Abstract
The Drosophila R7 photoreceptor provides an excellent model system with which to study how cells receive and “decode” signals that specify cell fate. R7 is specified by the combined actions of the receptor tyrosine kinase (RTK) and Notch (N) signaling pathways. These pathways interact in a complex manner that includes antagonistic effects on photoreceptor specification: RTK promotes the photoreceptor fate, whereas N inhibits. Although other photoreceptors are subject to only mild N activation, R7 experiences a high-level N signal. To counter this effect and to ensure that the cell is specified as a photoreceptor, a high RTK signal is transduced in the cell. Thus, there are two levels of RTK transduction in the photoreceptors: in R7 it is high, whereas in others it is low. Here, we address how this high-level RTK signal is transduced in R7 and find that, in addition to Ras, another small GTPase, Rap, is also engaged. Thus, when N activity is high, a robust RTK signal operates that uses both Ras and Rap, but when N activity is low, only a mild RTK signal is transduced and Ras alone suffices for the purpose.
The Drosophila compound eye is made from many hundreds of ommatidia, each containing a stereotypical arrangement of various cell types. The development of the ommatidium has become a model system for studying how cells receive and decode short-range fate-specifying signals. Particular attention has been paid to the specification of the R7 photoreceptor, which is directed to its fate by the receipt of two distinct signals from directly neighboring photoreceptors (1, 2). Its R1/6 neighbors express Delta (3–5), a membrane-bound ligand for the ubiquitously expressed Notch (N) receptor. Another neighbor, R8, expresses Bride of Sevenless (Boss), a transmembrane protein that is the ligand (6, 7) for Sevenless (Sev), a receptor tyrosine kinase (RTK) (8) expressed (among other cells) in the R7 precursor (9, 10). Thus, from its immediate neighbors, the R7 precursor receives signals that separately activate the N and RTK pathways.
Tramtrack (Ttk) is a transcription factor that represses the photoreceptor fate, and its removal requires the activation of the RTK pathway (11, 12). If RTK activity suffices to degrade Ttk, then a cell becomes a photoreceptor; if not, it becomes an ommatidial support cell, such as a lens cone cell. N plays a number of roles in this process, including antagonizing the ability of RTK activity to remove Ttk. The molecular nature of this antagonism is unknown, but its effect opposes the specification of photoreceptors (13). If N signaling is low, only a mild RTK signal is required for photoreceptor specification; however, if N activity is high, then robust RTK activity is needed. These two molecular states are found in the R1/6 and R7 precursors, respectively. The R1/6 precursors experience low N activity, receive RTK signaling through the Drosophila EGF receptor (DER) (14–16), degrade Ttk, and differentiate as photoreceptors. These cells then express high levels of Delta that induce a high-level N activity in the R7 precursor, which now experiences a potent block to its ability to degrade Ttk. N now performs a second role; it transcriptionally activates sev, supplying high levels of the Sev RTK to the R7 precursor (13). Activation of Sev by its ligand on the neighboring R8 cell supplies the robust RTK signal that degrades Ttk and specifies the cell as a photoreceptor. [The third function of N specifies the cell as the R7 rather than the R1/6 photoreceptor type (13).] Thus, in response to their levels of N activity, the R1/6 and R7 precursors differ dramatically in their levels of RTK signaling: it is mild in the former and robust in the latter.
RTK signaling typically activates the small GTPase Ras, which recruits Raf to the plasma membrane and initiates the phosphorylation cascade that leads to the translocation of MAPK to the nucleus. Rap is another member of the Ras GTPase superfamily; it was initially considered a Ras antagonist because it suppressed Ki-Ras oncogenesis (17) and inhibited Ras-dependent activation of MAPK (18). Roughened is a dominant mutant of Drosophila rap in which R7 photoreceptors are frequently absent from the ommatidia (19). Because Rap was at that time viewed as a Ras antagonist, Roughened was assumed to encode a dominant Ras antagonist, and Rap itself was assumed to not function normally in Drosophila eye development (19). Later, Rap was shown to cooperate with Ras to transduce Torso RTK signaling in the embryonic terminal system (20); both Rap and Ras were required for the full output of the pathway.
We have reexamined the role of Rap in Drosophila eye development and found that it is critically required along with Ras to specify the R7 precursor as a photoreceptor. The other photoreceptors do not require Rap for their specification, and we correlate the requirement for Rap with the level of N activity in the photoreceptor precursors: R7 (high N activity) requires Rap, and the other photoreceptors (low N activity) do not.
Results
Reduced Rap Activity Specifically Affects R7 Specification.
sev.rapN17 phenocopies Roughened.
