Abstract
A general approach for the computational design of enzymes to catalyze arbitrary reactions is a goal at the forefront of the field of protein design. Recently, computationally designed enzymes have been produced for three chemical reactions through the synthesis and screening of a large number of variants. Here, we present an iterative approach that has led to the development of the most catalytically efficient computationally designed enzyme for the Kemp elimination to date. Previously established computational techniques were used to generate an initial design, HG-1, which was catalytically inactive. Analysis of HG-1 with molecular dynamics simulations (MD) and X-ray crystallography indicated that the inactivity might be due to bound waters and high flexibility of residues within the active site. This analysis guided changes to our design procedure, moved the design deeper into the interior of the protein, and resulted in an active Kemp eliminase, HG-2. The cocrystal structure of this enzyme with a transition state analog (TSA) revealed that the TSA was bound in the active site, interacted with the intended catalytic base in a catalytically relevant manner, but was flipped relative to the design model. MD analysis of HG-2 led to an additional point mutation, HG-3, that produced a further threefold improvement in activity. This iterative approach to computational enzyme design, including detailed MD and structural analysis of both active and inactive designs, promises a more complete understanding of the underlying principles of enzymatic catalysis and furthers progress toward reliably producing active enzymes.
Keywords: computational protein design, de novo enzyme design, proton transfer
The high efficiency, chemoselectivity, regio- and stereospecificity, and biodegradability of enzymes make them extremely attractive catalysts. However, the finite repertoire of naturally occurring enzymes limits their applicability to broad problems in biotechnology. A general method for the computational design of enzymes that can efficiently catalyze arbitrary chemical reactions would allow the benefits of enzymatic catalysis to be applied to chemical transformations of interest that are currently inaccessible via natural enzymes. Bolon and Mayo provided important early evidence that such an approach is feasible (1), which motivated significant progress toward this goal in recent years. Using quantum mechanics-based active site design and the Rosetta software suite, Baker, Houk, and coworkers designed enzymes for three chemically unrelated nonnatural reactions in a variety of catalytically inert scaffolds (2–4).
In early incarnations of computational protein design, a strategy for methods development was put forth in terms of the so-called “protein design cycle” in which experimental evaluation of an initial design is used to inform adjustments to the design process for subsequent rounds of design (5, 6). Ideally, these steps would be continued iteratively until the protein sequences predicted by the algorithm exhibit the desired characteristics. However, there is little evidence that this strategy has been used for purposes other than force-field parameterization (5, 7–9). Proteins from failed computational design efforts are typically discarded without comment or investigation into the cause of failure. This situation is unfortunate, because valuable information is lost when only successful designs are reported. Without detailed computational and/or experimental analysis of failed designs, flaws in the design procedure cannot be identified and remedied to produce proteins with the desired characteristics (10, 11). In addition, a focus on reporting only successful designs can lead to the impression that current computational protein design methods are errorless.
The recent successes in designing enzymes show that the field is well on the way to its goal of developing a general method for designing protein catalysts (2–4). However, the catalytic rate enhancements of computationally designed enzymes are still well below those of natural enzymes, and the methods are dominated by false positives. In the case of the Kemp eliminase enzymes designed by Röthlisberger et al., 59 of the many individual sequences predicted to be active by their protein design methods were selected for experimental screening, and only eight of these turned out to be active. Although active enzymes were in fact produced, the need for a “shotgun” approach suggests an incomplete understanding of the details of the enzymatic system and/or inaccurate modeling by the protein design algorithm (12).
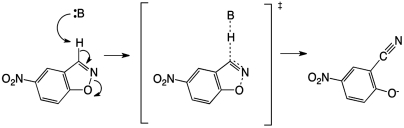
In this work, we focus on the development of a single designed enzyme to test our understanding of enzymatic catalysis and the applicability of the protein design cycle to computational enzyme design problems. We targeted our efforts on the Kemp elimination (KE) (Fig. 1), a well-studied model system for the deprotonation of carbon (13). The KE was selected as a model reaction for this study because catalysts for it have been reported in multiple protein scaffolds (2, 14–16). In addition, from a computational design perspective, the use of the KE allows a direct comparison to the eight enzymes that were computationally designed for this reaction by Röthlisberger et al. (2).
