping motifs and modifications to these sites affect the inter- and intramolecular interactions surrounding the motifs, thereby altering either F-BAR homodimer curvature (S76) and/or filament assembly through dimer–dimer interactions (T181). Our findings highlight a novel mechanism for the regulation of a F-BAR protein by phospho-regulatory control over helix interactions affecting stability within a folded domain.
Only two other proteins in the BAR domain superfamily, APPL1 (25) and Cdc15 (26), are known to contain phosphosites in its BAR domain; however, these phosphosites are not part of an N-capping motif. S76 is a newly identified syndapin I phosphosite, while T181 has been previously identified (27), but its physiological relevance was not explored. T181 phosphorylation was not confirmed, replicated, or demonstrated by metabolic labeling, as we have done. S76 and T181 are highly conserved amongst the F-BAR subfamily. Therefore, phosphorylation of these sites could represent a broader mechanism for regulation of F-BAR function if it occurred in other family members.
Helix-capping motifs contribute to the stability of an α-helix and to overall protein structure by providing hydrogen bonding partners in the first four backbone NH groups of an α-helix (21, 28). S76 and T181 are both located in an N-cap position within the F-BAR. Two pieces of evidence suggest that the S76 helix-capping motif regulates F-BAR homodimer curvature. Firstly, structural modeling suggested that replacing this Ser with Ala, Glu, or phospho-Ser could alter the bend angle of the overall F-BAR dimer. Secondly, our experimental analysis revealed that both phospho-deficient and phospho-mimetic S76 mutants presented similar phenotypes for in vitro and in-cell lipid-binding and tubulation. They bind and tubulate only larger, more shallow curvature liposomes. This suggests that phosphorylation of the S76 helix-capping motif is a molecular regulatory signal to control F-BAR homodimer curvature.
T181 is located at the beginning of helix α-4 in the syndapin I F-BAR domain crystal structure (Fig. 2B). The hydrogen bonds formed by T181 are typical of those in an N-capping motif (29) and homologous to those in other F-BAR crystal structures (Fig. S2). T165 in FBP17 is important for stabilizing filament assembly of the FBP17 dimers by forming hydrogen bonds with D168 (i.e., forming an N-cap motif). Its mutation to Ala abolished lipid binding and membrane tubulation in cells (19). Therefore, these end-to-end filaments formed by F-BAR dimers are important for binding and tubulating membranes (3, 9, 19). The phospho-mimetic mutant, T181E, has a dominant-negative phenotype by abolishing lipid binding and induction of tubulation. This suggests that phosphorylation has disrupted the N-cap motif and syndapin I F-BAR dimers to form end-to-end filaments and subsequent lipid binding and tubulation as previously reported (9, 19). Phosphorylation on α-helices either stabilizes (30, 31) or destabilizes (32) the structure, depending on the phosphosite position and the surrounding electrostatic interactions (33). Together with our data, this supports that phosphorylation can regulate protein function in cells via regulating inter- and intramolecular α-helix interactions and stability. Previous studies have demonstrated a role for F-BAR domains in cellular processes involving dynamic membrane changes and remodeling. Syndapin function in notochord development in the zebrafish (34) and formation of postsynaptic membrane system in Drosophila (35) have been attributed to its F-BAR. In mammalian cells, syndapin I can regulate neuronal morphogenesis (14, 15). Similarly, the F-BAR domain of slit-robo GTPase (srGAP2) regulates neuronal migration and morphogenesis primarily by inducing filopodia-like membrane protrusions (36). We extend these observations by showing that phosphorylation can control the syndapin I F-BAR structure and function in membrane tubulation, with at least T181 inhibiting its role in neurite morphogenesis.
Materials and Methods
Detailed materials and methods available in SI Text include: DNA plasmids and protein purification,  -labeling of synaptosomes and immunoprecipitation of syndapin I, phosphopeptide enrichment, mass spectrometry, protein sequence and structure alignment, CD analysis, liposome preparation, syndapin I knockdown in neurons, ADBE assay, fluorescence microscopy and imaging.
-labeling of synaptosomes and immunoprecipitation of syndapin I, phosphopeptide enrichment, mass spectrometry, protein sequence and structure alignment, CD analysis, liposome preparation, syndapin I knockdown in neurons, ADBE assay, fluorescence microscopy and imaging.
