Abstract
Protein synthesis on the ribosome requires translational GTPase factors to bind to the ribosome in the GTP-bound form, take individual actions that are coupled with GTP hydrolysis, and dissociate, usually in the GDP-bound form. The multiple copies of the flexible ribosomal stalk protein play an important role in these processes. Using biochemical approaches and the stalk protein from a hyperthermophilic archaeon, Pyrococcus horikoshii, we here provide evidence that the conserved C terminus of the stalk protein aP1 binds directly to domain I of the elongation factor aEF-2, irrespective of whether aEF-2 is bound to GTP or GDP. Site-directed mutagenesis revealed that four hydrophobic amino acids at the C terminus of aP1, Leu-100, 103, 106, and Phe-107, are crucial for the direct binding. P1 was also found to bind to the initiation factor aIF5B, as well as aEF-1α, but not aIF2γ, via its C terminus. Moreover, analytical ultracentrifugation and gel mobility shift analyses showed that a heptameric complex of aP1 and aP0, aP0(aP1)2(aP1)2(aP1)2, can bind multiple aEF-2 molecules simultaneously, which suggests that individual copies of the stalk protein are accessible to the factor. The functional significance of the C terminus of the stalk protein was also shown using the eukaryotic proteins P1/P2 and P0. It is likely that the conserved C terminus of the stalk proteins of archaea and eukaryotes can bind to translation factors both before and after GTP hydrolysis. This consistent binding ability of the stalk protein may contribute to maintaining high concentrations of translation factors around the ribosome, thus promoting translational efficiency.
Keywords: GTPase-associated center, ribosome protein P0, ribosome protein P1, hyperthermophilic archaeon
Major dynamic steps in translational initiation, elongation, and termination are promoted by the actions of several translational GTPase factors (1–3). During the elongation step, two elongation factors bind alternately to the ribosome in their GTP-bound states. After they have carried out their respective functions, which are linked to GTP hydrolysis, they then dissociate from the ribosome. The large ribosomal subunit contains an active center, termed the GTPase-associated center or factor-binding center, which interacts with the elongation factors (4). The GTPase-associated center plays a crucial role in the recruitment of translation factors, GTP hydrolysis, and the release of inorganic phosphate, which is required for the subsequent dissociation of the factor. Multiple copies of the acidic ribosomal protein, or so-called stalk protein, are key components of this functional center (5–9). The stalk proteins form homo- or heterodimers, and two or three dimers bind to the ribosome through an anchor protein, L10 in bacteria and P0 in eukaryotes (7, 9–12).
In the case of bacteria, the structure and function of the stalk protein L7/L12 (termed L12 hereafter) is well established. The L12 protein is composed of an N-terminal dimerization domain and a globular C-terminal domain, which is connected by a flexible hinge region and has a wide range of movement (7, 13). Recently, cryoelectron microscopy (cryo-EM) and X-ray crystallographic analyses have shown that, in the ribosome•elongation factor G (EF-G) complex, the globular C-terminal domain of L12 interacts directly with EF-G through the G′ domain, which is unique to EF-G (14–16). However, the interaction between L12 and EF-Tu and its contribution to GTP hydrolysis has not been clarified in the crystal structure of the ribosome•EF-Tu complex (17, 18). The analysis of NMR chemical shifts has demonstrated the presence of interactions between the C-terminal domain of purified L12 and the translational GTPases EF-G, EF-Tu, initiation factor 2 (IF2), and release factor 3 (RF3), which implies that L12 interacts with a structural feature shared by these factors (19). The detailed mechanisms of the interactions and their contributions to the functions of the factors remain unclear.
The archaeal L12 (termed aP1 hereafter) and the eukaryotic P1/P2 stalk proteins are related closely in terms of structure and function (20, 21). However, sequence comparisons and small-angle X-ray scattering analyses indicate that the archaeal/eukaryotic stalk proteins are not related structurally to bacterial L12, and might not be linked evolutionarily either (20). In fact, our recent crystal structure data showed that the mode of dimerization by the N-terminal domain of archaeal aP1 and its binding to aP0 are completely different from those of bacterial L12 (9). Whereas the C-terminal domain (CTD) of bacterial L12 is a relatively large globular structure that comprises 70 amino acids, the archaeal/eukaryotic stalk proteins contain a unique and compact C-terminal region that comprises only 20 amino acids (20). It is apparently impossible for this 20 amino acid C-terminal region to adopt a conformation that is similar to that of the CTD of the bacterial stalk protein. Therefore, even though the archaeal/eukaryotic and bacterial stalk proteins play analogous roles in the GTPase-associated events on the ribosome, they show remarkable structural differences. Thus, to gain a complete understanding of the ribosomal stalk, it is important to elucidate the structure-function relationships in the archaeal/eukaryotic proteins as well as in their bacterial counterparts.
The sequences of the C-terminal region of the stalk protein are highly conserved among archaea/eukaryotes; in particular, the Leu-Phe sequence at the C terminus is strictly conserved (Fig. S1). An additional characteristic feature of the archaeal/eukaryotic stalk proteins is that the conserved C-terminal sequence is shared by not only all stalk proteins (aP1 and P1/P2) but also the anchor proteins aP0 and P0 (20, 22). Although functional implications of the C-terminal segments of eukaryotic P0/P1/P2 (23–25) and archaeal aP0/aP1 (9) have been reported, their exact role is not yet well understood. Here we demonstrate the direct binding of the conserved C terminus of aP1 to elongation factors aEF-2 and aEF-1α, and initiation factor aIF5B. Surprisingly, the P1 stalk protein showed a similar ability to bind to both the GTP- and GDP-bound forms of aEF-2, and the P0•P1 complex could bind multiple aEF-2 molecules. These characteristics seem to be favorable for efficient translation.