Fig. 1.
The KE reaction scheme.
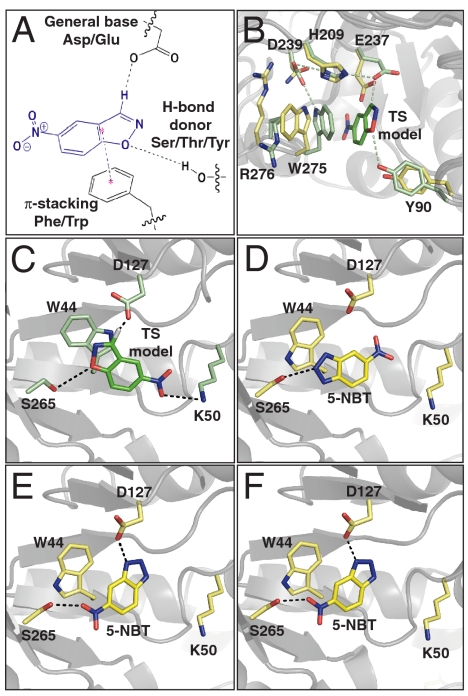
Our approach to KE enzyme design consisted of three steps, which are described in detail by Lassila et al. (17). First, we designed an idealized active site for the KE that included an ab initio calculated transition state (TS) and contacting catalytic residues oriented to facilitate binding and catalysis (Fig. 2A). Next, targeted ligand placement was used to simultaneously sample TS poses and catalytic amino acid positions and orientations within a poly-alanine–substituted binding pocket of a protein scaffold that does not naturally catalyze the KE. Active site configurations that fulfill all of the required catalytic contacts were identified. Finally, one of these active site configurations was selected, and the remaining binding pocket residues were designed to support the TS pose and the geometry of the catalytic residues.
Fig. 2.
KE enzyme design models and crystal structures. (A) KE idealized active site. (B) Overlay of HG-1 crystal structure active site residues (yellow) with design model (green). (C) The HG-2 design model. (D) and (E) Crystal structure of HG-2 active site, chain A. The two conformations of the TSA 5-NBT are shown separately for clarity. (F) Crystal structure of HG-2 active site, chain B with the single observed conformation of the TSA.
Our initial design, HG-1, showed no measurable KE activity. To identify deficiencies in the design procedure, we investigated possible causes of inactivity by using X-ray crystallography and molecular dynamics (MD) simulations. Two problems were identified: The active site was overly exposed to solvent, and critical active site residues showed a high degree of flexibility and orientations inconsistent with the design objectives. Iterating on the protein design process, we corrected these problems in subsequent rounds of computational design using the same protein scaffold. The design with the highest activity, HG-3, was found to have a kcat/Km of 430 M-1 s-1.
Results and Discussion
First-Generation Design.
An idealized active site configuration for the KE is shown in Fig. 2A: A glutamate or aspartate serves as the general base, an aromatic residue provides π-stacking interactions to support substrate binding, and a serine, threonine, or tyrosine residue donates a hydrogen bond to the isoxazolic oxygen of the 5-nitrobenzisoxazole substrate (5-NBZ), which develops a negative charge in the TS. These contacts were inspired by the active site residues observed in the catalytic antibody 34E4 (18); similar active site arrangements were used by Röthlisberger et al. to obtain eight active KE enzymes (2). The geometric constraints for the catalytic contacts are listed in SI Appendix, Table S1.
The active site search was carried out in the xylan binding pocket of the xylanase from Thermoascus aurantiacus (TAX) (19); multiple active site configurations were identified. One of the best scoring active sites included the wild-type glutamate at position 237, which is predicted to serve as a general base for the reaction and adopts a conformation similar to that seen in the crystal structure of wild-type TAX. This active site also has a wild-type tryptophan (W275) in its crystallographic conformation, which is predicted to make a π-stacking contact with the TS. Finally, a tyrosine at position 90 fulfills the hydrogen bond contact to the isoxazolic oxygen. Repacking of the active site gave the final HG-1 design, which differs from the wild-type TAX scaffold by a total of seven mutations (SI Appendix, Table S2).
