Abstract
Heme is an iron-coordinated porphyrin that is universally essential as a protein cofactor for fundamental cellular processes, such as electron transport in the respiratory chain, oxidative stress response, or redox reactions in various metabolic pathways. Parasitic kinetoplastid flagellates represent a rare example of organisms that depend on oxidative metabolism but are heme auxotrophs. Here, we show that heme is fully dispensable for the survival of Phytomonas serpens, a plant parasite. Seeking to understand the metabolism of this heme-free eukaryote, we searched for heme-containing proteins in its de novo sequenced genome and examined several cellular processes for which heme has so far been considered indispensable. We found that P. serpens lacks most of the known hemoproteins and does not require heme for electron transport in the respiratory chain, protection against oxidative stress, or desaturation of fatty acids. Although heme is still required for the synthesis of ergosterol, its precursor, lanosterol, is instead incorporated into the membranes of P. serpens grown in the absence of heme. In conclusion, P. serpens is a flagellate with unique metabolic adaptations that allow it to bypass all requirements for heme.
Keywords: cytochromes, respiration, sterols, protist
Heme is a tetrapyrrole molecule that consists of a porphyrin ring coordinated with the iron molecule. It interacts with various apoproteins giving rise to functional hemoproteins, which are ubiquitous in biological systems and exhibit a wide range of activities. The oxidation state of the iron is important for most biological roles of heme, but its exact function is ultimately determined by the properties of the polypeptide bound to it (1). Heme can exist in either the oxidized ferric (Fe3+) or reduced ferrous (Fe2+) state, which enables it to accept or donate electrons and to function in various redox reactions and electron transport.
The most abundant group of heme proteins are cytochromes (2). In aerobic organisms that produce energy mainly through oxidative phosphorylation, most of the synthesized heme is used for the formation of the cytochromes functioning in the electron transport respiratory chain. Other cytochromes, such as the members of the cytochrome b5 or cytochrome P450 family, are involved in various redox reactions of specific metabolic pathways, such as desaturation of fatty acids and sterol biosynthesis, and also in drug detoxification (3, 4). In catalases, heme functions in the degradation of hydrogen peroxide, whereas in peroxidases, it oxidizes a wide variety of organic and inorganic compounds in the presence of hydrogen peroxide. Through the consumption of hydrogen peroxide, these enzymes greatly contribute to the oxidative stress defense (5, 6). In addition to its function as an electron carrier, heme iron has the capacity to bind diatomic gases. Hemoglobin is well known as the oxygen transporter in animals, but members of the same protein family are widespread in all groups of organisms, including anaerobes. The original roles of globins might have been the responses to nitric oxide and nitrosative stress (7) or sensing of oxygen, which was highly toxic to cells before they managed to adapt to an aerobic environment (8). In soluble guanylyl cyclase, heme serves as the nitric oxide sensor, and thus plays an important role in signal transduction. Heme is also an important regulatory molecule because it reversibly binds to certain proteins, such as transcription factors and ion channels, and thus modulates their functions (9).
The central position of heme in a variety of cellular functions makes it essential for the viability of virtually all living systems. There are only a few examples of facultatively anaerobic or pathogenic bacteria that do not require heme (10–12), but no eukaryote that can survive without heme has been identified. Most aerobic organisms synthesize heme by a multistep pathway that is conserved in all three domains of life: bacteria, archaea, and eukaryotes. A few eukaryotes that lost this pathway are known to scavenge heme from external sources. For example, ticks have easy access to heme from blood (13), whereas parasitic nematodes uptake it either from their host or from endosymbiotic bacteria (14). The free-living nematode Caenorhabditis elegans lacks the capacity to synthesize heme but is able to take it from the bacteria it feeds on (15). Even the parasitic protists Entamoeba, Trichomonas, and Giardia, which dwell in an anaerobic environment and do not need heme for processes connected to oxidative metabolism, have retained a few hemoproteins, for which heme is likely obtained from their hosts (16).
Flagellates of the order Kinetoplastea, which includes major human parasites, depend on oxygen but are unable to produce heme. Media for their cultivation must therefore be supplemented with heme to support their growth (17). Members of the genus Trypanosoma lost the entire biosynthetic pathway and extract heme from host blood (18, 19), whereas Leishmania spp. have retained genes for the last three steps of the pathway, allowing them to synthesize heme from their host-derived precursors (20). Some kinetoplastids that parasitize insects obtain heme from their bacterial endosymbionts, which can be eliminated by antibiotic treatment, turning these protists into heme auxotrophs (17).
Kinetoplastid flagellates of the genus Phytomonas are important yet understudied parasites of plants with a major economic impact in Latin America and the Caribbean (21). They reside in carbohydrate-rich tissues, such as phloem, latex, fruits, and seeds; their ATP production is based on glycolysis (22). In the present study, we show that Phytomonas serpens does not require heme for viability and possesses unique metabolic properties that allow it to bypass all functions of this otherwise omnipresent molecule.
Results and Discussion
We cultivated P. serpens strain 9T in a chemically defined medium without heme (Table S1) continuously for over a year without noticeable decrease of the growth rate (generation time of ∼8 h), compared with parallel cultures supplemented with heme (Fig. 1A). Abrupt transfer of cells grown in heme-containing medium into one lacking it did not alter their growth. This is in contrast to related flagellates, such as Crithidia fasciculata, which requires heme for growth and was used as a control (Fig. 1A).
Fig. 1.