Results
Characteristics of Binding Between Archaeal aP1 and aEF-2.
Using mass spectrometry under nondenaturing conditions, we first checked whether purified aEF-2 (with His tag) was free of associated nucleotides (Fig. S2 A and D). The mass of the purified aEF-2 was determined to be 83,914 ± 1 Da, which is slightly larger than the calculated mass of aEF-2 on the basis of its amino acid sequence (83,823 Da). When the purified aEF-2 was incubated with GDP, a shift in the peaks for aEF-2 was observed that corresponded to a mass increase of 426 Da (Fig. S2 B and E), which gave a total mass of 84,340 ± 1 Da. Furthermore, when the nonhydrolyzable GTP analogue guanylyl 5′-(β,γ-methylenediphosphonate) (GMPPCP) was added, a shift in the peaks was again observed. In this case, the mass of the major component was 84,431 ± 5 Da (Fig. S2 C and F), which gave an estimated increase in mass of 517 Da. These results indicate that the isolated aEF-2 was free of nucleotides, and that GDP- and GMPPCP-associated complexes were formed upon incubations with the respective nucleotides.
Binding between aP1 and aEF-2 was analyzed by native gel electrophoresis. Specific amounts of aP1 were incubated with increasing amounts of aEF-2•GDP (Fig. 1A) and aEF-2•GMPPCP (Fig. 1B). The incubation of aP1 with aEF-2 led to the disappearance of the high-mobility band of free aP1 (Fig. 1 A and B, lane 1) and the appearance of a distinct band with lower mobility than that of free aEF-2 (Fig. 1 A and B, lanes 2–5). We confirmed that the shifted band with lower mobility was the aP1•aEF-2 complex by removing this band from the gel and analyzing it by SDS-PAGE and immunoblotting with an anti-aP1 antibody (Fig. S3 A and B). The complexes of aP1•aEF-2 with GDP (Fig. 1A) and GMPPCP (Fig. 1B) formed very similar patterns on the nondenaturing gel, which suggested that aP1 binds to aEF-2•GDP and aEF-2•GMPPCP with more or less the same affinity. We also confirmed that aP1 has a similar ability to bind to aEF-2 in the absence of any nucleotide as well as in the presence of GTP, 5′-guanylyl imidodiphosphate, or guanosine 5′-O-[γ-thio]triphosphate (Fig. S4). Therefore, the binding of aP1 to aEF-2 seems to be nucleotide independent.
Fig. 1.
Binding of the archaeal stalk protein aP1 to elongation factor aEF-2. In the presence of excess amounts (1 mM) of GDP (A) or GMPPCP (B), aP1 homodimer (200 pmol) was incubated without aEF-2 (lane 1) or with 200 pmol (lane 2), 400 pmol (lane 3), 600 pmol (lane 4), or 800 pmol (lane 5) of aEF-2 in 10 μL solution at 70 °C. aEF-2 (200 pmol) was also incubated alone (lane 6). Individual samples were subjected to a gel mobility shift assay, as described in Materials and Methods. In the presence of the same amounts of GDP (C) and GMPPCP (D) as in A and B, the complexes were formed by mixing 200 pmol of aP1 dimer and 600 pmol of aEF-2 (lane 2), in the presence of 1 nmol (lane 3), 2 nmol (lane 4), or 4 nmol (lane 5) of the peptide that comprised the C-terminal 18 amino acids of aP1. Gel analysis was as in A and B.
Structural Elements Involved in aP1•aEF-2 Binding.
Considering previous functional data on the archaeal/eukaryotic stalk (9, 23–25), it can be inferred that the C-terminal portion of aP1 contains the site for aEF-2 binding. To confirm this functionality, nondenaturing gel analysis was performed after aP1 had been preincubated with aEF-2 in the presence of increasing amounts of a peptide that comprised the 18 C-terminal amino acids of aP1 (Fig. 1 C and D). As the amount of peptide added was increased, the bands that corresponded to the complexes disappeared, whereas bands of free aP1 appeared, which indicated that the peptide prevented the formation of a complex between aP1 and aEF-2•GDP (Fig. 1C) and between aP1 and aEF-2•GMPPCP (Fig. 1D). To characterize the binding further, we used various variants of aP1 and aEF-2 in the binding experiments. The substitution in aP1 of Ser for highly conserved Leu106 (Fig. 2A) and Phe107 (Fig. 2B) completely disrupted binding to aEF-2. The disruption of the factor binding was also observed by using the L103S mutant (Fig. S5). A partial effect on the binding was detected in the L100S mutation, whereas no effect was detected in the other amino acid substitutions E97L, E98L, A99S, A101S, A105S, and G108D (Fig. S5). On the other hand, aP1 could bind to the aEF-2 fragment that comprised domain I (Fig. 2C), as well as the fragment that comprised both domains I and II (Fig. S6), and migrate faster than the free aEF-2 fragments, presumably because of the increase in acidity upon formation of the complex with aP1. These results suggest that the hydrophobic amino acid residues at the C terminus of the aP1 stalk protein and domain I of aEF-2 are responsible for the binding between aP1 and aEF-2.
