Abstract
Photoreceptor cell death accompanying many retinal degenerative disorders results in irreversible loss of vision in humans. However, the precise molecular pathway that executes cell death is not known. Our results from a Drosophila model of retinal degeneration corroborate previously reported findings that the developmental apoptotic pathway is not involved in photoreceptor cell demise. By undertaking a candidate gene approach, we find that players involved in the immune response against Gram-negative bacteria are involved in retinal degeneration. Here, we report that the NF-κB transcription factor Relish regulates neuronal cell death. Retinal degeneration is prevented in genetic backgrounds that block Relish activation. We also report that the N-terminal domain of Relish encodes unique toxic functions. These data uncover a unique molecular pathway of retinal degeneration in Drosophila and identify a previously unknown function of NF-κB signaling in cell death.
Keywords: apoptosis, retinitis pigmentosa, nuclear factor kappa B, innate immunity, norpA
Progressive retinal dystrophies attributable to genetic or environmental causes result in gradual yet severe impairment of vision in humans. The pathogenesis of retinal degeneration is complex and not very well understood. There is considerable heterogeneity not only in the underlying causes but in the mode of inheritance of disease genes and the broad spectrum of clinical severity (1). Conditions such as retinitis pigmentosa (RP) and macular degeneration present themselves as the most commonly encountered forms of retinal degeneration in patients. A number of disease loci have been mapped, and, to date, >150 genes have been implicated (http://www.sph.uth.tmc.edu/retnet/) (2). Despite diversity in the ascribed etiologies, the eventual death of photoreceptor neurons by apoptosis is the shared pathway in retinal degeneration and is the cause of irreversible vision loss. Thus, therapeutic strategies targeting photoreceptor cell death may present as a viable alternative to individual gene therapies. A major impediment to such an approach is lack of a clear understanding of precise molecular pathways engaged during photoreceptor cell death.
In Drosophila, as in humans, mutations in the visual transduction pathway result in retinal degeneration (3). Previously, we and others have uncovered a cell death pathway that is initiated by the excessive endocytosis of rhodopsin (4, 5). Several mutations in flies and in humans trigger retinal cell death by this mechanism. One such mutation, norpA, results in rapid and light-dependent retinal degeneration. The norpA locus encodes an eye-specific phospholipase C that is essential for the photoresponse. The retinal degeneration is attributed to excessive light-dependent endocytosis of rhodopsin (4, 6) and its subsequent accumulation in the late endosomes (7). Previous analysis of norpA retinal degeneration has revealed that the key regulators of the developmental programmed cell death (PCD) in Drosophila are not involved in the cell death pathway (8). These results suggest that photoreceptor cell death in norpA might use alternative signaling mechanism(s).
In an effort to uncover the molecular mechanism behind retinal cell death, we explored the involvement of the death effector domain-containing caspase (Dredd) in retinal degeneration. We observed that in norpA dredd double mutants, photoreceptor cell death was prevented. Based on the known functions of Dredd in the innate immune response against Gram-negative bacterial infection, we examined the possibility of an activated immunity pathway in photoreceptor cell death. Surprisingly, the key regulator of this pathway, relish, mediates the death of norpA photoreceptors. We also report that the aminoterminal domain of Relish can exert toxicity in several different cell types. Together, our findings uncover an alternative mechanism of cell death in Drosophila, assign pleiotropic roles to previously characterized genes, and provide the basis for exploring therapeutic intervention of NF-κB signaling in a subset of retinal degeneration.
Results
Developmental Apoptosis Plays a Minimal Role in norpA Photoreceptor Cell Death.
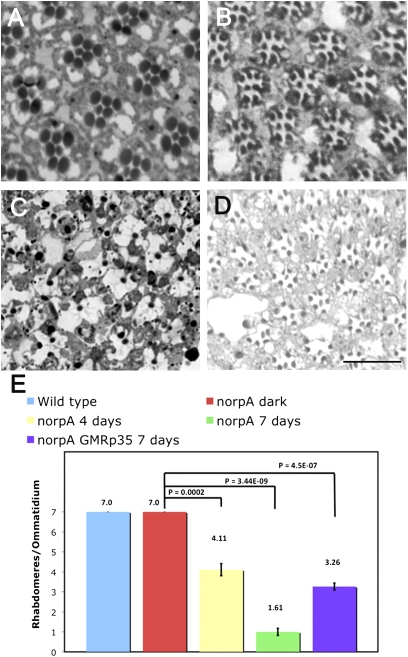
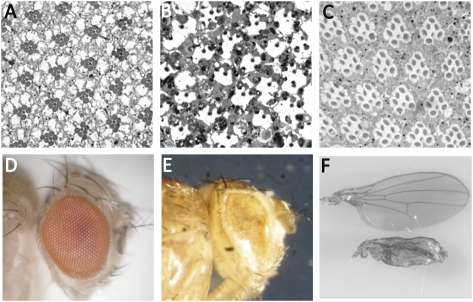
Ultrastructural studies of the degenerating norpA retina reveal that the dying photoreceptors display characteristic hallmarks of cells undergoing apoptosis, namely, loss of cell-cell contact, cytoplasmic condensation, accumulation of aberrant vesicles, and phagocytosis by the neighboring pigment cells (4). At a lower magnification, retinal degeneration is evident by the loss of outer (R1–R6) photoreceptors, which can be readily assayed by following the rhabdomere morphology, a widely accepted readout for retinal degeneration in flies (5). The R1–R6 cells express major rhodopsin (Rh1) that is photoactivable by wavelengths in the visible spectrum, whereas the R7 cell expresses UV-sensitive rhodopsin. The outer photoreceptors of dark-raised norpA flies do not undergo degeneration (Fig. 1A). Each ommatidium has seven compact rhabdomeres (Fig. 1 A and E). With the onset of light-dependent retinal degeneration, outer photoreceptor rhabdomeres become less compact, and by 4 d of light exposure, some photoreceptors are lost (discernable rhabdomeres/ommatidium: 4.1 ± 0.3) (Fig. 1 B and E). The degeneration is complete by 7 d of light exposure, when most of the photoreceptors are lost from the retina (discernable rhabdomeres/ommatidium: 1.6 ± 0.1) (Fig. 1 C and E).
Fig. 1.