Growth dependence on availability of heme and quantification of heme b in P. serpens and related flagellates. (A) Growth rate of P. serpens is the same without heme or when heme is supplied up to a concentration of 5 μM; 25 μM heme inhibits growth, likely attributable to toxic effects of free heme. Quite opposite dependence of growth on heme concentration is observed in the closely related C. fasciculata, which grows best when supplied with 25 μM heme and stops growing when heme concentration in the media is lowered to 1 μM. (B) Heme b extracted from equal numbers of cells from various kinetoplastids was separated by HPLC and detected by diode array detector. C. fasciculata was used as a related organism that possesses a complete set of respiratory complexes. It was grown in the same medium and supplemented with the same amount of heme (5 μM) as P. serpens. The bloodstream stage of T. brucei, which does not express its respiratory complexes III and IV in this life cycle stage, and thus functionally resembles P. serpens, was used as another control. The absence of respiratory complexes that normally consume most of heme is reflected in the much lower amount of extracted heme compared with C. fasciculata. The heme content in P. serpens is even lower than in T. brucei, which is in accordance with the lowest number of heme proteins found in the Phytomonas spp. genomes among all kinetoplastids (Table 1). Not even a trace amount of heme is detected in P. serpens grown without heme (dotted line). +H, with heme; −H, without heme.
We sought to test the heme biosynthetic capacity of P. serpens by measuring the amount of extractable heme. Even using a very sensitive HPLC assay, we failed to detect any traces of heme in cells grown in its absence (Fig. 1B). This indicates that P. serpens is able to survive without heme, which is further supported by the fact that, with the exception of ferrochelatase, no other genes for heme synthesis were found in the draft genome of P. serpens strain 9T obtained for this study. On the other hand, we found a small amount of heme in cells growing in the medium supplemented with heme (Fig. 1B), which implies that P. serpens is able to uptake this compound from the medium.
To find out how P. serpens can survive without the key heme-dependent activities and possibly identify any functions still using heme, we decided to test cellular processes experimentally in which heme is known to be involved. A screen for homologs of heme-containing proteins in the genome produced only a few hits, compared with the list of hemoproteins from related flagellates (Table 1 and Table S2). The same results were obtained for two other recently sequenced Phytomonas genomes (Table 1). Unlike other kinetoplastids, Phytomonas spp. have an apparent lack of respiratory cytochromes, heme-dependent peroxidases, and several enzymes that possess heme-binding domains, such as front-end fatty acid desaturases for the production of polyunsaturated fatty acids (23), a nitrate reductase, and two different ferric reductases, one of which was shown to be involved in the iron uptake of related Leishmania (24) (Table 1).
Table 1.
Heme proteins of kinetoplastid flagellates
| Heme protein | GenBank | TB | TC | Lei | CF | PS | PE | PH |
| Lanosterol 14α-demethylase (cytochrome P450) | CAJ02958 | + | + | + | + | + | + | + |
| Heme-binding subunit of the respiratory complex II | CAJ06243 | + | + | + | + | + | + | + |
| Soluble cytochrome c of the respiratory chain | AAY40864 | + | + | + | + | − | − | − |
| Cytochrome b subunit of the respiratory complex III | ABQ08756 | + | + | + | + | − | − | − |
| Cytochrome c1 subunit of the respiratory complex III | CAJ06965 | + | + | + | + | − | − | − |
| Heme a and heme a3 binding subunit of complex IV | ACJ47213 | + | + | + | + | − | − | − |
| Catalase | — | − | − | − | + | − | − | − |
| Ascorbate-dependent peroxidase | CAJ07706 | − | + | + | + | − | − | − |
| Heme-dependent plant peroxidase homolog 1 | CBZ12974 | + | + | + | + | − | − | − |
| Heme-dependent plant peroxidase homolog 2 | CAJ04972 | + | + | + | + | − | − | − |
| Δ9 Fatty acid desaturase (cytochrome b5 domain) | CAJ05408 | + | + | + | + | (−) | (−) | (−) |
| Δ4 Fatty acid desaturase (cytochrome b5 domain) | CAJ03208 | + | + | + | + | − | − | − |
| Δ5 Fatty acid desaturase (cytochrome b5 domain) | CAJ07076 | − | − | + | + | − | − | − |
| Δ6 Fatty acid desaturase (cytochrome b5 domain) | CAJ09677 | − | − | + | + | − | − | − |
| Nitrate reductase (cytochrome b5 domain) | CAJ05968 | + | + | + | + | − | − | − |
| Fumarate reductase-like (cytochrome b5 domain) | CAJ07044 | − | − | + | + | − | − | − |
| Ferric reductase (cytochrome b561) | CAJ06394 | + | + | + | + | − | − | − |
| Ferric reductase (flavocytochrome b558) | CAJ06274 | + | + | + | + | − | − | − |
| Globin domain of adenylate cyclase-like protein | CAJ04664 | − | − | + | + | − | − | − |
| Cytochromes P450 with unknown function* | 1 | 2 | 3 | 3 | − | − | 1 | |
| Cytochromes b5 with unknown function* | 7 | 19 | 16 | 14 | 12 | 11 | 7 |
The presence/absence data for PE and PH were kindly provided by Michel Dollet (CIRAD-BIOS, Montpellier, France) and Patrick Wincker (Genoscope, Evry, France). GenBank accession numbers are quoted for the proteins of Leishmania major. CF, Crithidia fasciculata; Lei, Leishmania spp.; PE, Phytomonas sp. strain EM1; PH, Phytomonas sp. strain Hart1; PS, Phytomonas serpens; TB, Trypanosoma brucei; TC, Trypanosoma cruzi.; (−), heme-binding domain is missing, but the rest of the protein is present.
*Proteins that do not have known function but were identified as either cytochrome P450 or cytochrome b5 are not listed individually. The number of these proteins is shown for each taxon.