Fig. 2.
Analyses of aP1•aEF-2 binding using the mutant proteins. (A and B) Homodimers of the aP1 point mutants, Leu106Ser (A) and Phe107Ser (B) (200 pmol each), were incubated without aEF-2 (lane 1) or with 400 pmol (lane 2), 800 pmol (lane 3), 1.2 nmol (lane 4), or 1.6 nmol (lane 5) of aEF-2. aEF-2 (400 pmol) was also incubated alone (lane 6). (C) 200 pmol of wild-type aP1 were incubated with domain I of aEF-2 as in A and B. Gel analysis was carried out as in Fig. 1.
Binding of aP1 to the Other GTPase Factors.
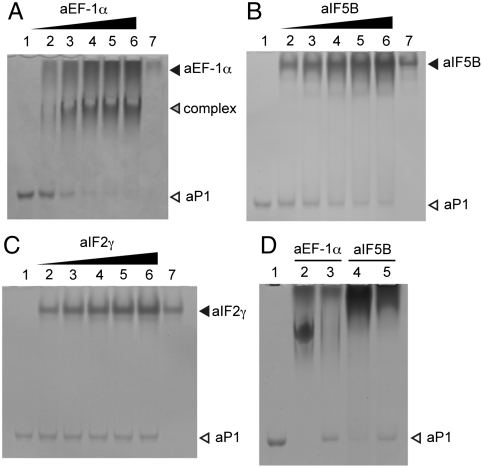
The ability of aP1 to bind to the other translational GTPase factors was checked by a gel mobility shift assay. In the case of aEF-1α (Fig. 3A), the purified aEF-1α did not enter the gel because of its basicity (Fig. 3A, lane 7). When aP1 (lane 1) was mixed with increasing amounts of aEF-1α (lanes 2–6), the band of free aP1 disappeared and a distinct band appeared, which corresponded to the aP1•aEF-1α complex, as shown by SDS-PAGE analysis of the shifted band (Fig. S3 C and D). For aIF5B (Fig. 3B), the intensity of the band of free aP1 (lane 1) decreased gradually as the amount of added aIF5B was increased (lanes 2–6), and a faint band appeared, which might correspond to the aP1•aIF5B complex. In contrast, aP1 did not bind to the GTPase aIF2γ at all (Fig. 3C). To test whether the C-terminal portion of aP1 also participates in the binding to aEF-1α and aIF5B, the C-terminal peptide of aP1 was added to the reactions as a binding competitor (Fig. 3D). The band for the aP1•aEF-1α complex that was formed by mixing aP1 and aEF-1α (Fig. 3D, lane 2) disappeared and the band for free aP1 appeared as the amount of peptide was increased (Fig. 3D, lane 3). Although a clear band for the aP1•aIF5B complex was not observed when aP1 and aIF5B were mixed, the band for free aP1 disappeared (Fig. 3D, lane 4). In the presence of an excess of the peptide, the band for aP1 reappeared (lane 5). These results indicate that the C-terminal portion of aP1 can bind to aEF-1α and aIF5B as well as aEF-2, but not to aIF2γ. However, the nucleotide dependency of the binding of aP1 to aEF-1α and aIF5B was not analyzed in the present study.
Fig. 3.
Binding of the aP1 stalk protein to GTPase factors other than aEF-2. (A–C) aP1 homodimer (200 pmol) was incubated without factor (lane 1) or with 400 pmol (lane 2), 800 pmol (lane 3), 1.2 nmol (lane 4), 1.6 nmol (lane 5), or 2.0 nmol (lane 6) of aEF-1α (A), aIF5B (B), or aIF2γ (C). aEF-1α, aIF5B, and a aIF2γ (400 pmol each) were also incubated alone (lane 7 of A–C, respectively). (D) The aP1•aEF-1α complex was formed by mixing 200 pmol of aP1 homodimer and 400 pmol of aEF-1α (lane 2), and 10 nmol of the C-terminal peptide of aP1 were also added (lane 3). The aP1•aIF5B complex was formed by mixing 200 pmol of aP1 dimer and 400 pmol of aIF5B (lane 4), and 10 nmol of the peptide were also added (lane 5). Gel analysis was carried out as in Fig. 1.
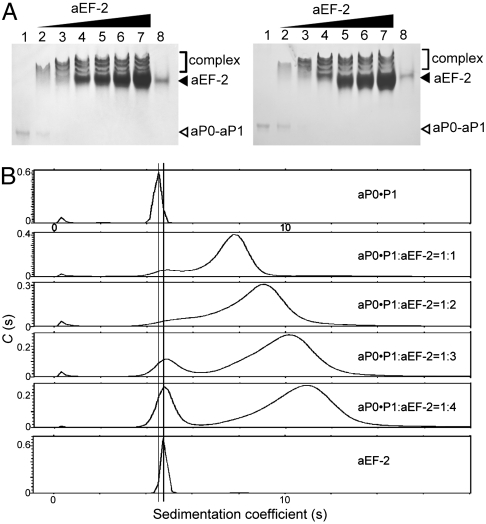
The Stalk Complex Can Bind Multiple Molecules of aEF-2.