Photoreceptor cell death in norpA. Cross-sections (0.5 μm) of retinas of 7-d dark-raised norpA flies (A), norpA flies exposed to 4 d of continuous light (B), norpA flies after 7 d of light exposure (C), and norpA GMRp35 flies exposed to 7 d of constant light (D) are shown. (Scale bar, 17.5 μm.) (E) Ratio of the total number of discernable rhabdomeres to the total number of ommatidia was used as a measure of photoreceptor cell death progression. Errors bars indicate SEM. Data from 7-d light-exposed WT (w1118) flies are included as a reference in the blue histogram bar. The number of ommatidia was counted for w1118 (light), 162 (n = 3 flies); for norpA (dark), 137 (n = 3 flies); for norpA (4 d of light), 107 (n = 4 flies); for norpA (7 d of light), 150 (n = 7 flies); and for norpA p35 (7 d of light), 134 (n = 6 flies).
Developmental apoptosis in Drosophila functions by the activation of the apical caspase Dronc, which, in turn, triggers the caspase-3 homolog Drice. Upstream activators of Dronc include the reaper (rpr), hid, and grim (RHG) genes, which are transcriptionally activated during PCD (9–12). A deficiency of these genes eliminates virtually all PCD during Drosophila embryogenesis (9). In addition, the initiator caspase Dronc relies on physical interaction with the Apaf-1–related killer (Ark) for its activation by formation of an apoptosome (13). Previous work has demonstrated that the deletion of RHG genes does not prevent norpA-mediated retinal degeneration (8). These results could mean that these genes might play a redundant role in norpA photoreceptor cell death or are not engaged at all. To analyze the role of these genes in cell death further, we performed real-time RT-PCR to detect if there is up-regulation of the transcripts from these loci. We did not detect any transcriptional activation of rpr and hid genes in light-exposed norpA (Fig. S1A). Further genetic and biochemical analysis demonstrated that caspases involved in PCD play a limited role in norpA-mediated cell death. Dronc is inhibited by the Drosophila Inhibitor of Apoptosis-1 protein (DIAP-1). DIAP-1 protein levels rapidly decline by ubiquitin/proteasome-mediated degradation after activation of RHG proteins (14–16). However, immunoblotting using the DIAP-1 antibody revealed that the steady-state protein level of monomeric, nonubiquitylated DIAP-1 did not decrease on light exposure in the norpA retina (Fig. S1B); instead, a slight light-dependent increase in DIAP-1 levels was observed. These data suggest that DIAP-1 is not the target of prodeath signaling in norpA. Moreover, we also found that Ark is not involved in the cell death pathway. We used the eyeless-FLP/FRT system (17) to generate mosaic clones of arkG8, a strong loss-of-function allele (18), and examined the role of this gene in cell death. The norpA+; arkG8 photoreceptors do not undergo light-dependent cell death (Fig. S1 C and E). However, this allele failed to rescue the norpA photoreceptors after 7 d of light exposure (Fig. S1 D and E). In contrast, we found a limited role for effector caspases in norpA-mediated cell death. We observed that the retina of norpA GMRp35 flies, which expresses the baculoviral caspase inhibitor p35 in an eye-specific manner, underwent degeneration after 7 d of light exposure (Fig. 1D). Although retinal degeneration was evident, many of the outer photoreceptors had discernable rhabdomeres and the retina resembled that of the norpA flies in the earlier stages of the degenerative process (discernable rhabdomeres/ommatidium: 3.2 ± 0.1) (Fig. 1E). Thus, expression of the baculoviral caspase inhibitor p35 provided partial protection by slowing the photoreceptor death, suggesting a minor role for effector caspases in the cell death pathway. Similar findings with p35 and DIAP-1 expression have been reported previously reported for another allele of norpA (8).
Together, these data corroborate previous results of degeneration in norpA, rule out allele-specific differences in the nature of cell death, and suggest that the developmental PCD pathway mediated by Dronc is not activated in photoreceptor cell death. In this light, the observation that the p35 caspase inhibitor slows retinal degeneration is interesting, because it suggests that caspase activation accounts for at least some of the observed death. We hypothesized that norpA degeneration might use an initiator caspase other than Dronc and that this caspase activates p35-sensitive and p35-insensitive substrates. Thus, we investigated the possibility of involvement of other initiator caspases in retinal degeneration.
norpA Photoreceptor Death Requires Dredd Caspase.
The Drosophila genome encodes three caspases with a long aminoterminal prodomain similar to the vertebrate initiator caspases. These are Strica, Dronc, and Dredd (19). Ectopic expression of Strica can induce death, but relatively little is understood about its participation in apoptosis. The dredd gene encodes a Drosophila homolog of mammalian caspase-8. dredd mRNA specifically accumulates in cells destined for PCD, and dredd mutants display defects in apoptosis (20, 21). Because the role played by dredd in apoptosis was more extensively studied, we explored if this gene participated in retinal degeneration.
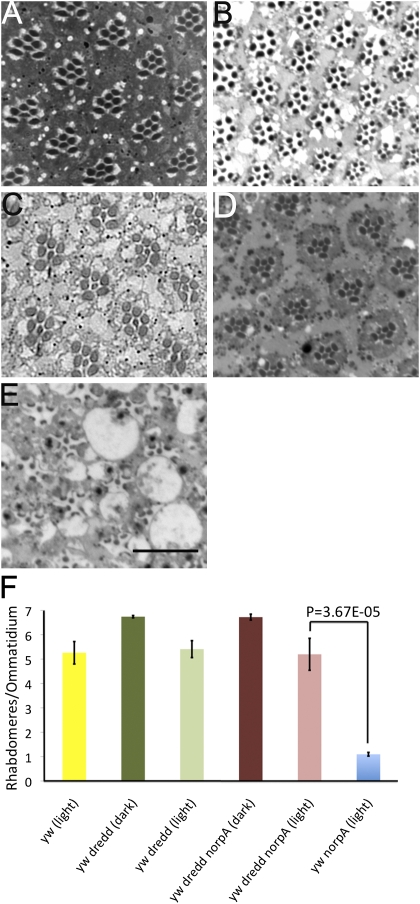
We used a previously characterized and extensively used null allele of dredd (dreddB118) (22) to examine its role in norpA. Dark-raised dredd flies display a WT retinal morphology, suggesting that this mutation does not affect PCD during eye development (Fig. 2 A and F). Continuous light exposure for 6 d results in a visible change in retinal morphology. The intraommatidial space increases, the rhabdomeres appear reduced in size, and some photoreceptors lose their rhabdomeres altogether (Fig. 2 B and F). The light-induced alteration in retinal morphology of norpA+ dredd flies (Fig. 2B) is likely attributable to the genetic background (y−) of these flies, because yw flies also display a loss of some photoreceptor cells after light treatment (Fig. 2F), which is not seen in w flies. Dark-adapted norpA dredd adult flies have a WT retinal morphology, indicating that norpA mutation in dredd background does not alter the normal developmental program (Fig. 2 C and F). Interestingly, norpA flies bearing the dreddB118 mutation did not undergo light-dependent retinal degeneration, as judged by the presence of intact rhabdomeres and a regular ommatidial array (Fig. 2 D and F), compared with norpA flies in the identical genetic background exposed to 6 d of light (Fig. 2 E and F). The quantitation of rhabdomeres per ommatidium indicates that a significant number of norpA dredd photoreceptors survive in light compared with norpA (Fig. 2F). These data suggest that Dredd is required for light-induced cell death but is dispensable during developmental apoptosis.