The absence of heme peroxidases in P. serpens, exceptional even among the kinetoplastids, most of which lack catalase (25) (Table 1), corresponds to our finding that heme added to the medium does not increase the resistance of P. serpens against oxidative stress induced by the superoxide generator paraquat (Fig. S1). This is the opposite of what was found for the evolutionarily related Trypanosoma brucei, which needs heme for oxidative stress defense (18).
Although P. serpens lacks the heme-containing respiratory complexes III and IV (26–28), the mitochondrial respiratory chain remains functional, serving to reoxidize NADH produced during glycolysis (22, 29). Complex I is present in P. serpens (27, 30), which, instead of cytochrome c reductase (complex III) and cytochrome c oxidase (complex IV), uses alternative oxidase to reduce oxygen to water (31). We found that succinate dehydrogenase (complex II) is also present (Fig. 2), with a conserved histidine residue in its SDH4 subunit, which supposedly binds heme in the related Trypanosoma cruzi and other kinetoplastids (32). Visualization of the P. serpens complex II by in-gel staining in clear-native gel revealed that its abundance is not influenced by the availability of heme in the medium (Fig. 2A). Moreover, its size of ∼600 kDa is unaltered in the heme-deprived cells, being almost the same as in T. cruzi (32) and the related C. fasciculata, used as a control (Fig. 2A), suggesting a proper assembly of complex II in the absence of heme. To assess the abundance of its subunits, we generated specific antiserum against one subunit of the kinetoplastid complex II, SDH1. The amount of the target protein was the same in cells grown with or without heme (Fig. 2B). Furthermore, the absence of heme did not affect the capacity of complex II to reduce ubiquinone (Fig. 2 C and D).
Fig. 2.
Respiratory complex II (succinate dehydrogenase) is assembled and active in P. serpens grown with (+H) or without (−H) heme. (A) Clear native gel (3–12%) after in-gel staining for succinate dehydrogenase activity; C. fasciculata (Cf) served as a control. Ferritin (monomeric and dimeric forms) was used as a molecular weight marker (M). (B) Lysates from the same cells as in A were analyzed by SDS/PAGE and immunoblotted with specific antiserum against the T. brucei subunit of complex II, SDH1. (C) Activity of succinate dehydrogenase in P. serpens grown without heme. The decrease in absorbance (A600) with time (curve 2) was caused by the addition of ubiquinone to the reaction, which mediated the electron transfer from succinate to 2,6-dichlorophenolindophenol. The activity was specifically inhibited using malonate (curve 3). Curve 1 represents the background without ubiquinone. (D) Activity did not significantly differ between P. serpens grown with (+H) or without (−H) heme. Medium values were calculated from three measurements.
These findings are in line with previous reports showing that heme is not universally indispensable for the function of complex II (33, 34). In mammalian cells, the absence of heme disrupts proper assembly and inhibits the activity of complex II (35). However, in yeast and Escherichia coli, the homologous complexes retain physiological activity even without heme (33, 34). It has been suggested that although heme does not participate in the electron transfer in complex II and is not necessarily required for the assembly of the complex, it may provide an electron sink to protect against free radical damage during periods of high electron flux (34). However, the presence of heme in the proton-pumping complexes III and IV is indispensable, because it directly mediates electron transport. However, when enough energy is produced by glycolysis, these heme-containing complexes, as well as the soluble cytochrome c, may be bypassed by using the alternative terminal oxidase, which utilizes nonheme iron to transfer electrons from ubiquinone directly to oxygen. This is also known for the bloodstream (mammalian) stage of T. brucei, which, similar to Phytomonas, dwells in a sugar-rich environment, whereas the T. brucei procyclic (insect) stage has a fully developed mitochondrion equipped with the heme-containing complexes (36). This metabolic switch is impossible in Phytomonas, which has lost the genes encoding the subunits of these complexes from its genome (26, 28) (Table 1).
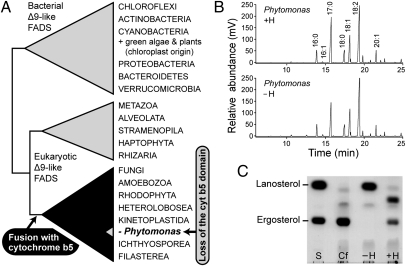
Because of its capacity to transfer electrons, heme participates in various redox reactions, some of which are virtually universal for eukaryotes. One of them is the desaturation of fatty acids. In eukaryotes, this reaction needs electron equivalents that are transferred from reduced cytochrome b5, and thus depends on heme (37, 38). Many desaturases contain cytochrome b5 as a domain conveniently fused to their N- or C-termini, including the most widespread one, which creates the double bond in the Δ9 position (23, 39). Our phylogenetic analyses revealed that this fusion took place only once in the evolution of eukaryotic Δ9 fatty acid desaturases, specifically at the base of a superclade comprising fungi, amoebozoans, rhodophytes, choanozoans, and excavates, including kinetoplastids (Fig. 3A and Fig. S2). Remarkably, Δ9 desaturase in P. serpens is the only member of this superclade that conspicuously lacks the cytochrome b5 domain, apparently as a consequence of its secondary loss, a singular event among all known eukaryotes. To assess the ability of P. serpens grown in the absence of heme to desaturate fatty acids, we analyzed their composition by gas chromatography. We found that P. serpens contains unsaturated fatty acids and that their composition is virtually the same regardless of the presence or absence of heme (Fig. 3B). These findings indicate that for the desaturation of fatty acids, P. serpens is able to use an electron donor other than cytochrome b5. It may likely be ferredoxin, which serves this role for the desaturases of some bacteria and plant plastids. For example, the plastid Δ12 fatty acid desaturase of plants and diatoms depends on ferredoxin as an electron donor, whereas a homologous desaturase with the same function in the endoplasmic reticulum of the same organisms, as well as in other eukaryotes, uses cytochrome b5 (40). The possibility that these redox molecules could substitute for each other has been experimentally demonstrated in E. coli and in yeast expressing cyanobacterial Δ6 fatty acid desaturase (41). Although ferredoxin is the natural electron donor for this desaturase, cytochrome b5 fully complemented its function when fused or coexpressed with the desaturase enzyme. Three different ferredoxin homologs were identified in the genomic sequences of P. serpens (Table S2).