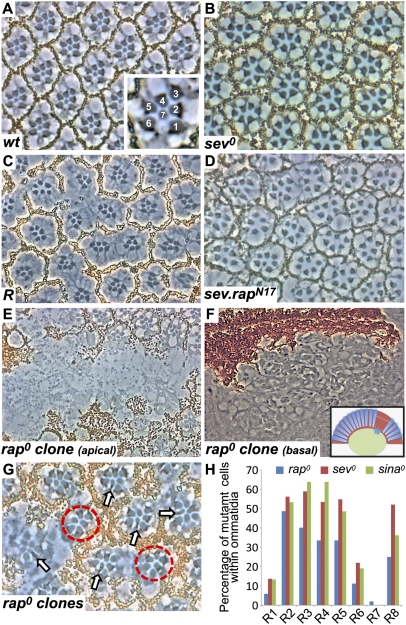
In the apical regions of the fly retina, the R7 photoreceptors have small rhabdomeres positioned centrally to those of the large outer photoreceptors (Fig. 1A). In mutations that remove R7 photoreceptors, such as sev (sev0), the central rhabdomeres are notably absent (Fig. 1B). Roughened mutants show the frequent loss of R7 photoreceptors from the ommatidia, with less frequent loss of other photoreceptors and pigment cells (19) (Fig. 1C).
Fig. 1.
rap is required for R7 specification. (A) A tangential section through a wild-type eye. Inset shows large rhabdomeres of R1–R6 surrounding the central small R7. (B) A tangential section through a sev0 eye shows only the six outer rhabdomere cells. (C) A tangential section through a Roughened eye. Many ommatidia lack R7 cells. (D) sev.rapN17 phenocopies Roughened; many R7 cells are absent. (E) A section through a large rap0 clone marked by the absence of pigment. (F) Apically, clones lack photoreceptors, which are found lying below the retina and above the lamina nuclei level. Inset shows a schematic representation with the upper white dotted box indicating the region shown in E and the lower box indicating the area shown in F. (G) A tangential section through an eye containing many late-induced rap0 clones. Normally constructed ommatidia containing a mixture of mutant and wild-type cells are found. R7 cells are almost invariably pigmented (rap+) (arrows). Additionally, many ommatidia lack R7 cells (red circles). (H) Histogram showing the percentage of mutant photoreceptors found in rap0 (blue bars), sev0 (red bars), and sina0 (green bars; adapted from ref. 27) mosaic ommatidia.
In the Ras superfamily, the substitution of serine at position 17 with an asparagine (N17) generates a dominant-negative protein (21), and we transformed flies with rapN17 under sev transcriptional control (22) (sev.rapN17). Sev is expressed at high levels in many cells of the developing ommatidia, including the R7 precursors, and this construct thus expresses a high level of a dominant-negative Rap protein in presumptive R7 photoreceptors. Eyes of these sev.rapN17 flies were rough when viewed from the exterior and, when sectioned, showed a Roughened-like phenotype; the majority of ommatidia lacked R7 photoreceptors, and other cells were lost at a lower frequency (Fig. 1D). The similarity of effects of Roughened and sev.rapN17 suggests that Roughened encodes a dominant-negative form of Rap and that Rap plays a critical role in R7 formation.
Mosaic analysis defines a requirement for rap gene function in R7.
Because Rap and Ras are related small GTPases, then it is possible that sev.rapN17 phenotypes result not from an interference with normal Rap function but from the sequestration of proteins required for Ras activity. To address this concern, we examined rap null eye clones [raprvB3 (19)] marked by the loss of the pigmentation gene white (w+). Clones induced early in larval life resulted in large retinal scars, rich in pigment cells but almost completely lacking photoreceptors (Fig. 1E). Deeper sections of these preparations showed a multitude of photoreceptors lying below the base of the retina, flattened above the outer nuclear layer of the lamina (the first layer of the optic lobes) (Fig. 1F). Thus, in the absence of rap gene function, photoreceptors fall out of the developing retina, which likely results from the role of Rap in regulating adherens junctions (23) through the PDZ protein Canoe (24). Indeed, canoe mutant clones show similar phenotypes with basally located photoreceptors (25). Adherens junctions “anchor” the cells together and maintain epithelial integrity, and when they fail, cells delaminate basally. The basally located photoreceptors were not all rap0 (most were unpigmented, but some were not), and we infer that the structural collapse that occurs in cells with compromised adherens junctions caused a similar effect on neighbors sharing the junctions. To prevent this photoreceptor delamination, late-induced clones (late second instar) were examined. Here, mutant photoreceptors survived in the retina, suggesting that perduring rap+ gene products allowed adherens junction maintenance. Thus, we do not consider these clones to represent the null condition; rather, they are the equivalent of a hypomorph in which some gene function persists.
Mosaic analysis was next performed on these clones. Normally constructed mosaic ommatidia containing pigmented (rap+) and unpigmented (rap0) cells were scored for the genotype of each photoreceptor (Fig. 1 G and H). If rap gene function were required in a specific photoreceptor type, then we would not expect to recover normal ommatidia in which that cell was mutant. Conversely, if rap gene function were not required in a photoreceptor type, then we would expect rap0 (w0) versions of these cells to be liberally present in the mosaic ommatidia. This analysis showed R2, R3, R4, R5, and R8 photoreceptors at high frequency, with many fewer R1/6 photoreceptors and very few R7 photoreceptors (Fig. 1H). To analyze this dataset, two issues need to be addressed: (i) the effects of gene product perdurance and (ii) the effects of lineage relationships of the cells.
(i) Perdurance effects: The R1/6/7 precursors undergo one more round of division than R2, R3, R4, R5, and R8 do (26); accordingly, we expect a further dilution of rap+ gene products in R1/6/7 with a consequential reduction of the survival of these cells in the retina.