Fig. 2.
Photoreceptor death requires dredd. Cross-sections (0.5 μm) of retinas of ydreddB118w flies raised in darkness for 6 d (A), ydreddB118w flies exposed to light for 6 d (B), dreddB118wnorpA mutants raised in the dark for 6 d (C), ydreddB118wnorpA flies exposed to 6 d of light (D), and ywnorpA flies exposed to 6 d of constant light (E) are shown. (Scale bar, 17.5 μm.) (F) Ratio of the total number of discernable rhabdomeres to the total number of ommatidia was used as a measure of photoreceptor viability in the indicated genotypes. Data from 6-d light-exposed yw flies are included as a reference in the yellow histogram bar to compare the effects of genetic background. Ratios for ydreddB118w (dark), 6.73 ± 0.047 (110 ommatidia, 4 flies); for ydreddB118w (light), 5.4 ± 0.34 (66 ommatidia, 3 flies); for ydreddB118wnorpA (dark), 5.19 ± 0.65 (58 ommatidia, 3 flies); for ydreddB118wnorpA (light), 6.68 ± 0.08 (114 ommatidia, 5 flies); and for ywnorpA (light), 1.09 ± 0.07 (92 ommatidia, 6 flies) are shown. Data are the mean ± SEM.
norpA Degeneration Is Independent of Immune Response Mediators Upstream of dredd
Although dredd mutants display defects in cell death, the known physiological function of dredd is in immune response to Gram-negative bacterial infection (22). In this pathway, Imd and dFadd proteins form an important connecting link between bacterial recognition and Dredd activation that ultimately leads to transcriptional up-regulation of antimicrobial peptide (AMP)-encoding genes (21). We investigated the role of the Imd/dFadd signaling complex in cell death using loss-of-function alleles in each of these genes. The norpA imd1 flies underwent light-dependent degeneration after 7 d of exposure (Fig. S2A). There is no significant difference in the extent of cell death between norpA and norpA imd1 flies, as judged by discernable rhabdomeres/ommatidium quantitation (Fig. S2B). The imd1 allele is a severe hypomorph (21). To examine if retinal degeneration depends on the Imd/dFadd complex, we used the null allele of dfadd (dfaddf02804) (23). Epistasis analysis shows that dfadd acts downstream of imd but upstream of dredd in immunity signaling (24). Because of the w+ marker associated with this allele, the norpA dfadd flies were red-eyed. This necessitated prolonged light exposure (Materials and Methods). Thus, we examined rhabdomere integrity in norpA dfadd flies exposed to 2, 3, and 4 wk of constant light (Fig. S2 C–E). We observed that the retinal morphology progressively deteriorated with light treatment. Comparison of discernable rhabdomeres/ommatidium of norpA dfadd flies with red-eyed norpA flies treated with a similar light regimen indicated that the photoreceptors were lost at a similar rate (Fig. S2F). Thus, norpA dfadd double mutants did not reveal any photoreceptor rescue. These results suggest that in photoreceptors, dredd-mediated cell death occurs via a pathway that is distinct from the canonical Imd-dFadd signaling.
Relish Triggers Photoreceptor Cell Death.
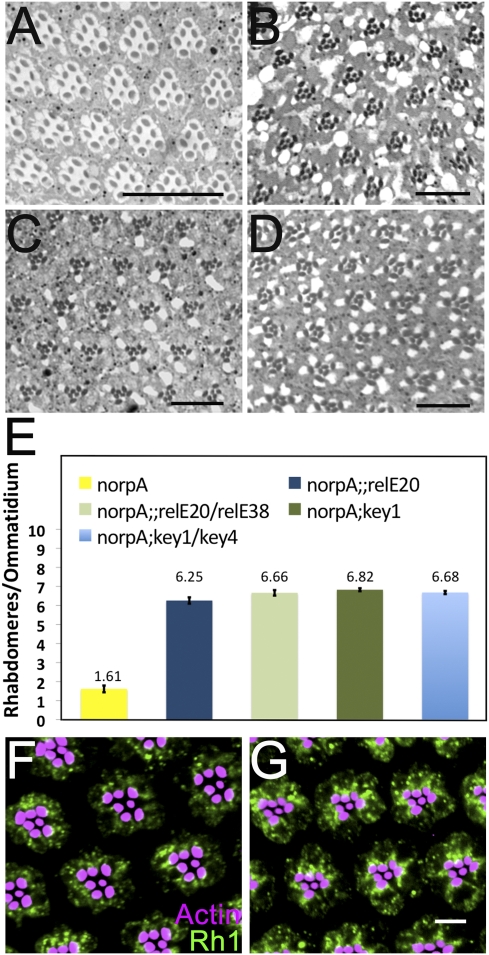
Next, we examined if activation of the downstream immune signaling pathway occurs in norpA and if this is involved in retinal degeneration. The innate immune response to Gram-negative bacteria in Drosophila requires activation of the NF-κB homolog encoded by the relish (rel) gene (25). Active Dredd mediates endoproteolytic cleavage of Relish to generate an N-terminal fragment and a C-terminal fragment (26, 27). N-terminal domain (NTD) of Relish is capable of serving as a transcriptional activator. Among the known nuclear targets of this domain are the conserved κB-like motifs in the promoter region of AMP genes. To examine if Relish plays a role in light-dependent retinal degeneration, we assessed the retinal morphology of light-exposed norpA relE20 flies (Fig. 3A). Interestingly, we observed that rel completely rescued the light-dependent photoreceptor death in norpA flies. The relE20 is a null allele obtained by imprecise P-element excision (25). To confirm that the rescue of norpA photoreceptors is a relish-dependent phenomenon, we used an allelic series of relE20 and relE38 in a norpA background. The norpA relE20/relE38 flies also did not undergo retinal degeneration on light exposure (Fig. 3B). These data suggest that Relish plays a key role in retinal degeneration.
Fig. 3.