We have shown previously that the archaeal ribosomal stalk proteins form a heptameric complex, aP0(aP1)2(aP1)2(aP1)2 (9). Given that aP1 and aP0 share a common C-terminal sequence, the stalk complex contains seven copies of this identical C-terminal sequence. Therefore, it could be imagined that the stalk complex in the ribosome might have the ability to interact simultaneously with multiple molecules of the GTPase factors. A gel mobility shift assay was performed with the aP0-aP1 heptameric complex (Fig. 4A). In the presence of either GDP (Fig. 4A, Left) or GMPPCP (Fig. 4A, Right), the isolated aP0•aP1 complex (lane 1) and aEF-2 (lane 8) migrated as single bands. However, when the aP0•aP1 complex was mixed with increasing amounts of aEF-2, at least three extra bands appeared, which migrated with lower mobility than aEF-2 (lanes 2–7). The binding of aEF-2 to the aP0•aP1 complex was also measured by sedimentation velocity (Fig. 4B). The aP0•aP1 complex and aEF-2 sedimented with an s value of 4.61S (f/f0 = 1.58) and 4.80S (f/f0 = 1.26), which gave an Mr of 102,000 and 76,500, respectively. On the basis of the calculated Mr for aEF-2 of 82,758, aEF-2 exists as a monomer in solution. The measured Mr for aP0•P1 agreed with the calculated Mr (117,207) of the heptameric complex aP0(aP1)2(aP1)2(aP1)2. When the aP0•aP1 complex was mixed with aEF-2 at a molar ratio of 1∶1, the complex sedimented with an s value of 7.85S. When the aP0•aP1 complex was mixed with increasing amounts of aEF-2 (two- to fourfold), the complex sedimented at 9.15–11.2S with a broad peak, which indicates that the peak corresponds to a reaction boundary that is composed of a mixture of both free aP0•P1 and aEF-2 species and their complexes. It is noteworthy that a significant amount of free aEF-2 (4.80S) was observed when the molar ratio of aP0•aP1:aEF-2 was 1∶4. From these results, we infer that the aP0•aP1 complex can interact simultaneously with multiple, presumably at least three, molecules of aEF-2, when excess amounts of the translation factors are present relative to the aP0•aP1 complex.
Fig. 4.
Binding of the P0-P1 stalk complex to multiple molecules of aEF-2. (A) In the presence of GDP (Left) or GMPPCP (Right), the aP0•aP1 heptameric complex (25 pmol) was incubated without aEF-2 (lane 1) or with 25 pmol (lane 2), 50 pmol (lane 3), 100 pmol (lane 4), 200 pmol (lane 5), 250 pmol (lane 6), or 380 pmol (lane 7) of aEF-2 at 70 °C. aEF-2 (25 pmol) was also incubated alone (lane 8). Gel analysis was carried out as in Fig. 1. (B) C(s) distributions from sedimentation velocity analytical ultracentrifugation of the following samples: The aP0•aP1 heptameric complex, which was preincubated without aEF-2 (Top) or with aEF-2 at a molar ratio of 1∶1 (Second Row), 1∶2 (Third Row), 1∶3 (Fourth Row), and 1∶4 (Fifth Row). aEF-2 was also incubated alone (Bottom).
Conserved Functional Feature of the C Terminus of Stalk.
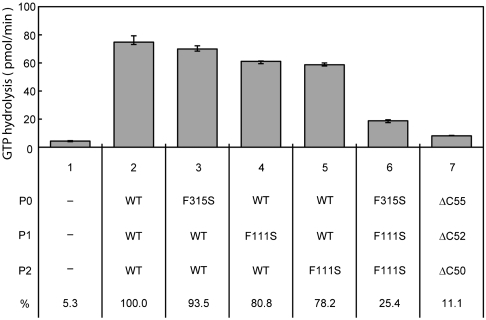
The ability of the C-terminal region of the stalk protein to bind the translation factors was also investigated with eukaryotic proteins. Formation of a stable complex between the isolated eukaryotic P1/P2 heterodimer and eEF-2 could not be detected by gel mobility shift assay. To investigate the functional contribution of the C termini in the eukaryotic P0/P1/P2 complex, we tested the effect of substituting the highly conserved Phe at the C terminus (Fig. S1) with Ser; we mutated Phe111 of both silkworm P1 and P2, which corresponds to Phe107 of Pyrococcus horikoshii aP1, as well as Phe315 of P0. The eukaryotic stalk complexes P0(P1–P2)2 were formed in vitro with mutants P0 (F315S), P1 (F111S), and P2 (F111S), as indicated in Fig. 5. Various complexes that included the mutant protein(s) were substituted for the Escherichia coli stalk complex L10(L12)4 in the 50S subunit of the E. coli ribosome, and the eEF-2-dependent GTPase activity of these complexes was measured. Although no significant effect was observed when a single protein in the P0(P1–P2)2 complex was mutated, the mutation of all three proteins caused a marked reduction in the GTPase activity. The activity when all three proteins were mutated was comparable with that of a complex composed of ΔC55-P0, ΔC52-P1, and ΔC50-P2, in which the C-terminal 55, 52, and 50 amino acids were truncated from P0, P1, and P2, respectively (25). These results indicate that the functionality of the C-terminal portion of the stalk protein is preserved from archaea to eukaryotes.
Fig. 5.