(ii) Lineage relationships: Within the R1/6/7 group, there is an unexpected lineage bias: R1/7 cells are more closely related than R1/6 are. In a neutral mosaic analysis of 325 ommatidia, mosaicism in the R1/6/7 cells was found in 142 (37%) ommatidia. In 76 (24%) of these, R1 and R7 were in the same clone, whereas R6 and R7 were found in 44 (13%) (P = 0.047). Thus, R1 cells are almost twice as likely to be in the same clone as R7 cells than are R6 cells. This clonal relationship predicts a bias in the number of R1 and R6 cells scored in a mosaic analysis using a mutation that selectively removes R7 cells. Consider two presumptive ommatidia, one of which has an R1 mutant and the other of which has an R6 mutant. The ommatidium with the R1 mutant is more likely to also have an R7 mutant than the ommatidium with R6 mutant is. Because the presence of an R7 mutant results in an aberrant ommatidium that is not scored, the ommatidium in which R6 is mutant is more likely to avoid this fate and contribute to a normal ommatidium that is scored. Therefore, because of this clonal relationship, we predict more mutant R6 cells in normally constructed ommatidia than there are mutant R1 cells.
Indeed, this is what we observe with the rap mosaic analysis: more R6 cells than R1 cells (Fig. 1H). To validate the reasoning above, we examined mosaic data from two other mutations that remove R7 cells: sev and seven in absentia (sina). For sev we generated and scored the clones, whereas for sina we adapted the mosaic analysis performed by Carthew et al. (27) (Fig. 1H). Both these mosaic analyses showed the same relationship: R6 cells were more frequently recovered than R1 cells were.
From the combined effects of perdurance and lineage relationships, the mosaic analysis suggests a pronounced requirement for Rap in R7 precursors. That is, although the decaying perdurance effects reduce the total number of R1/6/7 cells in the sample, within those that survive there is the key lineage relationship signature of a mutation that specifically removes R7 photoreceptors.
Analysis of rap mutant clones in eye discs.
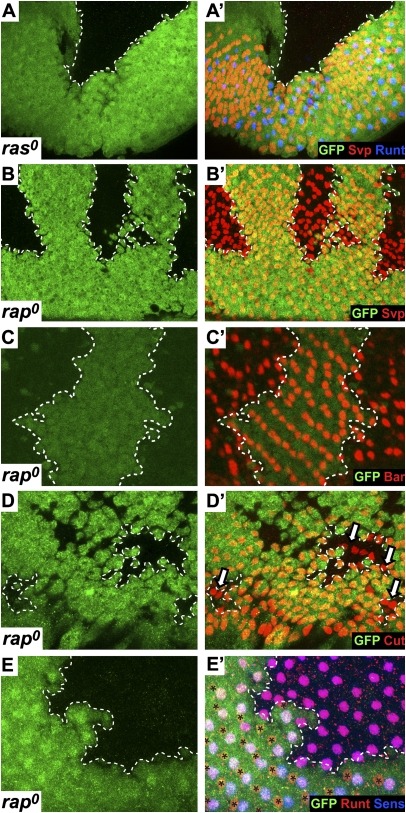
The analysis of the rap clones in the adult eye suggested a preferential requirement for Rap function in R7 precursors. We next examined eye imaginal disk rap clones and compared them with equivalent ras clones. To ensure survival of the clones, and to compare the two mutations under the same genetic conditions, both were induced in a minute background (the mutant clones were given a growth advantage over their surrounding wild-type tissue). As has been described previously (28), such ras0 clones show no differentiation of ommatidial cells (Fig. 2A). The rap0 clones, however, were very different; they showed extensive evidence of ommatidial differentiation. The analysis of these rap0 ommatidia was compromised by the effects on adherens junctions: the tissue in the clones was flattened, and the nuclei were somewhat squashed together basally. Despite these problems, ommatidial differentiation occurred relatively normally. R8 photoreceptors, marked by the expression of Senseless (Sens) were observed (Fig. 2E), and Seven-up (Svp), which labels R1/6/3/4 precursors, was expressed (Fig. 2B). R1/6 cells express Svp early in their differentiation but later turn on the transcription factor Bar, which was clearly detected (Fig. 2C). Later in ommatidial maturation, the lens cone cell precursors express the transcription factor Cut, which was also detected in the posterior regions of the rap0 clones (Fig. 2D). Notably absent from these clones was evidence for R7 specification. Runt is a good marker for R7 cells but also labels R8 cells. We therefore costained with Sens, which selectively labels R8 cells, and confirmed that all Runt-expressing cells in the rap0 clones were R8 cells (Fig. 2E). Thus, although ras0 clones showed no differentiation of photoreceptors or lens cone cells, the rap0 clones were dramatically different, showing the expression of photoreceptor and lens cone cell markers but the specific absence of R7 markers. Collectively, the adult and eye disk analyses suggest that Rap is critically required for the formation of R7 photoreceptors and appears dispensable in the other cells.
Fig. 2.