(A and B) Retinal cross sections (0.5 μm) of (A) norpA;; relE20 and (B) norpA relE20/relE38 (B) flies exposed to 7 d of constant light. (C and D) Retinal sections from 7 d light-treated (C) norpA; key1 and (D) norpA; key1/key4 flies reveal rescue of photoreceptor cells in these backgrounds. (Scale bars, 17.5 μm.) (E) Ratio of total number of discernable rhabdomeres to the total number of ommatidia was used as measure of photoreceptor viability in indicated genotypes. Ratio for norpA;;relE20, 6.25 ± 0.16 (82 ommatidia, 5 flies); for norpA;;relE20/relE38, 6.62 ± 0.144 (47 ommatidia, 3 flies); for norpA;key1, 6.82 ± 0.80 (70 ommatidia, 4 flies); for norpA;key1/key4, 6.68 ± 0.08 (77 ommatidia, 4 flies). Data are Mean ± SEM. (F and G) Rh1 accumulates in the cytoplasm of (F) norpA and (G) norpA relE20 retina in a light pulse-chase experiment. (Scale bar, 5 μm.)
Dredd regulates Relish activation in at least two ways. In addition to its suggested role as the responsible protease, Dredd is required for IκB kinase (IKK) complex activation (28). The Drosophila IKK complex consists of a catalytic subunit encoded by the ird5 gene and a regulatory subunit encoded by the kenny (key) gene (27, 29–31). In the absence of a functional IKK complex, flies fail to activate Relish. We reasoned that if Relish activation is responsible for retinal degeneration, failure to activate Relish should rescue photoreceptor cell death. Thus, we examined the effect of light exposure on norpA key1 flies (Fig. 3 C and E). We observed that norpA key1 flies did not undergo light-dependent retinal degeneration after 7 d of continuous light exposure. key1 is a null allele generated by ethyl methane sulfonate mutagenesis (30). To reduce the likelihood that the rescue of retinal degeneration was attributable to second-site mutations on the chromosome, we used a heterozygous combination of key1 and key4 alleles. This allelic combination also rescued photoreceptor cell death in norpA after 7 d of light exposure (Fig. 3 D and E). This analysis suggests a crucial role of Relish activation in triggering photoreceptor cell death.
We have previously shown that in norpA, massive light-induced endocytosis of Rh1 causes photoreceptor death (6). Preventing Rh1 internalization blocks photoreceptor death (6). This rapid internalization is marked by failure in timely degradation of Rh1 and can be assessed by the presence of persistent vesicles in a light pulse-chase experiment (7). In this assay, flies are exposed to 2 d of constant light to induce Rh1 endocytosis, followed by a 13-h chase period in darkness. The norpA photoreceptors show accumulated cytoplasmic Rh1 after the chase period (Fig. 3F). One possible explanation for the rescue of norpA-mediated cell death by rel is that this mutation also blocks Rh1 endocytosis or enhances clearance of accumulated Rh1. To examine if rel affects Rh1 trafficking, we examined the fate of internalized Rh1. In norpA rel flies, we observed that Rh1+ vesicles persisted in the cytoplasm after the recovery period (Fig. 3G), indicating that rel does not affect endocytosis and accumulation of Rh1 in the norpA background and that Relish activation occurs downstream of Rh1 accumulation.
Relish-Dependent Immunity Pathway Is Activated in Degenerating Photoreceptors.
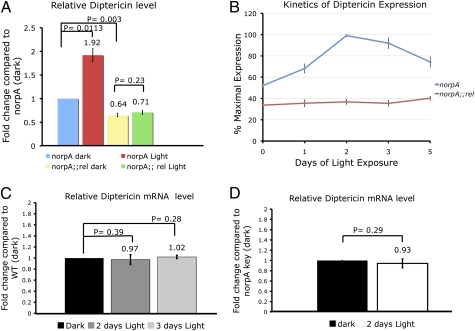
Next, we examined if Relish functions as a transcription factor in photoreceptors. We assessed the transcription from the diptericin (dipt) locus, which is a widely used signature for Relish activity (32). We measured the amount of dipt mRNA in norpA flies before and after 2 d of light treatment (Fig. 4A). Light exposure led to an approximately twofold up-regulation of dipt mRNA in norpA flies. In the norpA relE20 (norpA rel) dark-raised flies, the dipt transcript levels were ∼35% less than those in norpA dark-raised flies (Fig. 4A). In norpA rel flies exposed to 2 d of light, up-regulation of the dipt transcript level was not observed (Fig. 4A), indicating that light-induced up-regulation of dipt transcription was relish-dependent. We find that the dipt mRNA levels reached a maximum after 2 d in light and then declined by 5 d (Fig. 4B). In contrast, norpA rel flies failed to up-regulate dipt transcription even after prolonged light exposure (Fig. 4B). To determine if Relish activation in the eye is a general response to light, we measured dipt mRNA level in WT flies (Fig. 4C). We did not observe a significant increase in the transcript levels in WT flies after 2 d in light. Extending the continuous light treatment to 3 d did not significantly elevate the dipt transcript levels (Fig. 4C). Thus, relish activation appears to be a specific outcome of light stimulation in norpA mutants. Together, these data indicate that the immune signaling pathway mediated by relish is activated in the norpA retina in a light-dependent manner.
Fig. 4.
Relish is activated in norpA. (A) Data show fold change in the dipt transcript level in 2-d light-exposed norpA eyes relative to dark-reared norpA retinas examined by quantitative RT-PCR assay. The dipt mRNA levels in light-exposed norpA rel flies are not significantly different from those in their dark-reared siblings. (B) Kinetics of dipt expression in norpA and norpA rel flies. (C) Quantitative RT-PCR analysis shows the relative change in dipt expression in WT (w1118) flies. (D) Relative dipt expression in norpA key1 flies after light exposure. Error bars indicate SEM.
Next, we examined the effect of the key1 mutation on Relish activity in light-treated norpA flies. We observed that the norpA key1 flies failed to significantly up-regulate the dipt mRNA level after light treatment (Fig. 4D), suggesting that the key mutation diminishes Relish activity in norpA flies. In summary, a mutation in relish confers protection against retinal degeneration, photoreceptor death is prevented in genetic backgrounds that prevent Relish activation, and Relish transcriptional targets are up-regulated during cell death. Together, based on this genetic evidence, we conclude that Relish activation plays an important role in eliciting photoreceptor cell death.
NTD of Relish Has Unique Toxic Functions.
The preceding data suggest a central role of Relish activation in retinal degeneration, and the presence of transcriptionally active Relish in light-exposed norpA. Evidence from studies on the innate immune response indicates that the transcriptional activity resides in NTD and that the cytosolic C-terminal domain (CTD) has an as yet uncharacterized function. Thus, we wanted to know if Relish-mediated transcriptional activity or the cytosolic fragments play any role in the death of photoreceptor neurons. We reasoned that Relish activation in photoreceptors might generate NTD and CTD, either or both of which might prove toxic to the photoreceptors. This argument assumes toxicity to be innate to one or both of the domains but sequestered in the unprocessed, full-length Relish. Thus, we hypothesized that expression of NTD or CTD independent of any upstream Relish processing should uncover any toxic functions. We used the UAS/GAL4 system of Drosophila to express upstream activation sequence NTD (UAS-NTD) and CTD (UAS-CTD).