Fig. 3.
Heme is not needed for desaturation of fatty acids but is required for ergosterol biosynthesis in P. serpens. (A) Schematic phylogenetic tree of Δ9-fatty acid desaturases (FADS). P. serpens is the only organism that secondarily lost the cytochrome b5 domain. The full phylogenetic tree of Δ9-fatty acid desaturase is shown in Fig. S1. (B) Analyses of fatty acid composition by gas chromatography demonstrate that in P. serpens, the desaturation of fatty acids is not affected by the absence of heme. (C) Analysis of sterol composition by TLC. Ergosterol, which is the major membrane sterol of Trypanosomatida, and lanosterol, the precursor of heme-dependent demethylation, were used as standards (S). C. fasciculata (Cf) served as a control. P. serpens synthesized a sterol that corresponded to the ergosterol standard only when heme was added to the growth medium (+H). Cells grown without heme (−H) accumulated lanosterol.
The oxidative 14α-demethylation of lanosterol, another key reaction in the eukaryotic cell, fully depends on heme. Its substitution by means of analogous nonheme enzyme has never been documented. This reaction is a crucial step in the synthesis of sterols, such as cholesterol in animals or ergosterol in fungi, as well as in protists, including kinetoplastid flagellates (42). It is catalyzed by lanosterol 14α-demethylase (CYP51), which belongs to the cytochrome P450 family, found in most eukaryotes, including Phytomonas spp. (Table 1). No eukaryotic cell can function without sterols or their analogs in its membranes; inhibition of this enzymatic step is thus frequently lethal (43). Consequently, CYP51 is a popular target of fungicides and other drugs, which are also effective against kinetoplastids (44). Until now, the only kinetoplastid known to be naturally resistant to inhibitors of CYP51 is Leishmania braziliensis, a flagellate closely related to Phytomonas, which seems to be able to incorporate 14-methyl sterols into its membranes (45). We have found that P. serpens possesses this unique capability as well. Based on the TLC analysis, the cells synthesized a sterol that corresponded to the ergosterol standard only when heme was added to the growth medium. In contrast, they accumulated lanosterol in the absence of heme with no impact on cell viability (Fig. 3C). The fact that certain eukaryotes are able to use lanosterol but others are not is very interesting and implies the existence of some regulatory mechanism. Cholesterol-deficient human T cells can adapt to growth with lanosterol; the initial growth of these cells dropped 10-fold when cholesterol was depleted, yet their prolonged cultivation resulted in a growth rate ∼65% that of the cholesterol-supplemented cells (46). A study on yeast revealed that what regulates the incorporation of lanosterol in the membranes is the level of synthesized heme (47). The growth of P. serpens in the absence of heme precludes the activity of CYP51; thus, this flagellate meets the two conditions that are required in yeast for lanosterol utilization (low heme levels and CYP51 inhibition).
Overall, there are several cellular processes for which heme is crucial in a typical eukaryote, yet it is dispensable in P. serpens. Somewhat lower dependence of a typical kinetoplastid on heme has been noted when cystathionine-β-synthase, a hemoprotein of animals and amoebae that is essential for cysteine formation, was shown to lack heme in kinetoplastids (48). However, P. serpens is unique, because it lacks most hemoproteins that are present even in closely related protists. Moreover, the few retained in the P. serpens genome are not crucial for its survival, at least under culture conditions. In addition to CYP51 and the SDH4 subunit of respiratory complex II, we identified 13 proteins that supposedly bind heme, because they are homologous to cytochrome b5 (Table 1). Their functions are unknown, however, and 5 of them lack the HPGG heme-binding motif typical for cytochrome b5 (41). Thus, it is by no means certain that these proteins actually bind heme in vivo. One of them is a protein recently identified in the flagellar proteome of T. brucei, shown to be indispensable for the bloodstream stage but nonessential for the procyclic stage (49). Therefore, the only process for which heme, if present, was found to be actively used by P. serpens, is the 14α-demethylation of lanosterol in the ergosterol biosynthetic pathway (Fig. 3C). Surprisingly, however, in vitro growth remains unaffected by the lack of this activity.
It is conceivable that some anaerobic eukaryotes possessing only a few of the known hemoproteins may survive without heme as well; however, this will be hard to test, because, so far, none of these anaerobic protists can be grown in a chemically defined medium. Furthermore, anaerobic protists need to obtain some products of heme-dependent enzymes, such as cholesterol and fatty acids from their environment; thus, their existence cannot be considered to be independent of heme (50). To the best of our knowledge, P. serpens is the only eukaryote that can survive without heme and yet depends on oxidative metabolism. This unique metabolic property, a feature likely developed as an adaptation to the carbohydrate-rich environment of plant sap, makes it an ideal model to study different cellular functions in a heme-free background, which may shed further light on the exact roles and essentiality for life of the otherwise omnipresent heme.
Materials and Methods
Cultivation Conditions and Growth Curves.