Experimental Characterization and Analysis of an Inactive Design.
HG-1 showed no KE activity over background under any of the conditions tested (pH 5.0–9.0, 20–37 °C). Analysis by circular dichroism spectroscopy showed that the secondary structure of HG-1 is similar to that of the wild-type scaffold (SI Appendix, Fig. S1A), which indicates that the inactivity of the designed protein was not due to global unfolding or misfolding. The apparent melting temperature (Tm) for HG-1, 55 °C, is about 20 °C lower than that of the wild-type scaffold at pH 7.25 (SI Appendix, Fig. S1B). However, HG-1 remains fully folded under the conditions of the standard KE assay (27 °C, pH 7.25).
In an effort to elucidate the source of inactivity of HG-1, a combined experimental and computational strategy was explored: The X-ray crystal structure was solved to a resolution of 2.0 Å, and explicit solvent MD simulations were performed on the design model. The crystal structure showed a backbone atom root mean square deviation (rmsd) of 0.65 Å with respect to the wild-type scaffold, which confirms that the overall fold is indeed maintained. The active site residues, including the designed general base (E237) and the hydrogen bond donor (Y90), are in positions similar to the predicted side-chain conformations (Fig. 2B). The largest active site deviations are in the π-stacking residue (W275) and the associated arginine (R276)—both are rotated out of the active site with respect to the design, which thereby excludes the possibility of predicted binding interactions with the substrate and exposes the active site to solvent. The crystal structure also revealed six ordered water molecules in the active site, five of which would have to be displaced during substrate binding. The data collection and refinement statistics for this structure are summarized in SI Appendix, Table S3.
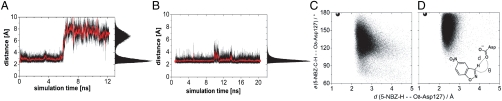
MD simulations for HG-1 were carried out for both substrate-bound (5-NBZ:HG-1) and unliganded (apoHG-1) structures. The trajectories show in both instances that the side chains of the active site residues are highly mobile and that their predicted orientations are lost when subjected to explicit solvent dynamics. This observation applies particularly to the loop that consists of residues 273–276 and most notably to W275. The active site opens up and allows an influx of water molecules in the 5-NBZ:HG-1 MD (SI Appendix, Fig. S2). The catalytic base E237 is coordinated by multiple water molecules that compete with 5-NBZ for interactions with the base and other polar active site residues. The additional loss of π-stacking and packing interactions with W275 allows the substrate to diffuse away from the catalytically competent position (Fig. 3A and SI Appendix, Fig. S2A). The apoHG-1 MD further underlines the flexibility of the 273–276 loop (SI Appendix, Fig. S2C). Here, W275 is rotated into the active site where it hydrogen bonds to E237 while assuming a conformation that directly competes with the catalytically competent binding pose of 5-NBZ.
Fig. 3.
MD-assisted design refinement: from HG-1 to HG-2. (A) and (B) MD base-substrate distance versus time plots (Asp OD to acidic hydrogen of 5-NBZ) for HG-1 (A) and HG-2 (B). (C) and (D) Angle versus distance scatter plots of the catalytic contact (as displayed in the Inset). Data points were taken from 20-ns MD trajectories of HG-2 with 5-NBZ bound in orientations O1 (C) and O2 (D). The coordinates of the TS geometry are displayed in filled discs.
Second-Generation Design.