A comparison of ras0 and rap0 mutant clones in third-instar eye discs. In all images, the clones are indicated by the absence of GFP (green) and are circumscribed by a dashed line. (A and A′) A ras0 clone shows no differentiating ommatidia. (B and B′) rap0 clones show extensive Svp staining (red), which labels the R1/3/4/6 photoreceptors. (C and C′) rap0 clones contain many cells expressing Bar (red), which selectively marks R1/6 cells. (D and D′) rap0 clones show the cone cell marker Cut (red; arrows). (E and E′) rap0 clones stained for Runt (red) and Sens (blue). Cells expressing Sens and Runt (R8 cells) populate the clone, but Runt-positive/Sens-negative R7 cells are only found in the wild-type tissue (asterisks).
Activation of Rap Induces Supernumerary R7 Cells.
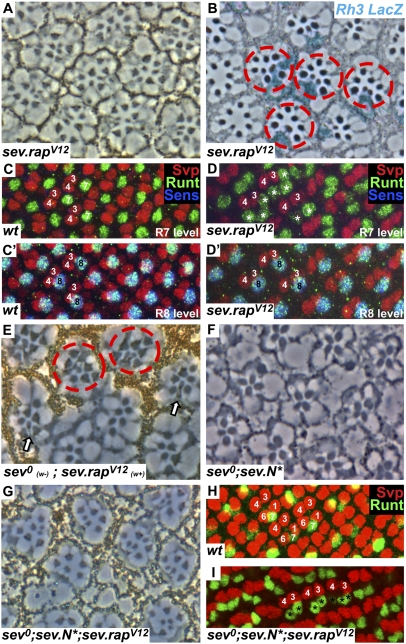
Substitution of glycine to valine at position 12 (V12) in members of the Ras superfamily inhibits the GTPase activity and renders the protein constitutively active (29). When RasV12 was expressed in the developing eye with the sev enhancer element, multiple R7 photoreceptors were induced in the ommatidia (30). To determine whether activated Rap would behave similarly, we constructed and transformed rapV12 under sev transcriptional control (sev.rapV12). These flies had rough eyes from the exterior and in tangential section showed ommatidia containing supernumerary small rhabdomere cells. The multiple-small rhabdomere phenotype is characteristic of transgenes that activate the RTK pathway, and although sev.rapV12 induces the RTK pathway, this activation appeared weaker than, for example, a sev.rasV12 transgene (30). However, when the chromosome carrying this transgene was made homozygous, the phenotype was stronger with an abundance of supernumerary small rhabdomere cells (Fig. 3A). Next, we recombined another copy of sev.rapV12 onto this chromosome, producing a single chromosome with two copies of sev.rapV12. The phenotype of this single chromosome (Fig. 4D) was as potent in inducing the multiple small rhabdomere phenotype as the original was when homozygous. This chromosome was subsequently used in the following analyses (except where stated otherwise).
Fig. 3.
Activated Rap specifies R7 photoreceptors. (A) A section through a sev.rapV12 homozygous eye shows multiple small rhabdomeres. (B) A tangential section through an X-Gal–stained sev.rapV12 eye in an Rh3–lacZ reporter background. Subsets of R7 cells and supernumerary R7 cells are stained in blue (red circles). (C and C′) Posterior region of a wild-type eye disk stained with Svp (red), Runt (green), and Sens (blue). Each ommatidium has a single apical Runt-labeled R7 (C) and a single basal Runt- and Sens-stained R8 (C′). (D and D′) A sev.rapV12 eye disk stained as in C showing multiple R7 cells (asterisks) in the apical level and a single R8 basally. (E) A tangential section through a sev0 mutant eye in which a clone of sev.rapV12 has been induced marked by pigment. In the pigmented region, ommatidia containing multiple R7 cells are evident (red circles), and at the mosaic interface normally constructed ommatidia form carrying pigmented (sev.rapV12) R7 cells (arrows). (F) A tangential section through a sev0;sev.N* eye (reproduced from ref. 13) showing the absence of R7 cells. (G) When RapV12 is added to this background (sev0;sev.N*;sev.rapV12) R7 cells are restored. (H) Image of a wild-type eye disk at the region where R1/6 precursors label with Svp (red). (I) In the sev0;sev.N*;sev.rapV12 background, R1/6 precursors no longer express Svp but now express Runt (green), indicating their differentiation into R7 photoreceptors (marked by asterisks).
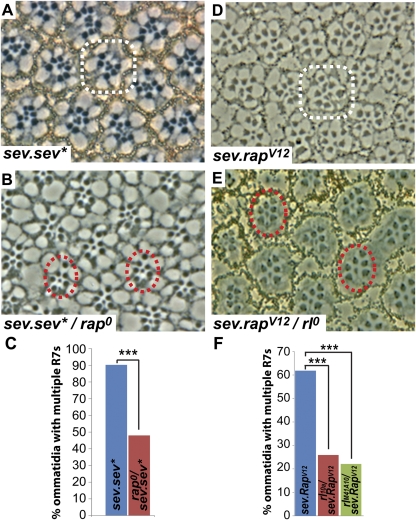
Fig. 4.