Both P. serpens and C. fasciculata were grown in a chemically defined medium (Table S1) supplemented with different concentrations of hemin at 27 °C and shaking at 80 rpm, daily diluted with fresh media to the density of 6 × 106 cells per milliliter. Cell concentration was measured daily using a Beckman Coulter Z2 counter.
Quantification of Heme b.
In total, 2 × 109 cells of P. serpens, C. fasciculata, and the bloodstream form of T. brucei were filtrated through a DEAE-cellulose column and washed five times with PBS buffer to remove all traces of heme from the media. The cell pellets were extracted with methanol/0.2% NH4OH, and heme was extracted from the delipidated cells with acetone/2% HCl (vol/vol) and separated by HPLC on a Nova-Pak C18 column (4-μm particle size, 3.9 × 150 mm; Waters) using linear gradient 25–100% (vol/vol) acetonitrile/0.1% trifluoroacetic acid at a flow rate of 1.1 mL/min at 40 °C. Heme b was detected by diode array detector (Agilent 1200; Agilent Technologies) and quantified using authentic hemin standard (Sigma–Aldrich) and extinction coefficient as described previously (51).
Analysis of Fatty Acids.
Lipids were extracted from P. serpens cell pellets by a modified method of Bligh and Dyer (52) with dichloromethane used instead of chloroform. The methyl esters were prepared by trans-esterifying the lipid extract with BF3-CH3OH at 85 °C for 1 h and analyzed using a gas chromatograph (HRGC 5300; Carlo Erba) equipped with a flame ionization detector and TR-FAME capillary column for the separation of Fatty Acid Methyl Esters (FAMEs) (60-m, 0.25-mm inner diameter and 0.25-μm film thickness; Thermo Scientific). Hydrogen was used as the carrier gas with a pressure of 200 kPa. The following temperature ramp was used: 140 °C to 240 °C with a rate of 4 °C per min−1 and holding at 240 °C for 10 min. The flame ionization detector was isothermal at 260 °C, and the injector was set to 250 °C. Separated fatty acids were identified by comparison of their retention times with known standards (37-component fatty acid methyl ester mix 47885-U, Supelco; polyunsaturated fatty acid no. 3, menhaden oil).
Analysis of Sterols.
Sterols were extracted and separated on TLC silica gel plates as described previously (43) and visualized by spraying the plates with a water solution of 0.05% ferric chloride/5% (vol/vol) acetic acid/5% (vol/vol) sulfuric acid and heating to 100 °C for 15 min.
Detection and Activity Measurements of Respiratory Complex II.
Mitochondria were isolated by hypotonic lysis as described previously (53). Protein lysates were prepared by digitonin lysis (4 mg of digitonin per 1 mg of proteins, 1 h on ice) for native gel electrophoresis and histochemical staining and by dodecylmaltoside lysis (40 μL of 0.5 M aminocaproic acid and 10 μL of 10% (wt/vol) dodecylmaltoside, 1 h on ice) for spectroscopic activity measurements and SDS/PAGE. Whole-complex II was detected in 3–12% (wt/vol) clear native gel (80 μg of proteins per line) by incubating in a staining solution [50 mM NaPi (pH 7.4), 84 mM sodium succinate, 0.2 mM N-methylphenazonium methyl sulfate, 4.5 mM EDTA (pH 8.5), 10 mM potassium cyanide, 2 mg/mL Nitrotetrazolium blue chloride) for 3 h at room temperature in dark. Nitrotetrazolium blue chloride changes color on accepting electrons from succinate via N-methylphenazonium methyl sulfate, a process catalyzed by complex II. SDH1 subunit of complex II was detected by Western blot analysis using 10% (wt/vol) SDS/PAGE and a specific polyclonal antiserum generated against the oligopeptide SHLSKAYPVIDHTFDC [SDH1 subunit of T. brucei (Tb927.8.6580) in a rabbit].
Specific succinate dehydrogenase activity was measured using the following protocol: 5 μL of mitochondrial protein lysate was incubated with 1 mL of succinate dehydrogenase solution [25 mM KPi (pH 7.2), 5 mM MgCl2, 20 mM sodium succinate] for 10 min at 30 °C. This mixture was transferred in the cuvette, and antimycin A (2 μg/mL), rotenone (2 μg/mL), potassium cyanide (2 mM), and 2,6-dichlorphenolindophenol (50 μM) were added. Background absorbance at 600 nm was then measured for 5 min. The reaction was triggered by adding 65 μM coenzyme Q2, and the absorbance at 600 nm was measured every 20 s for 5 min. Change in absorbance was caused by the electron transfer from succinate via coenzyme Q2 to 2,6-dichlorophenolindophenol. The activity was specifically inhibited by the addition of 1 mM sodium malonate.
Genome Sequencing, Assembly, and Protein Search.
P. serpens nuclear DNA fraction was sequenced using Illumina technology at BGI-Hong Kong (HiSeq 2000 sequencing system, average insert size of 500 bp, read length of 90 bp). A dataset of 1.62 Gbp was obtained after basic filtering of low-quality reads. Genome assembly with MIRA 3.4rc2 (54) produced 5,399 contigs longer than 500 bp (N50 contig size of 6,781 bp) with average coverage 60 (genome assembly deposited in National Center for Biotechnology Information BioProject database under accession no. PRJNA80957). Translated reads and contigs were screened using tblastn 2.2.24+ with e-value cutoffs at 10−3 and 10−10, respectively, against Leishmania major heme-binding proteins (Table 1) and heme-synthesis enzymes. Conserved protein domains were identified using InterPro database. Draft genome sequences of C. fasciculata were kindly provided by Stephen M. Beverley (Washington University School of Medicine, St. Louis, MO), produced by The Genome Center at Washington University School of Medicine in St. Louis, and can be obtained from tritryp database. Two of the heme-proteins of C. fasciculata (lanosterol 14α-demethylase and Δ6 fatty acid desaturase) were not identified in the draft genome sequences, but their partial sequences were amplified by PCR assay from C. fasciculata and sequenced (Table S2).