Functional effect of the point mutations at the C termini of eukaryotic P0, P1, and P2. The point mutants, Phe315Ser in P0, Phe111Ser in P1, and Phe111Ser in P2, were generated as described in Materials and Methods. The truncation mutants ΔC55-P0, ΔC52-P1, and ΔC50-P2, in which the C-terminal 55, 52, and 50 amino acids were deleted from P0, P1, and P2, respectively, were prepared as described previously (25). The P0•P1•P2 complexes were reconstituted (11) using the mutant proteins indicated below the bars. Each complex (10 pmol) was incorporated into the E. coli 50S core (2.5 pmol) with eL12, and eukaryotic eEF-2-dependent GTPase activity was assayed in the presence of E. coli 30S subunits (11).
Discussion
The aP1 stalk protein, which is present in multiple copies in the archaeal ribosome, and the eukaryotic homologs P1/P2 share a conserved C-terminal segment of approximately 20 amino acids, which shows no sequence similarity with the bacterial stalk protein L12 (20). The results of the present study with the archaeal proteins provide evidence that the translational GTPases aEF-1α, aEF-2, and aIF5B bind directly to the C terminus of aP1, and that the hydrophobic amino acid residues including the highly conserved Leu106 and Phe107 at the C terminus are responsible for the binding. We did not detect binding of aP1 to the initiation factor aIF2γ (Fig. 3C), which is involved in the binding of the initiator tRNA to the initiation codon on the 48S preinitiation complex; the preinitiation complex does not contain the large 60S subunit and thus does not contain the stalk (26). Regarding another GTPase, the RF3 homolog, which is absent from archaea, it has recently been clarified that, in archaea, aEF-1α plays an RF3-like role in addition to its usual role (27). Therefore, the aP1 stalk protein appears to interact directly with all the GTPases whose role in translation is coordinated by the 60S subunit.
Our results are compatible with previous data on the binding of eukaryotic P1 and P2 to eEF-2, which was detected by a yeast two-hybrid assay (10) and surface plasmon resonance experiments (28, 29), although these studies did not identify the exact binding site within P1/P2. We failed to detect direct binding between purified silkworm P1/P2 and eEF-2 under the conditions used in the present study for the archaeal proteins. However, we did determine, by mutagenesis, that the conserved Phe residues at the C termini of silkworm P1/P2 and P0 were required for eEF-2-dependent GTPase activity (Fig. 5). Therefore, it can be inferred that the conserved amino acid residues at the C termini of the stalk proteins participate in functional interactions with translational GTPases, although this interaction is very weak in mesophilic organisms. This view is supported by several lines of functional evidence, some of which have been published previously; for example, (i) removal of the C-terminal regions of eukaryotic P0/P1/P2 (Fig. 5) and of archaeal P0/P1 (9) markedly reduces ribosomal activity that depends on elongation factors; (ii) removal of the C-terminal portion of P0 in P1/P2-deficient yeast cells is lethal (24); and (iii) binding of a monoclonal antibody to the C terminus of the ribosomal stalk protein efficiently inhibits elongation factor-dependent events (23).
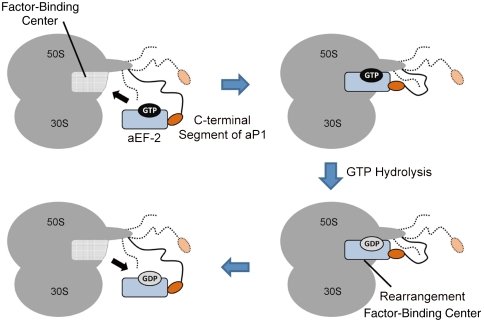
One of the most remarkable findings in the present study was that the ribosomal stalk protein aP1 bound to aEF-2 in a nucleotide-independent manner, which implies that the interaction between the C-terminal region of the stalk protein and aEF-2 remains constant before and after GTP hydrolysis. The results are unexpected, because it is generally accepted that the GTP-bound forms of translation factors have a high affinity for the ribosome, whereas their GDP-bound forms have a low affinity (1), and that the stalk proteins play an important role in the recruitment of translation factors to the ribosome and stimulate GTP hydrolysis (7). However, results obtained with the bacterial elongation factor EF-G were also inconsistent with the GTP/GDP affinity switch model. Kinetic experiments under saturating nucleotide conditions indicated that EF-G•GTP and EF-G•GDP bind to the ribosome with similar affinities (30). It can be inferred from current evidence that a conformational change in EF-G including the switch I region (31) occurs after GTP hydrolysis on the ribosome and induces the rearrangement of the ribosomal factor-binding center, which is composed of the sarcin/ricin loop and the L11 (E. coli terminology) binding region, including the H43/H44 domain, of the 23S/28S rRNA (15–17, 32). This structural rearrangement of the factor-binding center, which depends on GTP hydrolysis, seems to be responsible for a change in the affinity of binding of the factor to the ribosome and recycling of the factor. The evidence from the present study, namely that the C terminus of the stalk protein interacts with aEF-2 consistently in a nucleotide-independent manner, suggests that the stalk protein, at least in archaea/eukaryotes, does not participate in the GTP/GDP affinity switch of the ribosome/translation factor. Rather, it seems to interact both before and after GTP hydrolysis with a site on aEF-2 that is separate to the region that binds to the ribosomal factor-binding center and engages in the GTP/GDP affinity switch (Fig. 6). This interaction might help not only to recruit aEF-2•GTP to the ribosomal factor-binding center and promote GTP hydrolysis, but also to retain the factor in the GDP-bound or even nucleotide-free form near the ribosome after GTP hydrolysis. This nucleotide-independent binding of the stalk protein to translation factors would maintain a high concentration of translation factors around the ribosome, which would be favorable for efficient translation.