Epistasis experiments with rap. Examples of multiple R7 ommatidia (white circles) and normal ommatidia (red circles) are indicated. (A–C) Removing one copy of rap from sev.sev* causes a significant reduction in ommatidia with multiple R7 cells (P = 5.0 × 10−4). (D–F) Removing one copy of rl from sev.rapV12 causes a significant reduction in ommatidia with multiple R7 cells (rl10a, P = 1.5 × 10−4; rlM41A10, P = 3.0 × 10−4).
Both R7 and R8 photoreceptors have small rhabdomeres; to confirm that the supernumerary cells were indeed R7 cells, two assays were performed. First, adult eyes were stained for Rh3–lacZ [an opsin reporter that is expressed in a subset of R7 cells (31)], and a subset of supernumerary small rhabdomere cells were highlighted (Fig. 3B). Second, we examined sev.rapV12eye discs. In wild-type developing ommatidia, a single R7 forms apically marked by Runt (Fig. 3C), and a single R8 forms basally labeled with Runt and Sens (Fig. 3C′). In sev.rapV12eye discs, multiple Runt-positive (Sens-negative) cells are present at the R7 level (Fig. 3D), and single R8 photoreceptors expressing both Runt and Sens are detected basally. From these results, we infer that the supernumerary small rhabdomere cells induced by sev.rapV12 represent bona fide R7 photoreceptors.
Mystery cells are early occupants of the precluster that, along with their R3/4 neighbors, express Sev. It is thought that the R3/4 cells occlude the mystery cells from the DER-activating signals released by the R2/5/8 precluster cells, and, in the absence of RTK activity, these cells leave the cluster and join the surrounding pool of dividing cells. If the RTK pathway is ectopically activated in these cells, they can then differentiate as photoreceptors (32). Thus, the mystery cells appear as protocluster cells able to be specified as photoreceptors if their RTK pathways are activated. Although sev.rasV12 induces the incorporation of some mystery cells, in sev.rapV12 eyes, we see no evidence of it. This difference may represent a difference in the potency of the two transgenes. That is, mystery cells may require a strong RTK signal for their incorporation, and sev.rasV12 may provide this level but sev.rapV12 does not.
Epistasis Experiments to Indicate Where Rap May Operate in the RTK Pathway.
sev.rapV12 compensates for the absence of Sev.
The experiments to date indicate that Rap is required for R7 development, and its ectopic activation can induce supernumerary R7 cells. sev0 mutant flies specifically lack R7 cells (Fig. 1B), and, if Rap lies downstream of Sev in the RTK transduction pathway, then an activated form of Rap should substitute for the absence of Sev and restore R7 cells. sev.rapV12 was crossed in into a sev0 background (sev0;sev.rapV12), and the multiple R7 phenotype was still present, indicating the potency of sev.rapV12 to activate the RTK pathway even in the absence of Sev (circles in Fig. 3E). The next experiments asked whether sev.rapV12 can specifically substitute for the absence of sev gene function in the R7 precursor itself. We induced clones of sev.rapV12 (marked by the presence of pigment) in an otherwise sev0 (unpigmented) eye tissue. We examined ommatidia at the clonal interface and observed normally constructed ommatidia (containing a single R7 in its correct location; arrows in Fig. 3E). The R7 cells in these ommatidia were invariably pigmented (indicating the presence of the sev.rapV12 transgene) and demonstrated the cell-autonomous rescue of sev0 in the R7 precursor by activated Rap.
A second assay for sev gene function lies in R1/6 precursors in which N is activated (sev.N*). In an otherwise wild-type background, sev.N* causes R1/6 cells to differentiate as R7 cells (2). However, in the absence of Sev (sev0;sev.N*), R1/6 cells differentiate as cone cells (13), and, in the adult eyes, only large rhabdomere cells are evident in the outer regions (R7 level) of the eye (Fig. 3F). Thus, removal of Sev from sev.N* ommatidia prevents the specification of the R1/6 cells as photoreceptors. The absence of Sev here can be rescued by the presence of activated Ras (sev0;sev.rasV12;sev.N*), and R7 cells are restored (13). We therefore tested whether activated Rap could similarly rescue the ectopic R7 cells of sev0;sev.N* eyes. Indeed it could: adult eyes of sev0;sev.rapV12;sev.N* showed the restoration of the R7 cells (Fig. 3G). In wild-type eye discs, the R1/6 cells express Svp, and the Runt-labeled R7 is found between them (Fig. 3H). In sev0;sev.rapV12;sev.N* eye discs, cells in the R1/6 positions do not express Svp and instead express the R7 marker Runt (Fig. 3I). Thus, from two separate assays, we find that an activated form of Rap is able to substitute for the absence of Sev, which is consistent with Rap normally lying downstream of Sev in its transduction pathway.
sev.sev* is suppressed by the loss of one copy of rap.
We next assayed whether an activated form of Sev was sensitive to rap gene dosage. We constructed and transformed an activated form of Sev (sev.sev*) following the process in ref. 32. These flies have rough eyes that show the multiple R7 photoreceptors in tangential section (Fig. 4A). When one copy of rap was removed from these eyes (sev.sev*/+;rap0/+), a decrease in the eye roughening occurred, and there was a significant reduction in the multiple R7 phenotype (from 90% of the ommatidia containing multiple R7 cells to 48%) (Fig. 4 A–C). Because activated Sev is sensitive to rap gene dosage, this finding is consistent with Rap lying downstream of Sev in the RTK transduction cascade.
sev.rapV12 is suppressed by the loss of one copy of rolled (rl).