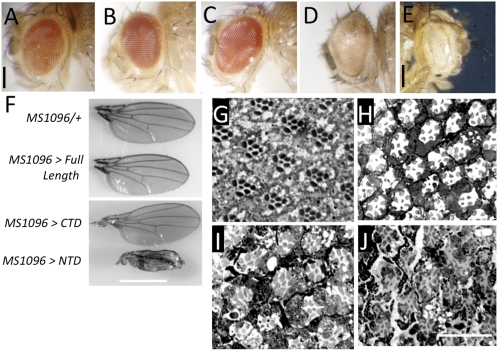
First, we used the glass multimer reporter (GMR)-Gal4 driver, which drives expression in all the differentiated cells posterior to the morphogenetic furrow during eye development. GMR-GAL4 control flies had no visible eye developmental defect, and the eyes of these flies resemble a WT eye (Fig. 5A). Overexpression of full-length Relish (Fig. 5B) and CTD (Fig. 5C) with the GMR driver had no obvious effect on eye morphology. In contrast, expression of NTD resulted in a glassy appearance with visibly reduced ommatidial facets and nonuniform and diminished pigmentation (Fig. 5D). Increasing the dosage with two copies of UAS-NTD resulted in complete ablation of the eye (Fig. 5E). From these results, we conclude that ectopic expression of NTD is toxic to the cells in the eye, whereas overexpressed CTD and the full-length version have no effect.
Fig. 5.
(A–E) Eye morphology of (A) GMR-GAL4/+ control flies and those expressing (B) full-length Relish (C), C-terminal domain (D) or the N-terminal domain using the GMR-GAL4 driver. (E) Eye depicting the effect of increased dosage of NTD in the photoreceptors, accomplished by two copies of the UAS-NTD transgene using the GMR-GAL4 driver. (Scale bar, 150 μm.) (F) MS1096-GAL4-driven expression of full-length Relish, CTD or the NTD in the developing wing. Wing of MS1096-GAL4/+ driver line included as a control. (Scale bar, 1 mm.) (G–J) Effect of NTD expression in adult photoreceptors. (G) UAS-NTD tubGAL80(ts) flies were reared at 30 °C for 4 wk and served as a control in this experiment. (H) Retinal degeneration was evident after 1 wk at 30 °C in UAS-NTD GAL80(ts)/LGMR-GAL4 flies. Extending the time period to (I) 2 wk or (J) 4 wk exacerbated the retinal degeneration. (Scale bar, 20 μm.)
To assess NTD-induced toxicity in other tissues, we first examined the effects on the developing wing. We expressed full-length Relish, CTD, and NTD using the MS1096-GAL4 driver that is expressed in the wing pouch. MS1096-GAL4 driver control flies did not have any observable defects in adult wing (Fig. 5F). Overexpression of CTD or full-length relish did not affect the wing development; the wing morphology, size, and vein pattern were indistinguishable from those of the control (Fig. 5F). However, expression of NTD in the wing pouch resulted in a stark wing phenotype evident as a dramatic reduction in size with 100% penetrance (n = 86 flies) (Fig. 5F). In addition, expression of NTD resulted in a total absence of the characteristic pattern of longitudinal and transverse veins. In contrast to an otherwise flat wing blade in control flies, we noted that NTD-expressing wings had a downward curvature.
To examine if Relish NTD is toxic to the fully differentiated photoreceptor cells, as might be the case in norpA flies, we expressed this domain in the adult photoreceptor cells. The spatiotemporal expression was achieved by using the temperature-sensitive Gal80 in conjunction with the UAS/Gal4 system. We used the longGMR-GAL4 (LGMR) driver because it has been reported to be more photoreceptor-specific (33). The control flies not expressing NTD did not display any signs of retinal degeneration (Fig. 5G). At permissive temperatures for Gal4 activity, retinal degeneration was evident 1 wk after the onset of NTD expression (Fig. 5H) and progressively deteriorated with time (Fig. 5 I and J). These data indicate that NTD-mediated toxicity can be manifested in fully differentiated neurons. The data depicted in Fig. 5 I and J do not precisely phenocopy those observed in light-treated norpA flies. This is to be expected, because the NTD is overexpressed using the LGMR driver. Therefore, we assume that there is a difference in the amount of NTD in these two backgrounds, and we would expect a difference in the onset and severity of the degeneration.
To examine the toxicity of NTD in selected cell types further, we investigated the effect of its expression on neurons. We used the pan-neuronal elav-GAL4 driver to express full-length Relish, CTD, and NTD. For this analysis, the crosses were designed such that only the female progeny would express from the UAS transgene. We reasoned that if any of the domains prove toxic to the neurons, this would result in organismal lethality and skew the normal progeny male/female sex ratio of 1:1 in favor of males. The percentage of female progeny from the control elav-GAL4 cross was 45.23% (of 167 flies scored). The percentage of females eclosing after full-length Relish expression was 46.42% (n = 209). CTD expression resulted in a 54.91% (n = 275) eclosion rate for females. We observed that expression of CTD or full-length Relish did not alter the sex ratios to a significant extent (P > 0.05) compared with elav-GAL4 control flies. However, expression of NTD in neurons resulted in drastic lethality, as judged by significantly fewer eclosing females (10.79% of 343 flies; P < 0.005). We noticed that the surviving females displayed a defect in unfurling of the wings but otherwise normal locomotor behavior. We corroborated these data by expressing NTD and CTD in dopamine/serotonin neurons using the Ddc-GAL4 driver. The percentage of female progeny from the control Ddc-GAL4 cross was 46.57% (n = 74). We observed that expression of NTD (0.69% eclosion rate, n = 435; P < 0.005) but not of CTD (42.94% eclosion rate, n = 319; P > 0.05) in these neurons caused a dramatic decrease in the survival of female flies.