Protein Structure Modeling.
Modeling of the S76A/E mutations and S76 phosphorylation were completed in vacuo with Sybyl-X 1.2 (www.tripos.com), using the MMFF94 forcefield and default parameters. Figures were prepared with PyMOL (www.pymol.org).
In Vitro Liposome Binding and Fluorescence Tubulation Assays.
These assays were performed as previously described (37) with modifications.
In-Cell Membrane Tubulation assay.
COS7 cells were transfected with GFP-syndapin I F-BAR constructs (1 μg). After 24 h transfection, cells were fixed with 4% paraformaldehyde in PBS, pH 7.4 and counterstained with DAPI, then analyzed by fluorescence microscopy collecting Z stacks.
Hippocampal Neurons and Transfection.
Cultured neurons were prepared using brains from Wistar rats at embryo day 18 (E18). For neuronal development morphological analysis, neurons were transfected with GFP-syndapin I FL constructs (1 μg) by calcium phosphate precipitation at day 4 in vitro (DIV4). Fixed at DIV6, immunostained with anti-MAP2 and analyzed by fluorescence microscopy.
Syndapin I T181 Phosphorylation in Brain and Neurons.
Phospho-T181 specific antibody was generated (full details in SI Materials and Methods) and used in Western blotting to analyze phosphorylation changes to T181 in brain lysates from different aged rats and stimulation-dependent changes in cultured neurons.
Supplementary Material
Acknowledgments.
We wish to thank our colleagues at the Children’s Medical Research Institute (CMRI) for providing reagents and technical assistance. This work is supported by grants from the Australian National Health and Medical Research Council, the Ramaciotti Foundation, Cancer Institute N.S.W., and the Ian Potter Foundation. A.Q. is supported by a CMRI postgraduate research scholarship.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108294109/-/DCSupplemental.
References
- 1.Frost A, Unger VM, De Camilli P. The BAR domain superfamily: Membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peter BJ, et al. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 3.Frost A, et al. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plomann M, et al. PACSIN, a brain protein that is upregulated upon differentiation into neuronal cells. Eur J Biochem. 1998;256:201–211. doi: 10.1046/j.1432-1327.1998.2560201.x. [DOI] [PubMed] [Google Scholar]
- 5.Qualmann B, Roos J, DiGregorio PJ, Kelly RB. Syndapin I a synaptic dynamin-binding protein that associates with the neural Wiskott-Aldrich syndrome protein. Mol Biol Cell. 1999;10:501–513. doi: 10.1091/mbc.10.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Modregger J, Ritter B, Witter B, Paulsson M, Plomann M. All three PACSIN isoforms bind to endocytic proteins and inhibit endocytosis. J Cell Sci. 2000;113:4511–4521. doi: 10.1242/jcs.113.24.4511. [DOI] [PubMed] [Google Scholar]
- 7.Braun A, et al. EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol Biol Cell. 2005;16:3642–3658. doi: 10.1091/mbc.E05-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh T, et al. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, et al. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci USA. 2009;106:12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao Y, et al. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc Natl Acad Sci USA. 2010;107:8213–8218. doi: 10.1073/pnas.1003478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anggono V, et al. Syndapin I is the phosphorylation-regulated dynamin I partner in synaptic vesicle endocytosis. Nat Neurosci. 2006;9:752–760. doi: 10.1038/nn1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch D, et al. Proper synaptic vesicle formation and neuronal network activity critically rely on syndapin I. EMBO J. 2011 doi: 10.1038/emboj.2011.339. 10.1038/emboj.2011.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clayton EL, et al. The phospho-dependent dynamin-syndapin interaction triggers activity-dependent bulk endocytosis of synaptic vesicles. J Neurosci. 2009;29:7706–7717. doi: 10.1523/JNEUROSCI.1976-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dharmalingam E, et al. F-BAR proteins of the syndapin family shape the plasma membrane and are crucial for neuromorphogenesis. J Neurosci. 2009;29:13315–13327. doi: 10.1523/JNEUROSCI.3973-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwintzer L, et al. The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J. 2011;30:3147–3159. doi: 10.1038/emboj.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilton JM, et al. Phosphorylation of a synaptic vesicle-associated protein by an inositol hexakisphosphate-regulated protein kinase. J Biol Chem. 2001;276:16341–16347. doi: 10.1074/jbc.M011122200. [DOI] [PubMed] [Google Scholar]