MAPK in Drosophila is encoded by the rl gene (33), and removal of one copy of this gene suppresses the phenotype of activated Ras (33). We therefore tested whether activated Rap was similarly sensitive to rl gene dosage. Indeed it was: the eye roughening and multiple R7 phenotype of sev.rapV12 was significantly suppressed by the loss of one copy of rl (+/+;sev.rapV12/+ versus rl0/+;sev.rapV12/+) (from 62% of ommatidia containing multiple R7 cells to 26%) (Fig. 4 D–F). This analysis used the null rl10a allele. Another null allele (rlM41A10) showed a similar suppression (from 62% of ommatidia containing multiple R7 cells to 22%) (Fig. 4F).
Discussion
Work in the Drosophila embryonic terminal system defined a role for Rap in RTK transduction in which it acted cooperatively with Ras (20). tailless and huckebein are target genes of the Torso RTK pathway, and, when either Rap or Ras was removed, there was a decrease in their transcription. However, removal of Raf (which lies immediately downstream of both the GTPases) removed all transcriptional output from the target genes. Thus, Torso signaling was seen to be relayed to Raf partially by Ras and partially by Rap. In the Drosophila eye, we found a similar but interestingly different use of these small GTPases. In the general specification of the ommatidial cells, Ras is critically required but Rap is not. Consider the ras clones in the eye disk at the border with the wild-type tissue (Fig. 2A). Here, mosaic ommatidia are formed, and ras mutant cells can enter otherwise wild-type ommatidia. Even under these conditions, there is no response from the mutant cells; there is no evidence of even mild responses to RTK signaling. In comparison, the rap mutant cells show liberal evidence of general differentiation, and the only decrement is the absence of any R7 differentiation. In R7 cells, both Ras and Rap appear to be required to relay the high RTK signal, but, in the other cells, Ras alone is critically required.
We argue that the requirement for Rap in R7 likely stems from the high N activity in this cell. The antagonism of the photoreceptor fate generated by N requires a potent RTK activation to overcome it. This activation is provided by Sev, which is expressed at high levels in the R7 precursor, and the potent transduction of the RTK pathway requires both Ras and Rap. In comparison, cells that experience low N activity are readily specified by DER, requiring only Ras function for this purpose. We do not argue that there is any qualitative difference between the RTK pathways activated by Sev versus DER. Rather, we see Sev as simply supplying a high-level activation of the pathway (up and above the level already achieved by DER in the R7 precursor).
In all assays tested here, Rap and Ras appear to perform the same function in R7 specification. Both appear to lie downstream of Sev, dominant-negative forms of each induce the loss of R7 cells, and ectopic activation of either induces ectopic R7 cells. These observations raise the question of whether Ras and Rap behave identically or whether they engage different effectors. Our data do not distinguish between these models, and we do not rule out the possibility that Ras and Rap may have distinct activities in R7 specification. However, Mishra et al. (20), in their study of Torso signaling, demonstrated that Rap binds Raf, and that Torso activates MAPK in a raf-dependent manner. Hence, Rap may act similarly in the Drosophila eye: it may relay the RTK signals in a similar manner to Ras by binding and activating Raf and triggering the RTK phosphorylation cascade within the cell. If this model is correct, then Rap simply substitutes for a limitation in the Ras activity. One might imagine, for example, that there is not enough endogenous Ras protein to transduce the signal and simply supplying more may remove the need for Rap. However, it need not be the level of the Ras itself that is limiting; overactivity of Ras GTPase activating proteins or underactivity of Ras guanine exchange factors, among other possibilities (including combinations of these effects), could account for the limitation in the Ras pathway. Thus, it is not necessarily easy to predict and rescue the possible Ras limitation to resolve this issue.
We note that sev.rapV12 is not as potent in inducing ectopic R7 photoreceptors as sev.rasV12 is. Furthermore, sev.rasV12 transforms mystery cells into developing photoreceptors, whereas sev.rapV12 does not. Both these observations suggest that RapV12 is a weaker activator of the photoreceptor RTK pathways than RasV12 is and indicate that the native Ras is a more potent transducer of the RTK pathway than the native Rap is. Consider the ras0 loss-of-function clones in the disk (Fig. 2A) where there is no differentiation of photoreceptors even though Rap is present. This finding provides evidence that Rap cannot substitute for Ras, and the two are not redundant. In contrast, the rap0 mutant clones (Fig. 2 B–E) show extensive evidence of photoreceptor differentiation (excepting R7), indicating that Ras can potently specify photoreceptor fates in its absence. Neither are the two redundant in R7 specification, but for a different reason. Here, both Ras and Rap are required to transduce the Sev signal; loss of either leads to a failure of the cell-fate specification. We see Rap as supplying the extra transduction load for the high-level RTK signal, up and above the level performed by Ras.
Materials and Methods
Immunohistochemistry and Histology.