A key observation from the crystal structure and the MD simulations is that a significant number of water molecules are present in the active site of the first-generation HG-1 design. This finding suggests a substantial desolvation barrier for substrate binding and a bulk, solvent-like pKa of the base (E237). The high flexibility of the active site side chains and low degree of preorganization may further add to the observed inactivity. On the basis of early work by Kemp and coworkers, who showed that a nonpolar environment is best suited for the base-catalyzed KE (13, 20), increasing the hydrophobic character of the HG-1 active site is expected to facilitate the binding of the hydrophobic 5-nitrobenzisoxazole substrate and also elevate the pKa of the base. We therefore sought a more embedded active site pocket in order to maximize these effects.
Manual inspection identified native D127 as a promising candidate for the catalytic base. This aspartate forms a salt bridge with R81 and defines the bottom of a well-packed, narrow solvent-accessible pocket in the core of the (α/β)8 barrel, well-removed from the native TAX binding pocket. Using a computational approach, we sought to increase the size of this pocket to accommodate the substrate and the additional catalytic residues. This area also contains polar and charged residues, which do not provide the ideal environment for the KE. Substantial modifications of R81, N130, N172, T236, and E237 would be necessary to allow the substrate access to the base and to form a hydrophobic binding pocket to facilitate proton abstraction.
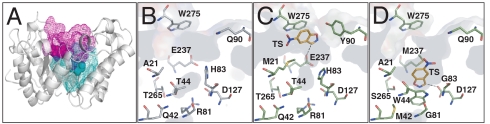
By focusing the design on the native D127 as the general base, an active site search was carried out in a manner similar to that for HG-1 using identical geometric constraints. Compared to the HG-1 calculation, the active site search for this design was shifted 7 Å further into the barrel of the scaffold (Fig. 4A).
Fig. 4.
Relative active site locations in the TAX scaffold and the designs. (A) The locations of the active sites of HG-1 (magenta mesh) and HG-2 (cyan mesh) in the TAX scaffold. The active sites of (B) the wild-type TAX scaffold, (C) the design model of HG-1, and (D) the design model of HG-2. The TS model in the two designs is shown in orange.
The final catalytic configuration consisted of D127 as the general base, T44W as the π-stacking residue, and T265S as the hydrogen bond donor (Fig. 2C). The isoxazole ring of the TS points into the back of the active site pocket and is well shielded from solvent. Active site repacking produced the second-generation design HG-2, whose sequence differs by 12 mutations from wild-type TAX (SI Appendix, Table S2) and 19 mutations from HG-1. As expected, the design model shows major changes in the size and hydrophobicity of the active site residues relative to wild-type TAX. Fig. 4 demonstrates the variation of the active sites basis on this scaffold. Of note, R81, which forms a buried salt bridge with D127 in TAX, was mutated to a glycine in the design, making room for the substrate to access the base. Nearby H83 and N130 were also mutated to glycine to further open up space in the active site for the substrate and the catalytic residues. Q42, T84, N172, T236, and E237 were mutated to large hydrophobic residues, which increases the overall hydrophobicity of the active site and promotes packing around the TS and catalytic residues.
Characterization of Second-Generation Design.
As for HG-1, MD simulations were carried out for the unliganded and liganded HG-2. The latter was carried out with the 5-NBZ substrate bound in the computationally predicted orientation in which the 5-NBZ nitro group points toward the solvent (O1). Analysis of the apoHG-2 and 5-NBZ:HG-2 MD trajectories suggested that HG-2 should be active: The active site remains intact in both simulations, and the substrate stays bound throughout the 20-ns 5-NBZ:HG-2 MD simulation. The base-substrate contact is well-defined with a narrow distance distribution centered at 2.5 Å (Fig. 3B). The simulations also show that the binding site residues pack well around the substrate, which prevents invasion of any water molecules into the active site to compete for interactions with the base and suggests an elevation in the pKa of D127.
Analysis of the active site rmsd with respect to the initial O1 configuration shows that at least two distinct active site conformational states are explored (states O1.1 and O1.2, with all-atom rmsds of 1.9 and 2.3 Å versus the design model, respectively) (SI Appendix, Figs. S3 and S4). Structurally, state O1.1 has greater similarity to the design than state O1.2 (SI Appendix, Fig. S5). Overall, the contact between the base and the substrate remains intact in both conformational states. The difference between the states is most apparent in the secondary structure elements and the mode of substrate binding.