Fig. 6.
A schematic representation of the nucleotide-independent interaction between the ribosomal stalk protein aP1 and elongation factor aEF-2. Shown in orange is the C-terminal segment of a single copy of aP1 among the six copies bound to aP0. The C-terminal segment interacts with aEF-2 (blue) that is bound to GTP or GDP. The curved line between the C-terminal segment and the main body of the ribosome represents the flexible hinge region of aP1, which connects the C-terminal segment with the N-terminal domain (9).
Another interesting finding in the present study was that the stalk complex aP0(aP1)2(aP1)2(aP1)2 had the ability to bind multiple molecules of aEF-2 simultaneously, and the binding was, again, independent of the type of nucleotide bound (Fig. 4). Given the gel mobility shift of the P. horikoshii 50S subunit that occurred upon the addition of excess amounts of aEF-2 (Fig. S7), it seems likely that multiple aEF-2 molecules bind to the ribosome, although quantitative estimation of the number of copies of bound aEF-2 remains to be done. We could detect the binding of multiple molecules, presumably because the samples used were from a hyperthermophilic organism. Even so, the present study suggests that each aP1 protein bound to aP0 can interact independently with aEF-2. This view is consistent with our previous finding that each individual aP1 dimer shows a partial but significant factor-dependent GTPase activity, when assayed with hybrid ribosomes in which variants of the archaeal stalk complex are introduced into E. coli ribosomal cores that lack L10 and L12 (9). Therefore, all aP1 proteins within the heptameric complex seem to have the potential ability to recruit the elongation factor aEF-2 to the ribosome, and stimulate GTP hydrolysis at the factor-binding center, as discussed above. This tentative theory might explain the long-standing question of why the ribosome has multiple copies of the stalk protein. The combined action of multiple copies of the stalk protein seems to be required for efficient and accurate translation.
It is interesting to compare the present data on the archaeal (and eukaryotic) stalk with previous data on the bacterial stalk protein. The C-terminal domain of the bacterial L12 stalk protein, to which translation factors bind, is a globular form composed of three α-helices (α4–α6) and a triple-stranded β-sheet. The individual sites for interaction with IF2, EF-Tu, EF-G, and RF3 have been mapped to the α4-loop-α5 region of purified L12 by NMR (19), and the results are consistent with the functional findings from site-directed mutagenesis (8, 33). The binding of the C-terminal domain of L12 to EF-G was also detected in the ribosome•EF-G complex by cryo-EM (14, 15) and in recent crystal structure data (16). Although crystal structure data are not available for the C-terminal portion of the archaeal/eukaryotic ribosomal stalk protein, some structural features have been deduced by NMR analysis of peptides that comprise the 13 C-terminal amino acids from human and Leishmania brazililiensis P1/P2 (34). At low temperatures, the hydrophobic amino acid residues, including the conserved Leu–Phe motif, form a hydrophobic core within the peptide, which is unstable at higher temperatures. The present study showed that the four hydrophobic amino acids, which are located at one side of the putative α-helical structure of the C-terminal region (Fig. S5), are crucial for the factor binding. These amino acids residues seem to constitute an area, which might be important for its hydrophobic interaction with the translation factors. Therefore, the structure of the archaeal/eukaryotic stalk and the mode with which it binds translation factors are apparently different from those of the bacterial stalk described above. It is noteworthy that, despite the marked structural differences between the archaeal/eukaryotic and bacterial stalks, they can accomplish analogous factor-dependent functions, which might have been acquired through different and independent evolutionary pathways.
Materials and Methods
Plasmid Constructs.
The plasmids for the expression of P. horikoshii P1 and P0 were constructed as described previously (9). The coding sequences for P. horikoshii aEF-1α and aEF-2 were amplified by PCR and inserted into pET22b. The coding sequences for Sulforobus solfataricus aIF5B and Pyrococcus furiosus aIF2γ were amplified by PCR, inserted into pET28b, and cloned. We also generated a construct that encoded a His-tagged version of aEF-2 using pET26M. The plasmids for (i) P1ΔC3, which lacked the three amino acids at the C terminus of aP1, (ii) aP1 (L106S), in which Leu106 was replaced with Ser, and (iii) aP1 (F107S), in which Phe107 was replaced with Ser, were constructed using a QuikChange Site-Directed Mutagenesis Kit (Stratagene) in accordance with the manufacturer’s protocol. The DNA fragment that encoded domain I of aEF-2, namely amino acids 1–265, was also cloned into pET22b. The plasmids for the expression of silkworm P0, P1, and P2 were constructed as described previously (11). Mutants of P0 (F315S), P1 (F111S), and P2 (F111S), in which a Ser residue was substituted for Phe315 of silkworm P0, Phe111 of P1, and Phe111 of P2, were generated as described above with a QuikChange Site-Directed Mutagenesis Kit. The C-terminal truncation mutants of P0, P1, and P2 were constructed as described previously (25).
Preparation of Proteins and a Peptide.