Protocol for adult eye sections has been previously described (2). Protocol for antibody staining has been previously described (13). Primary antibodies used were as follows: rabbit α-GFP, mouse α-GFP IgG2a (Molecular Probes), guinea pig α-Runt (gift from John Reinitz,Stony Brook University, Stony Brook, NY), rabbit α-Runt (gift from Andrea Brand, Gurdon Institute, Cambridge, UK), guinea pig α-Sens (gift from Hugo Bellen, Baylor College of Medicine, Houston, TX), rabbit α-Bar (gift from Kaoru Saigo, Kobe University, Kobe, Japan), mouse α-Svp (gift from Yash Hiromi, National Institute of Genetics, Shizuoka, Japan), rat α-ELAV, and mouse α-Cut (Developmental Studies Hybridoma Bank). Secondary antibodies used were Alexa Fluor 488, 555, and 647 (Molecular Probes).
Stocks.
The following fly stocks were used: sev[d2] and sev.N[ECN] (34), sev.rasV12 (30), raprvB3 (19), rasx7b (28), and Df(2R)rl10a and Df(2R)M41A10 (Bloomington Drosophila Stock Center).
Generation of sev.rapV12 and sev.rapN17 Flies.
Wild-type cDNA was obtained from Drosophila Genomics Resource Center. Then, 5′ PCR primers that included the single-nucleotide substitutions required to transform G to V at position 12 and S to N at position 17 were designed and used with a wild-type 3′ primer. PCR products were cloned into an attB vector containing 2× sev enhancer elements and the sev promoter element. Transformation of the resulting attB plasmids was carried out by BestGene Inc. following standard protocols.
Clonal Analysis.
To make rap0 clones, the raprvB3 allele was recombined to Frt80 and crossed to hsflp;Frt80, M(3)i55, ubi.GFP for larvae or w, hsflp;Frt80, w+ for adult analysis. To make ras0 clones in larvae, the rasx7b allele was recombined to Frt82 and crossed to hsflp;Frt82, M(3)w124, ubi.GFP. To make sev.rapV12 clones in the sev0 background, we recombined Frt82 with sev.rapV12 then crossed to w, sev0, hsflp;Frt82, y+. To make sev0 clones in the adult eyes, w, sev[d2] were recombined to Frt19A then crossed to w, hsflp, Frt19A, w+ flies. To induce clones, larvae were heat-shocked for 1 h at 37 °C between 24 and 48 h after egg laying.
Acknowledgments
We thank Veronika Brundula Sodja for contributions to early parts of this work; Richard Axel for his comments on the manuscript; Hugo Bellen, Andrea Brand, Yash Hiromi, John Reinitz, and Kaoru Saigo for their generous donation of antibodies; the Bloomington Drosophila Stock Center for fly stocks; and BestGene Inc. for generating fly stocks. This work was funded by National Institutes of Health Grant R01 EY012536 (to A.T.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Cooper MT, Bray SJ. R7 photoreceptor specification requires Notch activity. Curr Biol. 2000;10:1507–1510. doi: 10.1016/s0960-9822(00)00826-5. [DOI] [PubMed] [Google Scholar]
- 2.Tomlinson A, Struhl G. Delta/Notch and Boss/Sevenless signals act combinatorially to specify the Drosophila R7 photoreceptor. Mol Cell. 2001;7:487–495. doi: 10.1016/s1097-2765(01)00196-4. [DOI] [PubMed] [Google Scholar]
- 3.Miller AC, Lyons EL, Herman TG. cis-Inhibition of Notch by endogenous Delta biases the outcome of lateral inhibition. Curr Biol. 2009;19:1378–1383. doi: 10.1016/j.cub.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parks AL, Turner FR, Muskavitch MA. Relationships between complex Delta expression and the specification of retinal cell fates during Drosophila eye development. Mech Dev. 1995;50(2-3):201–216. doi: 10.1016/0925-4773(94)00336-l. [DOI] [PubMed] [Google Scholar]
- 5.Tsuda L, Nagaraj R, Zipursky SL, Banerjee U. An EGFR/Ebi/Sno pathway promotes Delta expression by inactivating Su(H)/SMRTER repression during inductive Notch signaling. Cell. 2002;110:625–637. doi: 10.1016/s0092-8674(02)00875-9. [DOI] [PubMed] [Google Scholar]
- 6.Krämer H, Cagan RL, Zipursky SL. Interaction of bride of sevenless membrane-bound ligand and the sevenless tyrosine-kinase receptor. Nature. 1991;352(6332):207–212. doi: 10.1038/352207a0. [DOI] [PubMed] [Google Scholar]
- 7.Reinke R, Zipursky SL. Cell-cell interaction in the Drosophila retina: The bride of sevenless gene is required in photoreceptor cell R8 for R7 cell development. Cell. 1988;55:321–330. doi: 10.1016/0092-8674(88)90055-4. [DOI] [PubMed] [Google Scholar]
- 8.Hafen E, Basler K, Edstroem JE, Rubin GM. Sevenless, a cell-specific homeotic gene of Drosophila, encodes a putative transmembrane receptor with a tyrosine kinase domain. Science. 1987;236(4797):55–63. doi: 10.1126/science.2882603. [DOI] [PubMed] [Google Scholar]
- 9.Tomlinson A, Bowtell DD, Hafen E, Rubin GM. Localization of the sevenless protein, a putative receptor for positional information, in the eye imaginal disc of Drosophila. Cell. 1987;51(1):143–150. doi: 10.1016/0092-8674(87)90019-5. [DOI] [PubMed] [Google Scholar]