The salt bridge between D127 and R81 connects two neighboring β-strands in the TAX scaffold. In HG-2, R81 is mutated to a glycine, which removes this buried ionic contact and facilitates a hydrogen bond between D127 and the backbone-NH of G81 (SI Appendix, Fig. S5A). In the HG-2 MD simulation, this contact forms at 10 ns and leads to the bimodal distribution that is representative of the two observed conformational states (SI Appendix, Figs. S4 and S5).
In state O1.1, the substrate lies in the same plane as in the design. In state O1.2, the substrate assumes a binding mode that is rotated by approximately 90° about an axis perpendicular to the plane of the substrate. The contact between the base and substrate is well-defined in both states but is significantly weakened during the transition between states (Fig. 3B). Analysis of the path between the two conformational states identifies M42, M84, M172, and M237 as the main contributors to the transition. These four methionines are within 4 Å of the bound substrate. In light of the recent work by Tawfik and coworkers showing that increasing active site rigidity and preorganization coincided with increased activity for their designed KE catalysts (21), the activity of HG-2 may be adversely affected by the flexibility of its active site.
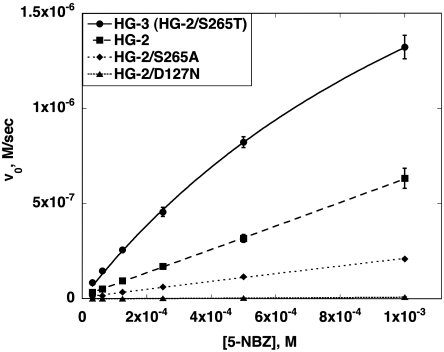
As predicted by the MD simulations, experimental analysis indicates that HG-2 is an active catalyst for the KE, with a reaction rate far above that of the buffer-catalyzed reaction (Fig. 5 and Table 1). Wild-type TAX was shown to have no KE activity above background. Substrate saturation was not reached for HG-2, which prevents reliable determination of kcat and Km. By using the slope of the initial rate vs. initial substrate concentration plot, kcat/Km was determined to be 122 M-1 s-1, which makes the efficiency of this enzyme comparable to the best KE enzymes designed by Röthlisberger et al. (2). Knockout mutations of the base (D127N) and the hydrogen bond donor (S265A) led to a significant decrease in activity compared to HG-2, with the base knockout losing almost all activity (Fig. 5 and Table 1). The loss of activity upon mutation of the putative catalytic residues indicates that these residues are, in fact, important for catalysis and supports the proposed mode of action for this enzyme.
Fig. 5.
Kinetic characterization of Kemp elimination enzymes. Michaelis-Menten plots of HG-2 and point mutants. The enzyme concentration for all reactions was 5 μM.
Table 1.
Kinetic parameters of designed KE enzymes
| Design | Mutation | Scaffold | kcat, s-1 | Km, mM | kcat/Km, s-1 M-1 | kcat/kuncat* | MD prediction |
| HG-1 | — | 1GOR | no activity detected | inactive | |||
| HG-2 | — | 1GOR | NA | NA | 123.2 ± 0.8 | NA | active |
| HG-2 | D127N | 1GOR | NA | NA | 1.6 ± 0.2 | NA | ND |
| HG-2 | S265A | 1GOR | NA | NA | 41.0 ± 1.1 | NA | ND |
| HG-2 | K50A | 1GOR | NA | NA | 37.7 ± 0.4 | NA | active† |
| HG-2 | G81A | 1GOR | NA | NA | 17.2 ± 0.5 | NA | active† |
| HG-3§ | S265T | 1GOR | 0.68 ± 0.04 | 1.6 ± 0.1 | 425.0 ± 36.5 | 5.9 × 105 | active‡ |
| 1A53-1 | — | 1A53 | no activity detected | inactive | |||
| 1A53-2 | — | 1A53 | 0.09 ± 0.01 | 1.4 ± 0.2 | 64.2 ± 11.6 | 7.8 × 104 | active |
| 1A53-3 | — | 1A53 | NA | NA | 20.6 ± 0.3 | NA | active |
| 1THF-1 | — | 1THF | no activity detected | active | |||
| 1THF-2 | — | 1THF | NA | NA | 7.9 ± 0.08 | NA | active |
General assay conditions: 25 mM Hepes, 100 mM NaCl pH 7.25, 2% acetonitrile. ND, not determined; NA, not applicable.