- 17.Cousin MA, Robinson PJ. The dephosphins: Dephosphorylation by calcineurin triggers synaptic vesicle endocytosis. Trends Neurosci. 2001;24:659–665. doi: 10.1016/s0166-2236(00)01930-5. [DOI] [PubMed] [Google Scholar]
- 18.Greengard P, Valtorta F, Czernik AJ, Benfenati F. Synaptic vesicle phosphoproteins and regulation of synaptic function. Science. 1993;259:780–785. doi: 10.1126/science.8430330. [DOI] [PubMed] [Google Scholar]
- 19.Shimada A, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 20.Henne WM, et al. Structure and analysis of FCHo2 F-BAR domain: A dimerizing and membrane recruitment module that effects membrane curvature. Structure. 2007;15:839–852. doi: 10.1016/j.str.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Aurora R, Rose GD. Helix capping. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan WY, Milner-White EJ. A recurring two-hydrogen-bond motif incorporating a serine or threonine residue is found both at alpha-helical N termini and in other situations. J Mol Biol. 1999;286:1651–1662. doi: 10.1006/jmbi.1999.2551. [DOI] [PubMed] [Google Scholar]
- 23.Halbach A, et al. PACSIN 1 forms tetramers via its N-terminal F-BAR domain. FEBS J. 2007;274:773–782. doi: 10.1111/j.1742-4658.2006.05622.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Z, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gant-Branum RL, Broussard JA, Mahsut A, Webb DJ, McLean JA. Identification of phosphorylation sites within the signaling adaptor APPL1 by mass spectrometry. J Proteome Res. 2010;9:1541–1548. doi: 10.1021/pr901043e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts-Galbraith RH, et al. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol Cell. 2010;39:86–99. doi: 10.1016/j.molcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tweedie-Cullen RY, Reck JM, Mansuy IM. Comprehensive mapping of post-translational modifications on synaptic, nuclear, and histone proteins in the adult mouse brain. J Proteome Res. 2009;8:4966–4982. doi: 10.1021/pr9003739. [DOI] [PubMed] [Google Scholar]
- 28.Seale JW, Srinivasan R, Rose GD. Sequence determinants of the capping box, a stabilizing motif at the N-termini of alpha-helices. Protein Sci. 1994;3:1741–1745. doi: 10.1002/pro.5560031014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson JS, Richardson DC. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- 30.Hurley JH, Dean AM, Thorsness PE, Koshland DE, Jr, Stroud RM. Regulation of isocitrate dehydrogenase by phosphorylation involves no long-range conformational change in the free enzyme. J Biol Chem. 1990;265:3599–3602. doi: 10.2210/pdb4icd/pdb. [DOI] [PubMed] [Google Scholar]
- 31.Pullen K, et al. Phosphorylation of serine-46 in HPr, a key regulatory protein in bacteria, results in stabilization of its solution structure. Protein Sci. 1995;4:2478–2486. doi: 10.1002/pro.5560041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szilak L, Moitra J, Krylov D, Vinson C. Phosphorylation destabilizes alpha-helices. Nat Struct Biol. 1997;4:112–114. doi: 10.1038/nsb0297-112. [DOI] [PubMed] [Google Scholar]
- 33.Andrew CD, Warwicker J, Jones GR, Doig AJ. Effect of phosphorylation on alpha-helix stability as a function of position. Biochemistry. 2002;41:1897–1905. doi: 10.1021/bi0113216. [DOI] [PubMed] [Google Scholar]
- 34.Edeling MA, et al. Structural requirements for PACSIN/Syndapin operation during zebrafish embryonic notochord development. PLoS ONE. 2009;4(12):e8150. doi: 10.1371/journal.pone.0008150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar V, et al. Syndapin promotes formation of a postsynaptic membrane system in Drosophila. Mol Biol Cell. 2009;20:2254–2264. doi: 10.1091/mbc.E08-10-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerrier S, et al. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsujita K, et al. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol. 2006;172:269–279. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.