- 10.Tomlinson A, Ready DF. Neuronal differentiation in Drosophila ommatidium. Dev Biol. 1987;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Li Y, Carthew RW, Lai ZC. Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell. 1997;90:469–478. doi: 10.1016/s0092-8674(00)80507-3. [DOI] [PubMed] [Google Scholar]
- 12.Tang AH, Neufeld TP, Kwan E, Rubin GM. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- 13.Tomlinson A, Mavromatakis YE, Struhl G. Three distinct roles for Notch in Drosophila R7 photoreceptor specification. PLoS Biol. 2011;9:e1001132. doi: 10.1371/journal.pbio.1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freeman M. The spitz gene is required for photoreceptor determination in the Drosophila eye where it interacts with the EGF receptor. Mech Dev. 1994;48(1):25–33. doi: 10.1016/0925-4773(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 15.Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 16.Kumar JP, et al. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development. 1998;125:3875–3885. doi: 10.1242/dev.125.19.3875. [DOI] [PubMed] [Google Scholar]
- 17.Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. A ras-related gene with transformation suppressor activity. Cell. 1989;56(1):77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- 18.Cook SJ, Rubinfeld B, Albert I, McCormick F. RapV12 antagonizes Ras-dependent activation of ERK1 and ERK2 by LPA and EGF in Rat-1 fibroblasts. EMBO J. 1993;12:3475–3485. doi: 10.1002/j.1460-2075.1993.tb06022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hariharan IK, Carthew RW, Rubin GM. The Drosophila Roughened mutation: Activation of a rap homolog disrupts eye development and interferes with cell determination. Cell. 1991;67:717–722. doi: 10.1016/0092-8674(91)90066-8. [DOI] [PubMed] [Google Scholar]
- 20.Mishra S, Smolik SM, Forte MA, Stork PJ. Ras-independent activation of ERK signaling via the Torso receptor tyrosine kinase is mediated by Rap1. Curr Biol. 2005;15:366–370. doi: 10.1016/j.cub.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 21.Feig LA. Tools of the trade: Use of dominant-inhibitory mutants of Ras-family GTPases. Nat Cell Biol. 1999;1:E25–E27. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- 22.Basler K, Siegrist P, Hafen E. The spatial and temporal expression pattern of sevenless is exclusively controlled by gene-internal elements. EMBO J. 1989;8(8):2381–2386. doi: 10.1002/j.1460-2075.1989.tb08367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knox AL, Brown NH. Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science. 2002;295:1285–1288. doi: 10.1126/science.1067549. [DOI] [PubMed] [Google Scholar]
- 24.O'Keefe DD, et al. Rap1 maintains adhesion between cells to affect Egfr signaling and planar cell polarity in Drosophila. Dev Biol. 2009;333(1):143–160. doi: 10.1016/j.ydbio.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuo T, Takahashi K, Suzuki E, Yamamoto D. The Canoe protein is necessary in adherens junctions for development of ommatidial architecture in the Drosophila compound eye. Cell Tissue Res. 1999;298:397–404. doi: 10.1007/s004419900107. [DOI] [PubMed] [Google Scholar]
- 26.Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53(2):217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- 27.Carthew RW, Rubin GM. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell. 1990;63:561–577. doi: 10.1016/0092-8674(90)90452-k. [DOI] [PubMed] [Google Scholar]
- 28.Halfar K, Rommel C, Stocker H, Hafen E. Ras controls growth, survival and differentiation in the Drosophila eye by different thresholds of MAP kinase activity. Development. 2001;128:1687–1696. doi: 10.1242/dev.128.9.1687. [DOI] [PubMed] [Google Scholar]
- 29.Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 30.Fortini ME, Simon MA, Rubin GM. Signalling by the sevenless protein tyrosine kinase is mimicked by Ras1 activation. Nature. 1992;355:559–561. doi: 10.1038/355559a0. [DOI] [PubMed] [Google Scholar]
- 31.Fortini ME, Rubin GM. Analysis of cis-acting requirements of the Rh3 and Rh4 genes reveals a bipartite organization to rhodopsin promoters in Drosophila melanogaster. Genes Dev. 1990;4:444–463. doi: 10.1101/gad.4.3.444. [DOI] [PubMed] [Google Scholar]
- 32.Basler K, Christen B, Hafen E. Ligand-independent activation of the sevenless receptor tyrosine kinase changes the fate of cells in the developing Drosophila eye. Cell. 1991;64:1069–1081. doi: 10.1016/0092-8674(91)90262-w. [DOI] [PubMed] [Google Scholar]
- 33.Brunner D, et al. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell. 1994;76:875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 34.Fortini ME, Rebay I, Caron LA, Artavanis-Tsakonas S. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]