P. horikoshii aP1 and its variants, aP0, and the aP0-aP1 complex were prepared as described previously (9). Individual translation factors were expressed in E. coli cells. After the cell extract had been heat treated at 70 °C for 30 min, the individual factors were prepared using the liquid chromatography system (GE Healthcare). aEF-2 (and domain I of aEF-2), IF5B, and IF2γ were purified with HiTrap Q-sepharose, followed by HiLoad 26/60 Superdex columns. For aEF-1α, HiTrap SP-sepharose was used instead of HiTrap Q-sepharose. Silkworm P0, P1, and P2 proteins and their variants were purified as described by Shimizu et al. (35). The silkworm stalk complex was reconstituted by mixing these components (11). A peptide that comprised the 18 amino acids of the C terminus of aP1, residues 91–108, was synthesized and purified by Hokkaido System Science.
Gel Mobility Shift Assay for aP1-Factor Binding.
aP1 dimers (200 pmol) or the aP0(P1)2(P1)2(P1)2 heptameric complex (25 pmol) were mixed with increasing amounts of individual translation factors in 10 μL of solution that contained 20 mM KCl, 10 mM MgCl2, and 20 mM Tris•HCl, pH 7.5. After preincubation at 70 °C for 10 min, each sample was subjected to gel electrophoresis using 5% or 6% polyacrylamide (acryalamide/bisacrylamide ratio 39/1) at 12.5 V/cm with a 192 mM glycine and 25 mM Tris buffer system for 1 h at room temperature. The gels were stained with Coomassie brilliant blue G-250.
Analytical Ultracentrifugation.
Sedimentation velocity experiments were carried out using an Optima XL-I analytical ultracentrifuge (Beckman-Coulter) with an eight-hole An50Ti rotor at 20 °C. Before centrifugation, aP0(aP1)2(aP1)2(aP1)2 heptameric complexes were mixed with increasing amounts of aEF-2 while maintaining a value of absorbance at 250 nm (A250) of 1.0 in a solution that contained 20 mM KCl, 10 mM MgCl2, and 20 mM Tris•HCl, pH 7.5. After preincubation at 70 °C for 10 min, each sample was transferred into a standard double sector cells with optical path length of 12 mm and centrifuged at a rotor speed of 40,000 rpm. The concentrations were monitored at 250 nm. The sedimentation velocity data were analyzed using the SEDFIT program (36).
Mass Spectrometry Under Nondenaturing Conditions.
Nucleotide-free aEF-2 (1 nmol) was mixed with 2 mM GDP or GMPPCP in 50 μL of a solution that contained 20 mM Tris•HCl, pH 8.0, 100 mM KCl, 1 mM dithiothreitol. After preincubation at 70 °C for 10 min, each sample was diluted twice, the buffer exchanged quickly to 100 mM ammonium acetate pH 8.0, and analyzed by using a Synapt High Definition Mass Spectrometer (Waters) under nondenaturing conditions at room temperature (37).
eEF-2-Dependent GTPase.
The silkworm stalk complex was used to replace the E. coli L10–L12 complex in the bacterial ribosome, and the GTPase activity that depended on eukaryotic eEF-2 was assayed, as described by Shimizu et al. (35).
Supplementary Material
Acknowledgments.
We thank Makoto Kimura (Kyushu University) for providing cells of P. horikoshii, Kosuke Itoh (Niigata University), and Tatsuaki Miwa (Hokkaido University) for helpful assistance with the research. This work was supported by a Grant-in-Aid for Scientific Research (B) (21370078 to T.U.), (A) (20247007 to I.T.), and on Innovative Areas (23121521 to S.U.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112934109/-/DCSupplemental.
References
- 1.Kaziro Y. The role of guanosine 5′-triphosphate in polypeptide chain elongation. Biochim Biophys Acta. 1978;505:95–127. doi: 10.1016/0304-4173(78)90009-5. [DOI] [PubMed] [Google Scholar]
- 2.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 3.Liljas A. Structural Aspects of Protein Synthesis. Singapore: World Scientific; 2004. pp. 99–158. [Google Scholar]
- 4.Wilson DN, Nierhaus KH. Ribosomal proteins in the spotlight. Crit Rev Biochem Mol Biol. 2005;40:243–267. doi: 10.1080/10409230500256523. [DOI] [PubMed] [Google Scholar]
- 5.Wahl MC, Möller W. Structure and function of the acidic ribosomal stalk proteins. Curr Protein Pept Sci. 2002;3:93–106. doi: 10.2174/1389203023380756. [DOI] [PubMed] [Google Scholar]
- 6.Mohr D, Wintermeyer W, Rodnina MV. GTPase activation of elongation factors Tu and G on the ribosome. Biochemistry. 2002;41:12520–12528. doi: 10.1021/bi026301y. [DOI] [PubMed] [Google Scholar]
- 7.Diaconu M, et al. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naganuma T, et al. Structural basis for translation factor recruitment to the eukaryotic/archaeal ribosomes. J Biol Chem. 2010;285:4747–4756. doi: 10.1074/jbc.M109.068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lalioti VS, Pérez-Fernández J, Remacha M, Ballesta JP. Characterization of interaction sites in the Saccharomyces cerevisiae ribosomal stalk components. Mol Microbiol. 2002;46:719–729. doi: 10.1046/j.1365-2958.2002.03179.x. [DOI] [PubMed] [Google Scholar]