Phylogenetic Analysis.
Amino acid sequences of Δ9 fatty acid desaturases from different eukaryotic lineages and bacteria were aligned using MAFFT 6.717b (55) and manually edited using BioEdit (56). A maximum likelihood tree was constructed with RAxML 7.0.3 using the PROTGAMMALG model (57) (1,000 replications). The bootstrap supports of individual branches were calculated using the same model after 1,000 iterations.
Oxidative Stress Assay.
The sensitivity of cells to oxidative stress was measured by exposing them to paraquat added to the cultivation medium in a wide range of concentrations, ranging from 10−8 to 100 mM. After 44 h of incubation, resazurin was added to each culture, and after 4 h, the viability of cells was established by measurement of fluorescence. Obtained data were analyzed by GraphPad Prism software using nonlinear regression (curve fit) with a sigmoidal dose–response analysis (58, 59).
Supplementary Material
Acknowledgments
We thank Martin Lukeš for his help with gas chromatography analysis of fatty acids. Michel Dollet and Patrick Wincker kindly provided the absence/presence data for selected genes in the Phytomonas EM1 and Hart1 genomes. This work was supported by the Grant Agency of the Czech Republic (Grants 204/09/1667, 206/08/1423, and P305/11/2179), a Praemium Academiae award (to J.L.), the Algatech Project (CZ.1.05/2.1.00/03.0110), and the Scientific Grant Agency of the Slovak Ministry of Education and the Academy of Sciences (Grant 1/0393/09).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201089109/-/DCSupplemental.
References
- 1.Frankenberg N, Moser J, Jahn D. Bacterial heme biosynthesis and its biotechnological application. Appl Microbiol Biotechnol. 2003;63:115–127. doi: 10.1007/s00253-003-1432-2. [DOI] [PubMed] [Google Scholar]
- 2.Panek H, O'Brian MR. A whole genome view of prokaryotic haem biosynthesis. Microbiology. 2002;148:2273–2282. doi: 10.1099/00221287-148-8-2273. [DOI] [PubMed] [Google Scholar]
- 3.Schenkman JB, Jansson I. The many roles of cytochrome b5. Pharmacol Ther. 2003;97(2):139–152. doi: 10.1016/s0163-7258(02)00327-3. [DOI] [PubMed] [Google Scholar]
- 4.Anzenbacher P, Anzenbacherová E. Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci. 2001;58:737–747. doi: 10.1007/PL00000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chelikani P, Fita I, Loewen PC. Diversity of structures and properties among catalases. Cell Mol Life Sci. 2004;61:192–208. doi: 10.1007/s00018-003-3206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifacio A, et al. Role of peroxidases in the compensation of cytosolic ascorbate peroxidase knockdown in rice plants under abiotic stress. Plant Cell Environ. 2011;34:1705–1722. doi: 10.1111/j.1365-3040.2011.02366.x. [DOI] [PubMed] [Google Scholar]
- 7.Poole RK, Hughes MN. New functions for the ancient globin family: Bacterial responses to nitric oxide and nitrosative stress. Mol Microbiol. 2000;36:775–783. doi: 10.1046/j.1365-2958.2000.01889.x. [DOI] [PubMed] [Google Scholar]
- 8.Green J, Crack JC, Thomson AJ, LeBrun NE. Bacterial sensors of oxygen. Curr Opin Microbiol. 2009;12(2):145–151. doi: 10.1016/j.mib.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Hou S, Reynolds MF, Horrigan FT, Heinemann SH, Hoshi T. Reversible binding of heme to proteins in cellular signal transduction. Acc Chem Res. 2006;39:918–924. doi: 10.1021/ar040020w. [DOI] [PubMed] [Google Scholar]
- 10.Brooijmans R, et al. Heme and menaquinone induced electron transport in lactic acid bacteria. Microb Cell Fact. 2009;8(1):28. doi: 10.1186/1475-2859-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lechardeur D, et al. Using heme as an energy boost for lactic acid bacteria. Curr Opin Biotechnol. 2011;22:143–149. doi: 10.1016/j.copbio.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Sambri V, Cevenini R, La Placa M. Susceptibility of iron-loaded Borrelia burgdorferi to killing by hydrogen peroxide and human polymorphonuclear leucocytes. FEMS Microbiol Lett. 1991;65(1):67–71. doi: 10.1016/0378-1097(91)90473-n. [DOI] [PubMed] [Google Scholar]
- 13.Braz GRC, Coelho HSL, Masuda H, Oliveira PL. A missing metabolic pathway in the cattle tick Boophilus microplus. Curr Biol. 1999;9:703–706. doi: 10.1016/s0960-9822(99)80312-1. [DOI] [PubMed] [Google Scholar]
- 14.Ghedin E, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao AU, Carta LK, Lesuisse E, Hamza I. Lack of heme synthesis in a free-living eukaryote. Proc Natl Acad Sci USA. 2005;102:4270–4275. doi: 10.1073/pnas.0500877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, Embley TM. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos Trans R Soc Lond B Biol Sci. 2010;365:713–727. doi: 10.1098/rstb.2009.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang KP, Chang CS, Sassa S. Heme biosynthesis in bacterium-protozoon symbioses: Enzymic defects in host hemoflagellates and complemental role of their intracellular symbiotes. Proc Natl Acad Sci USA. 1975;72:2979–2983. doi: 10.1073/pnas.72.8.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanhollebeke B, et al. A haptoglobin-hemoglobin receptor conveys innate immunity to Trypanosoma brucei in humans. Science. 2008;320:677–681. doi: 10.1126/science.1156296. [DOI] [PubMed] [Google Scholar]