Discussion
RP is a heterogeneous family of inherited retinal disorders that cause irreversible vision loss attributable to photoreceptor death. An interesting feature of RP is that many identified mutations affect genes that are solely expressed in rods, the cells required for night vision. Thus, these cells are nonfunctional or partially functional in patients with RP, and they eventually die because of yet unknown mechanisms. However, the death of rods eventually results in the nonautonomous death of cones, the cells required for daytime, high-acuity, and color vision. Thus, understanding the mechanism of rod cell death is the first step in devising therapies targeting vision loss attributable to photoreceptor death. In this respect, the norpA mutants are interesting because they undergo light-dependent retinal degeneration despite being blind. We have previously shown that norpA flies undergo retinal degeneration because of rapid endocytosis of Rh1, followed by its accumulation in the cell body (4, 6, 7). Accumulation of rhodopsin in endosomes is also observed in models associated with particularly aggressive forms of RP (34, 35). However, the mechanisms by which rhodopsin accumulation might trigger photoreceptor cell death were unclear. Because the features of photoreceptor cell death resemble those of apoptosis, we explored the possibility of a PCD mechanism in retinal degeneration. The results from this study indicate that the developmental PCD pathway involving the Dronc caspase is not involved in photoreceptor cell death. Instead, cell death is mediated by the Drosophila NF-κB homolog Relish. We observe Relish activity only in norpA photoreceptors after light exposure. Absence of Relish activity, such as in relish mutants or in backgrounds that prevent its activation, rescues photoreceptors from death. We have found that overexpression of an activated form of Relish (NTD) is toxic in developing and adult photoreceptors.
The NF-κB signaling pathway mediating cell death in photoreceptor neurons was surprising to us for two reasons. First, Relish activation in fat bodies as a response to bacterial invasion does not lead to the death of these cells. Second, overexpression of NTD in the malignant blood neoplasm (mbn-2) cell line is not toxic (36). These data indicate that only some cells are susceptible to Relish-mediated toxicity. The precise nature of the susceptibility determinants and elucidation of the downstream prodeath effectors of Relish are outside the scope of the current work but are important for gaining a complete understanding of the cell death pathway.
Interestingly, in norpA rel (dark-adapted) flies, a ∼35% decrease in dipt expression is observed compared with norpA (dark) flies (Fig. 4A). This could be indicative of basal processing of Relish in norpA (dark) flies. The presence of processed Relish fragments in Drosophila cells and larvae in an unstimulated state has been observed previously (26) and might suggest a basal level of processing. We also observe an increase in relish-dependent transcription of the target gene dipt in light-treated norpA flies. These data suggest that there is an increased ability of Relish to act as a transcription factor after light treatment. This can be attributed to increased Dredd-mediated proteolytic processing, increased phosphorylation of Relish by the IKK complex, or both. NTD is the only known Relish-derived protein known to act as a transcription factor. Although the presence of full-length Relish is observed in the nuclear extracts (26), its ability to influence transcription has not been reported. Also, the cytosolic functions, if any, for NTD are not known. Thus, in our gain-of-function studies using expression of NTD, we ascribe the effects to its transcriptional activity. We note that toxicity is observed when NTD is expressed and not when CTD or full-length Relish is overexpressed in different tissues (Fig. 5). The observed phenotypes could arise as a result of NTD killing the cells by directly activating the death pathway(s). It is equally likely that, NTD being a transcription factor, developmental programs might be altered, resulting in the observed phenotypes. However, the GMR-, elav-, and ddc-GAL4 drivers used in this study express in postmitotic cells; thus, reduced cell proliferation giving rise to these effects can be ruled out. Because there is no discernable effect of CTD or full-length version overexpression, we conclude that only NTD expression is toxic. In addition, NTD expression in the adult retina results in cell death (Fig. 5 H–J), evident as atrophic photoreceptors. Thus, we infer that the relish-dependent death pathways are most likely regulated at a transcriptional level.
The dredd, kenny, and relish flies are viable and fertile, indicating that developmental programs of cell death are executed normally in these backgrounds. However, these genes are required for photoreceptor cell death during retinal degeneration. This separation of developmental and nondevelopmental cell death uncovers an interesting aspect of biology by showing that genes dispensable for developmental PCD play a pivotal role in nondevelopmental cell death. Although the dredd, kenny, and relish genes are critical for both the antimicrobial response and retinal degeneration in norpA flies, we would like to highlight three key differences in the immunity and degeneration pathways. First, in the immune response, Dredd activation requires the Imd/dFadd complex. However, photoreceptor cell death seemingly occurs in the absence of this complex, indicating that Dredd is capable of being activated in an Imd/dFadd-independent manner. Second, in norpA flies, the fold up-regulation of dipt is approximately twofold, much less than the observed up-regulation during the immune response. This relatively small but significant fold up-regulation might be indicative of the fact that Relish expression in the photoreceptors is less than in the fat body cells or that only a fraction of Relish might be getting activated in norpA. Third, the kinetics of dipt expression are very different in norpA retinas (maximal expression after 2 d) vs. in the innate immune response (peak expression achieved within hours). We hypothesize that in norpA flies, Relish activation is a signaled by a sensing mechanism that detects Rh1 accumulation beyond a critical threshold. Thus, Relish maximal activation will occur only after a majority of cellular Rh1 accumulates in the cell body. Because there is no cytoplasmic Rh1 accumulation on light exposure in WT flies (7), Relish activation fails to occur.
Our model of cell death in norpA and contrasting features with the characterized pathway of Imd/dFADD signaling in the immune response is depicted in Fig. S3. This model speculates that Dredd is capable of getting activated via two pathways. The Imd/dFadd/Dredd complex is involved in the antimicrobial response, whereas Rh1 accumulation is relayed to Dredd that is not part of this complex. This model explains the Imd/dFadd-independent but Dredd-dependent nature of norpA cell death. Dredd in the Imd/dFadd complex might be optimally primed for rapid and efficient activation of its downstream effectors, thus resulting in robust activation of Relish during the immune response.
In this regard, the partial protection provided by p35 in norpA is confounding. Expression of p35 from the transgene used in our experiments completely rescues the striking eye ablation phenotype observed with rpr expression (Fig. S4 A and B), suggesting that the p35 levels are not limiting. One model to explain our observations is that p35 acts after Relish-mediated transcriptional activation of the apoptotic program. Relish might activate multiple prodeath genes, thus initiating parallel pathways of cell death. We hypothesize that only a subset of these pathways is susceptible to p35 inhibition. This might explain why p35 slows down but does not completely inhibit retinal degeneration. Interestingly, p35 expression does not completely rescue cell death observed with Relish NTD expression (Fig. S4 C and D).