- 11.Hagiya A, et al. A mode of assembly of P0, P1, and P2 proteins at the GTPase-associated center in animal ribosome: In vitro analyses with P0 truncation mutants. J Biol Chem. 2005;280:39193–39199. doi: 10.1074/jbc.M506050200. [DOI] [PubMed] [Google Scholar]
- 12.Krokowski D, et al. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol Microbiol. 2006;60:386–400. doi: 10.1111/j.1365-2958.2006.05117.x. [DOI] [PubMed] [Google Scholar]
- 13.Liljas A, Gudkov AT. The structure and dynamics of ribosomal protein L12. Biochimie. 1987;69:1043–1047. doi: 10.1016/0300-9084(87)90004-6. [DOI] [PubMed] [Google Scholar]
- 14.Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK. Interaction of the G’ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol Cell. 2005;20:723–731. doi: 10.1016/j.molcel.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 15.Harms JM, et al. Translational regulation via L11: Molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helgstrand M, et al. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J Mol Biol. 2007;365:468–479. doi: 10.1016/j.jmb.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 20.Grela P, et al. Structural relationships among the ribosomal stalk proteins from the three domains of life. J Mol Evol. 2008;67:154–167. doi: 10.1007/s00239-008-9132-2. [DOI] [PubMed] [Google Scholar]
- 21.Nomura T, et al. In vitro reconstitution of the GTPase-associated centre of the archaebacterial ribosome: the functional features observed in a hybrid form with Escherichia coli 50S subunits. Biochem J. 2006;396:565–571. doi: 10.1042/BJ20060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rich BE, Steitz JA. Human acidic ribosomal phosphoproteins P0, P1, and P2: Analysis of cDNA clones, in vitro synthesis, and assembly. Mol Cell Biol. 1987;7:4065–4074. doi: 10.1128/mcb.7.11.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uchiumi T, Traut RR, Kominami R. Monoclonal antibodies against acidic phosphoproteins P0, P1, and P2 of eukaryotic ribosomes as functional probes. J Biol Chem. 1990;265:89–95. [PubMed] [Google Scholar]
- 24.Santos C, Ballesta JP. The highly conserved protein P0 carboxyl end is essential for ribosome activity only in the absence of proteins P1 and P2. J Biol Chem. 1995;270:20608–20614. doi: 10.1074/jbc.270.35.20608. [DOI] [PubMed] [Google Scholar]
- 25.Naganuma T, Shiogama K, Uchiumi T. The N-terminal regions of eukaryotic acidic phosphoproteins P1 and P2 are crucial for heterodimerization and assembly into the ribosomal GTPase-associated center. Genes Cells. 2007;12:501–510. doi: 10.1111/j.1365-2443.2007.01067.x. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt E, Naveau M, Mechulam Y. Eukaryotic and archaeal translation initiation factor 2: A heterotrimeric tRNA carrier. FEBS Lett. 2010;584:405–412. doi: 10.1016/j.febslet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Saito K, et al. Omnipotent role of archaeal elongation factor 1 alpha (EF1α) in translational elongation and termination, and quality control of protein synthesis. Proc Natl Acad Sci USA. 2010;107:19242–19247. doi: 10.1073/pnas.1009599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bargis-Surgey P, et al. Interaction of elongation factor eEF-2 with ribosomal P proteins. Eur J Biochem. 1999;262:606–611. doi: 10.1046/j.1432-1327.1999.00434.x. [DOI] [PubMed] [Google Scholar]
- 29.Smulski CR, et al. Interaction map of the Trypanosoma cruzi ribosomal P protein complex (stalk) and the elongation factor 2. J Mol Recognit. 2011;24:359–370. doi: 10.1002/jmr.1089. [DOI] [PubMed] [Google Scholar]
- 30.Baca OG, Rohrbach MS, Bodley JW. Equilibrium measurements of the interactions of guanine nucleotides with Escherichia coli elongation factor G and the ribosome. Biochemistry. 1976;15:4570–4574. doi: 10.1021/bi00666a004. [DOI] [PubMed] [Google Scholar]
- 31.Ticu C, Nechifor R, Nguyen B, Desrosiers M, Wilson KS. Conformational changes in switch I of EF-G drive its directional cycling on and off the ribosome. EMBO J. 2009;28:2053–2065. doi: 10.1038/emboj.2009.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moazed D, Robertson JM, Noller HF. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 33.Kothe U, Wieden HJ, Mohr D, Rodnina MV. Interaction of helix D of elongation factor Tu with helices 4 and 5 of protein L7/12 on the ribosome. J Mol Biol. 2004;336:1011–1021. doi: 10.1016/j.jmb.2003.12.080. [DOI] [PubMed] [Google Scholar]
- 34.Soares MR, Bisch PM, Campos de Carvalho AC, Valente AP, Almeida FC. Correlation between conformation and antibody binding: NMR structure of cross-reactive peptides from T. cruzi, human and L. braziliensis. FEBS Lett. 2004;560:134–140. doi: 10.1016/S0014-5793(04)00088-2. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu T, et al. Interaction among silkworm ribosomal proteins P1, P2 and P0 required for functional protein binding to the GTPase-associated domain of 28S rRNA. Nucleic Acids Res. 2002;30:2620–2627. doi: 10.1093/nar/gkf379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.