- 19.Lara FA, et al. Heme requirement and intracellular trafficking in Trypanosoma cruzi epimastigotes. Biochem Biophys Res Commun. 2007;355:16–22. doi: 10.1016/j.bbrc.2006.12.238. [DOI] [PubMed] [Google Scholar]
- 20.Korený L, Lukeš J, Oborník M. Evolution of the haem synthetic pathway in kinetoplastid flagellates: An essential pathway that is not essential after all? Int J Parasitol. 2010;40:149–156. doi: 10.1016/j.ijpara.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Muller E, et al. Variability in the phloem restricted plant trypanosomes (Phytomonas spp) associated with wilts of cultivated crops; Isoenzyme comparison with the lower trypanosomatids. Eur J Plant Pathol. 1994;100:425–434. [Google Scholar]
- 22.Sánchez-Moreno M, Lasztity D, Coppens I, Opperdoes FR. Characterization of carbohydrate metabolism and demonstration of glycosomes in a Phytomonas sp. isolated from Euphorbia characias. Mol Biochem Parasitol. 1992;54:185–199. doi: 10.1016/0166-6851(92)90111-v. [DOI] [PubMed] [Google Scholar]
- 23.Tripodi KE, Buttigliero LV, Altabe SG, Uttaro AD. Functional characterization of front-end desaturases from trypanosomatids depicts the first polyunsaturated fatty acid biosynthetic pathway from a parasitic protozoan. FEBS J. 2006;273:271–280. doi: 10.1111/j.1742-4658.2005.05049.x. [DOI] [PubMed] [Google Scholar]
- 24.Flannery AR, Huynh C, Mittra B, Mortara RA, Andrews NW. LFR1 ferric iron reductase of Leishmania amazonensis is essential for the generation of infective parasite forms. J Biol Chem. 2011;286:23266–23279. doi: 10.1074/jbc.M111.229674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vonlaufen N, Kanzok SM, Wek RC, Sullivan WJ., Jr Stress response pathways in protozoan parasites. Cell Microbiol. 2008;10:2387–2399. doi: 10.1111/j.1462-5822.2008.01210.x. [DOI] [PubMed] [Google Scholar]
- 26.Nawathean P, Maslov DA. The absence of genes for cytochrome c oxidase and reductase subunits in maxicircle kinetoplast DNA of the respiration-deficient plant trypanosomatid Phytomonas serpens. Curr Genet. 2000;38(2):95–103. doi: 10.1007/s002940000135. [DOI] [PubMed] [Google Scholar]
- 27.González-Halphen D, Maslov DA. NADH-ubiquinone oxidoreductase activity in the kinetoplasts of the plant trypanosomatid Phytomonas serpens. Parasitol Res. 2004;92:341–346. doi: 10.1007/s00436-003-1058-4. [DOI] [PubMed] [Google Scholar]
- 28.Maslov DA, Zíková A, Kyselová I, Lukeš J. A putative novel nuclear-encoded subunit of the cytochrome c oxidase complex in trypanosomatids. Mol Biochem Parasitol. 2002;125:113–125. doi: 10.1016/s0166-6851(02)00235-9. [DOI] [PubMed] [Google Scholar]
- 29.Chaumont F, Schanck AN, Blum JJ, Opperdoes FR. Aerobic and anaerobic glucose metabolism of Phytomonas sp. isolated from Euphorbia characias. Mol Biochem Parasitol. 1994;67:321–331. doi: 10.1016/0166-6851(94)00141-3. [DOI] [PubMed] [Google Scholar]
- 30.Cermáková P, Verner Z, Man P, Lukeš J, Horváth A. Characterization of the NADH:ubiquinone oxidoreductase (complex I) in the trypanosomatid Phytomonas serpens (Kinetoplastida) FEBS J. 2007;274:3150–3158. doi: 10.1111/j.1742-4658.2007.05847.x. [DOI] [PubMed] [Google Scholar]
- 31.Van Hellemond JJ, Simons B, Millenaar FF, Tielens AG. A gene encoding the plant-like alternative oxidase is present in Phytomonas but absent in Leishmania spp. J Eukaryot Microbiol. 1998;45:426–430. doi: 10.1111/j.1550-7408.1998.tb05094.x. [DOI] [PubMed] [Google Scholar]
- 32.Morales J, et al. Novel mitochondrial complex II isolated from Trypanosoma cruzi is composed of 12 peptides including a heterodimeric Ip subunit. J Biol Chem. 2009;284:7255–7263. doi: 10.1074/jbc.M806623200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. Escherichia coli succinate dehydrogenase variant lacking the heme b. Proc Natl Acad Sci USA. 2007;104:18007–18012. doi: 10.1073/pnas.0707732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oyedotun KS, Sit CS, Lemire BD. The Saccharomyces cerevisiae succinate dehydrogenase does not require heme for ubiquinone reduction. Biochim Biophys Acta. 2007;1767:1436–1445. doi: 10.1016/j.bbabio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Lemarie A, Grimm S. Mutations in the heme b-binding residue of SDHC inhibit assembly of respiratory chain complex II in mammalian cells. Mitochondrion. 2009;9:254–260. doi: 10.1016/j.mito.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Bringaud F, Rivière L, Coustou V. Energy metabolism of trypanosomatids: Adaptation to available carbon sources. Mol Biochem Parasitol. 2006;149:1–9. doi: 10.1016/j.molbiopara.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 37.Jeffcoat R, Brawn PR, Safford R, James AT. Properties of rat liver microsomal stearoyl-coenzyme A desaturase. Biochem J. 1977;161:431–437. doi: 10.1042/bj1610431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uttaro AD. Biosynthesis of polyunsaturated fatty acids in lower eukaryotes. IUBMB Life. 2006;58:563–571. doi: 10.1080/15216540600920899. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell AG, Martin CE. A novel cytochrome b5-like domain is linked to the carboxyl terminus of the Saccharomyces cerevisiae delta-9 fatty acid desaturase. J Biol Chem. 1995;270:29766–29772. doi: 10.1074/jbc.270.50.29766. [DOI] [PubMed] [Google Scholar]