The data presented here open interesting avenues by bringing up two important questions. First, how is Relish activated by endosomal accumulation of Rh1 in an Imd/dFadd-independent manner? Currently, we do not have candidate molecules to speculate on a mechanism. However, it has recently been reported that in Drosophila, endocytosis plays an essential role in activation of another NF-κB signaling arm mediated by Dif and Dorsal proteins (37, 38). If endocytosis also plays an important role in activation of the Relish-mediated arm, one can imagine that the persistent presence of Rh1 vesicles in the cytoplasm of norpA photoreceptors might be conducive for signaling to Relish, whereas rapid turnover of these vesicles in a WT situation might prevent Relish activation. Second, how is Relish-mediated transcriptional activity toxic to the cells? It is likely that Relish might have multiple prodeath transcriptional targets, which, when activated, might initiate death using different mechanisms. Forward genetic screens using ectopic expression of proapoptotic hid and rpr genes have proven highly informative. We envision a similar approach using the NTD overexpression to elucidate the mechanism of Relish-mediated cell death.
To summarize, this work uncovers a previously unknown mechanism of death signaling in Drosophila involving the NF-κB homolog Relish. The data also suggest transcriptional control of cell death during retinal degeneration and provide potential unique therapeutic targets for photoreceptor cell death.
Materials and Methods
Drosophila Genetics.
The norpAEE5 allele was used in this work. The norpA flies were crossed into a w1118 background to eliminate eye pigment. Thus, w1118 served as a WT control. The MS1096-GAL4, LGMR-GAL4, elav-GAL4, and ddc-GAL4 were obtained from the Bloomington Stock Center. For expression of NTD in adult photoreceptors, UAS-NTD/+; tubGAL80(ts)/+; LGMR-GAL4/+ flies were shifted from 18 °C to 30 °C. UAS-NTD/+; tubGAL80(ts);+/+ flies at 30 °C served as a control. Crosses involving driver expression were carried out at 24 °C. For studying organismal lethality, a sex ratio distortion assay was carried out as follows. For panneuronal expression, elav-GAL4 (transgene on X chromosome) males were crossed with UAS-NTD (transgene on X) and UAS-CTD (on X chromosome) females. The UAS-Relish (full-length) (on chromosome 2) females were crossed with elav-GAL4 (on X chromosome) males. For expression in the dopaminergic/serotonergic neurons, UAS-NTD or UAS-CTD males were crossed with ddc-GAL4 (on chromosome 2) females. This scheme ensured that the transgenes were expressed only in the F1 females. For driver control experiments, elav-GAL4 (male) or ddc-GAL4 (female) flies were crossed with Canton-S and F1 progeny was scored. Every single F1 progeny was counted after eclosion. Data are the means from three independent experiments. The norpA dredd double mutants were generated by recombining norpAEE5 with ywdreddB118 using standard genetic schemes. The flies used in this work and their sources are tabulated in Table 1.
Table 1.
Fly lines and their sources
Retinal Degeneration, Histology, Immunohistochemistry, and Immunoblotting.
Flies were light-treated, and retinal tissue was processed for plastic sectioning and whole-mount immunochemistry as described previously (7). The dFADDf02804 flies have a PiggyBac insertion with a w+ marker that gives a red eye color. The red pigment effectively screens the incident light; hence, the light exposure of these flies was increased to 4 wk; over this period of time, the red-eyed norpA flies undergo complete degeneration. For quantifying photoreceptor cell death, rhabdomeres per ommatidium were used as the criterion. Rhabdomeres were visualized either by toluidine blue staining of plastic sections or by visualizing rhabdomeric F-actin using fluorescent phalloidin. In a WT retina, the rhabdomeres project into the intraommatidial space. Between the photoreceptor cell body and the rhabdomere is the stalk membrane. During degeneration, the rhabdomeres gradually diminish in radius and are progressively internalized into the cell body of the dying photoreceptor. The rhabdomeres that were clearly extending into the intraommatidial space and were held by a stalk membrane were counted. Photoreceptors that lacked a clear “neck” connecting the rhabdomeres with the cell body were excluded even if they had a dark-staining rhabdomere in them. Thus, only photoreceptors with a dark-staining rhabdomere connected by a lighter colored stalk membrane were counted. For consistency, only the ommatidia with a clearly visible inner photoreceptor rhabdomere were taken into account. Immunoblotting on retina devoid of brain and cornea using anti-DIAP1 antibody (1:3,000; gift from Kristin White, Massachusetts General Hospital, Charlestown, MA) was carried out essentially as described earlier (39). The light pulse-chase experiment was carried out as described by Chinchore et al. (7). Briefly, flies were exposed to light for 2 d and shifted to darkness for 13 h. The retinae were dissected in darkness using safelight illumination to prevent induction of new Rh1 endocytosis. The retinae were fixed in dark and processed for immunohistochemistry and confocal microscopy as described by Chinchore et al. (7). Alexa 568-conjugated phalloidin (Molecular Probes) was used at 5 U/mL with the primary antibody. Mouse monoclonal anti-Rh1 antibody (4C5; Developmental Studies Hybridoma Bank) was diluted 1:30. Secondary antibody, Alexa 647-conjugated Goat anti-Mouse (Molecular Probes), was used at a dilution of 1:500.
Real-Time Quantitative RT-PCR.
Around 50–75 flies (10–15 flies per vial) per genotype were light-exposed for each biological replicate. Anesthetized flies were decapitated, and the heads were rapidly submerged in RNAlater RNA stabilization solution (Qiagen). Total RNA from fly heads was isolated using the TRIzol reagent (Invitrogen). cDNA was synthesized using SuperScript III first-strand synthesis Supermix (Invitrogen), and a quantitative real-time PCR assay using SYBR Premix Ex Taq II (Perfect Real Time; TaKaRa) was performed. Expression was normalized to RP49 values and calculated using the cycle threshold (ΔΔCt) method. The primers used were RP49-qRTF (5′-TACAGGCCCAAGATCGTGAAG-3′), RP49-qRTR (5′-GACGCACTCTGTTGTCGATACC-3′), DiptF (5′-GTTCACCATTGCCGTCGCCTTAC-3′), and DiptR (5′-CCCAAGTGCTGTCCATATCCTCC-3′). Each biological replicate was subjected to three technical replicates.
Supplementary Material
Acknowledgments
We thank Svenja Stöven, Andreas Bergmann, John Abrams, Edan Foley, Dominique Ferrandon, Jean-Marc Reichhart, Louisa Wu, Neal Silverman, Bruno Lemaitre, Kristin White, Tony Ip, Barbara Conradt, Thomas Jack, Yashi Ahmed, Sharon Bickel, Richard Binari, Norbert Perrimon, Connie Cepko, and Jongkyeong Chung for reagents, discussions, and technical help. This work was funded by a grant from the National Institutes of Health. The confocal microscope was supported, in part, by National Science Foundation Grant DBI-9970048 (to Roger D. Sloboda).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 3612 (volume 109, number 10).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110666109/-/DCSupplemental.
References
- 1.Sullivan LS, Daiger SP. Inherited retinal degeneration: Exceptional genetic and clinical heterogeneity. Mol Med Today. 1996;2:380–386. doi: 10.1016/s1357-4310(96)10037-x. [DOI] [PubMed] [Google Scholar]
- 2.Pacione LR, Szego MJ, Ikeda S, Nishina PM, McInnes RR. Progress toward understanding the genetic and biochemical mechanisms of inherited photoreceptor degenerations. Annu Rev Neurosci. 2003;26:657–700. doi: 10.1146/annurev.neuro.26.041002.131416. [DOI] [PubMed] [Google Scholar]
- 3.Ranganathan R, Malicki DM, Zuker CS. Signal transduction in Drosophila photoreceptors. Annu Rev Neurosci. 1995;18:283–317. doi: 10.1146/annurev.ne.18.030195.001435. [DOI] [PubMed] [Google Scholar]
- 4.Alloway PG, Howard L, Dolph PJ. The formation of stable rhodopsin-arrestin complexes induces apoptosis and photoreceptor cell degeneration. Neuron. 2000;28:129–138. doi: 10.1016/s0896-6273(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 5.Kiselev A, et al. A molecular pathway for light-dependent photoreceptor apoptosis in Drosophila. Neuron. 2000;28:139–152. doi: 10.1016/s0896-6273(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 6.Orem NR, Dolph PJ. Loss of the phospholipase C gene product induces massive endocytosis of rhodopsin and arrestin in Drosophila photoreceptors. Vision Res. 2002;42:497–505. doi: 10.1016/s0042-6989(01)00229-2. [DOI] [PubMed] [Google Scholar]
- 7.Chinchore Y, Mitra A, Dolph PJ. Accumulation of rhodopsin in late endosomes triggers photoreceptor cell degeneration. PLoS Genet. 2009;5:e1000377. doi: 10.1371/journal.pgen.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu CD, et al. Limited role of developmental programmed cell death pathways in Drosophila norpA retinal degeneration. J Neurosci. 2004;24:500–507. doi: 10.1523/JNEUROSCI.3328-02.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White K, et al. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 10.Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 11.Chen P, Nordstrom W, Gish B, Abrams JM. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- 12.Robinow S, Draizen TA, Truman JW. Genes that induce apoptosis: Transcriptional regulation in identified, doomed neurons of the Drosophila CNS. Dev Biol. 1997;190:206–213. doi: 10.1006/dbio.1997.8696. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez A, et al. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- 14.Yoo SJ, et al. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- 15.Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol. 2002;4:432–438. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- 16.Hays R, Wickline L, Cagan R. Morgue mediates apoptosis in the Drosophila melanogaster retina by promoting degradation of DIAP1. Nat Cell Biol. 2002;4:425–431. doi: 10.1038/ncb794. [DOI] [PubMed] [Google Scholar]
- 17.Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- 18.Srivastava M, et al. ARK, the Apaf-1 related killer in Drosophila, requires diverse domains for its apoptotic activity. Cell Death Differ. 2007;14:92–102. doi: 10.1038/sj.cdd.4401931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- 20.Chen P, Rodriguez A, Erskine R, Thach T, Abrams JM. Dredd, a novel effector of the apoptosis activators reaper, grim, and hid in Drosophila. Dev Biol. 1998;201:202–216. doi: 10.1006/dbio.1998.9000. [DOI] [PubMed] [Google Scholar]
- 21.Georgel P, et al. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- 22.Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naitza S, et al. The Drosophila immune defense against gram-negative infection requires the death protein dFADD. Immunity. 2002;17:575–581. doi: 10.1016/s1074-7613(02)00454-5. [DOI] [PubMed] [Google Scholar]
- 24.Leulier F, Vidal S, Saigo K, Ueda R, Lemaitre B. Inducible expression of double-stranded RNA reveals a role for dFADD in the regulation of the antibacterial response in Drosophila adults. Curr Biol. 2002;12:996–1000. doi: 10.1016/s0960-9822(02)00873-4. [DOI] [PubMed] [Google Scholar]
- 25.Hedengren M, et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 26.Stöven S, Ando I, Kadalayil L, Engström Y, Hultmark D. Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoven S, et al. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc Natl Acad Sci USA. 2003;100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou R, et al. The role of ubiquitination in Drosophila innate immunity. J Biol Chem. 2005;280:34048–34055. doi: 10.1074/jbc.M506655200. [DOI] [PubMed] [Google Scholar]
- 29.Silverman N, et al. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rutschmann S, et al. Role of Drosophila IKK gamma in a toll-independent antibacterial immune response. Nat Immunol. 2000;1:342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- 31.Lu Y, Wu LP, Anderson KV. The antibacterial arm of the drosophila innate immune response requires an IkappaB kinase. Genes Dev. 2001;15:104–110. doi: 10.1101/gad.856901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dijkers PF, O'Farrell PH. Drosophila calcineurin promotes induction of innate immune responses. Curr Biol. 2007;17:2087–2093. doi: 10.1016/j.cub.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wernet MF, et al. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell. 2003;115:267–279. doi: 10.1016/s0092-8674(03)00848-1. [DOI] [PubMed] [Google Scholar]
- 34.Chen J, et al. Stable rhodopsin/arrestin complex leads to retinal degeneration in a transgenic mouse model of autosomal dominant retinitis pigmentosa. J Neurosci. 2006;26:11929–11937. doi: 10.1523/JNEUROSCI.3212-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chuang JZ, Vega C, Jun W, Sung CH. Structural and functional impairment of endocytic pathways by retinitis pigmentosa mutant rhodopsin-arrestin complexes. J Clin Invest. 2004;114(1):131–140. doi: 10.1172/JCI21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiklund ML, Steinert S, Junell A, Hultmark D, Stöven S. The N-terminal half of the Drosophila Rel/NF-kappaB factor Relish, REL-68, constitutively activates transcription of specific Relish target genes. Dev Comp Immunol. 2009;33:690–696. doi: 10.1016/j.dci.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Huang HR, Chen ZJ, Kunes S, Chang GD, Maniatis T. Endocytic pathway is required for Drosophila Toll innate immune signaling. Proc Natl Acad Sci USA. 2010;107:8322–8327. doi: 10.1073/pnas.1004031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lund VK, DeLotto Y, DeLotto R. Endocytosis is required for Toll signaling and shaping of the Dorsal/NF-kappaB morphogen gradient during Drosophila embryogenesis. Proc Natl Acad Sci USA. 2010;107:18028–18033. doi: 10.1073/pnas.1009157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alloway PG, Dolph PJ. A role for the light-dependent phosphorylation of visual arrestin. Proc Natl Acad Sci USA. 1999;96:6072–6077. doi: 10.1073/pnas.96.11.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]