- 40.Domergue F, et al. New insight into Phaeodactylum tricornutum fatty acid metabolism. Cloning and functional characterization of plastidial and microsomal δ12-fatty acid desaturases. Plant Physiol. 2003;131:1648–1660. doi: 10.1104/pp.102.018317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hongsthong A, et al. Revealing the complementation of ferredoxin by cytochrome b (5) in the Spirulina- (6)-desaturation reaction by N-terminal fusion and co-expression of the fungal-cytochrome b (5) domain and Spirulina- (6)-acyl-lipid desaturase. Appl Microbiol Biotechnol. 2006;72:1192–1201. doi: 10.1007/s00253-006-0407-5. [DOI] [PubMed] [Google Scholar]
- 42.Zhou W, Lepesheva GI, Waterman MR, Nes WD. Mechanistic analysis of a multiple product sterol methyltransferase implicated in ergosterol biosynthesis in Trypanosoma brucei. J Biol Chem. 2006;281:6290–6296. doi: 10.1074/jbc.M511749200. [DOI] [PubMed] [Google Scholar]
- 43.Lamb DC, et al. Plant sterol 14 α-demethylase affinity for azole fungicides. Biochem Biophys Res Commun. 2001;284:845–849. doi: 10.1006/bbrc.2001.5010. [DOI] [PubMed] [Google Scholar]
- 44.Lepesheva GI, et al. Sterol 14α-demethylase as a potential target for antitrypanosomal therapy: Enzyme inhibition and parasite cell growth. Chem Biol. 2007;14:1283–1293. doi: 10.1016/j.chembiol.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rangel H, Dagger F, Hernandez A, Liendo A, Urbina JA. Naturally azole-resistant Leishmania braziliensis promastigotes are rendered susceptible in the presence of terbinafine: Comparative study with azole-susceptible Leishmania mexicana promastigotes. Antimicrob Agents Chemother. 1996;40:2785–2791. doi: 10.1128/aac.40.12.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buttke TM, Van Cleave S. Adaptation of a cholesterol deficient human T cell line to growth with lanosterol. Biochem Biophys Res Commun. 1994;200:206–212. doi: 10.1006/bbrc.1994.1435. [DOI] [PubMed] [Google Scholar]
- 47.Gachotte D, et al. A yeast sterol auxotroph (erg25) is rescued by addition of azole antifungals and reduced levels of heme. Proc Natl Acad Sci USA. 1997;94:11173–11178. doi: 10.1073/pnas.94.21.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nozaki T, Shigeta Y, Saito-Nakano Y, Imada M, Kruger WD. Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, Trypanosoma cruzi. Isolation and molecular characterization of cystathionine β-synthase and serine acetyltransferase from Trypanosoma. J Biol Chem. 2001;276:6516–6523. doi: 10.1074/jbc.M009774200. [DOI] [PubMed] [Google Scholar]
- 49.Farr H, Gull K. Functional studies of an evolutionarily conserved, cytochrome b5 domain protein reveal a specific role in axonemal organisation and the general phenomenon of post-division axonemal growth in trypanosomes. Cell Motil Cytoskeleton. 2009;66(1):24–35. doi: 10.1002/cm.20322. [DOI] [PubMed] [Google Scholar]
- 50.Das S, et al. Lipid metabolism in mucous-dwelling amitochondriate protozoa. Int J Parasitol. 2002;32:655–675. doi: 10.1016/s0020-7519(02)00006-1. [DOI] [PubMed] [Google Scholar]
- 51.Rieske JS. The quantitative determination of mitochondrial hemoproteins. Methods Enzymol. 1967;10:488–493. [Google Scholar]
- 52.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 53.Horváth A, et al. Downregulation of the nuclear-encoded subunits of the complexes III and IV disrupts their respective complexes but not complex I in procyclic Trypanosoma brucei. Mol Microbiol. 2005;58:116–130. doi: 10.1111/j.1365-2958.2005.04813.x. [DOI] [PubMed] [Google Scholar]
- 54.Chevreux B, Wetter T, Suhai S. Proceedings of German Conference on Bioinformatics, GCB ‘99. Braunschweig, Germany: German Research Centre for Biotechnology; 1999. Genome sequence assembly using trace signals and additional sequence information; pp. 45–56. [Google Scholar]
- 55.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 56.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 57.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 58.Gould MK, Vu XL, Seebeck T, de Koning HP. Propidium iodide-based methods for monitoring drug action in the kinetoplastidae: Comparison with the Alamar Blue assay. Anal Biochem. 2008;382(2):87–93. doi: 10.1016/j.ab.2008.07.036. [DOI] [PubMed] [Google Scholar]
- 59.Räz B, Iten M, Grether-Bühler Y, Kaminsky R, Brun R. The Alamar Blue assay to determine drug sensitivity of African trypanosomes (T.b. rhodesiense and T.b. gambiense) in vitro. Acta Trop. 1997;68(2):139–147. doi: 10.1016/s0001-706x(97)00079-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.