*Under the assay conditions, kuncat was determined to be 1.16 × 10-6 s-1 by Röthlisberger et al. (1).
†This variant was predicted to be less active than HG-2.
‡This variant was predicted to be more active than HG-2.
§HG-3 exhibits somewhat higher activity in 50 mM phosphate, 100 mM NaCl, pH 7.0, 2% acetonitrile: kcat = 1.7 ± 0.1, Km = 2.4 ± 0.2, kcat/Km = 708 ± 70 M-1 s-1, kcat/kuncat = 1.5 × 106.
A 1.2-Å resolution X-ray crystal structure of HG-2 with the transition state analog (TSA) 5-nitrobenzotriazole (5-NBT) bound in the active site provides direct evidence of catalytically competent substrate interaction with the putative base (Fig. 2 D–F). The protein crystallized with two molecules in the asymmetric unit, which allows for observation of two active sites. Ligand density in chain A was modeled in two orientations (Fig. 2 D and E). The dual orientations may reflect the conformational flexibility of the engineered active site, some of which was observed in the MD simulations. Unambiguous density for a single TSA orientation appears in chain B (Fig. 2F). This orientation (O2) differs from that of the design (O1) in that the TSA is flipped from the designed position, which places the nitro group in contact with S265 rather than K50. In both O1 and O2, the TSA contacts the putative base (D127) in a catalytically relevant manner.
The cocrystal structure of an evolved variant of the Röthlisberger design KE70 with 5-NBT was recently reported (21). In that structure the position of the TSA is inconsistent with the required catalytic base-TS contact. In contrast, the structure of HG-2 with a TSA provides direct crystallographic confirmation of catalytically relevant ligand binding. The data collection and refinement statistics for the HG-2 holostructure are summarized in SI Appendix, Table S3.
MD simulations were carried out to further investigate the alternative substrate orientation O2. The substrate-base contact in the O2 MD is centered at 2.4 Å (SI Appendix, Fig. S3A), which compares well to the narrow distance distribution centered at 2.5 Å in the O1 MD. The oxazole-oxygen is coordinated to K50 and the nitro group contacts S265 throughout the trajectory (SI Appendix, Fig. S3 B and C). From these distance distributions, both O1 and O2 appear to be equally capable of deprotonating the substrate. However, because hydrogen bonds are closely related to the transition states of proton-transfer reactions and have a preference for linearity, their strengths can be more accurately assessed graphically with angle versus distance scatter plots (22). Scatter plots of the HG-2 trajectories show that in orientation O2 the base-substrate pair can assume a more transition-state-like arrangement than in O1 (Fig. 3 C and D, respectively). The plots suggest that O2 is not only a viable alternative orientation of the substrate but quite possibly a more potent one. Head-Gordon et al. made similar conclusions on the basis of the study of a computationally designed retroaldolase, pointing out that the consideration of alternative substrate orientations could be a beneficial strategy for computational enzyme design (23).
Third-Generation Design.
MD analysis of HG-2 suggested that an S265T mutation in the HG-2 background would improve catalysis, because the larger volume and lower conformational flexibility of threonine relative to serine was predicted to provide better packing around the substrate. S265T is a reversion to the wild-type amino acid at this position. MD analysis of HG-3 (HG-2/S265T) predicted that this variant would be more active than HG-2, because it lacks the active site conformational heterogeneity seen in the HG-2 simulation. Experimental evaluation confirmed that this design has a kcat/Km that is about threefold higher than HG-2 or any of the previously designed KE enzymes (Table 1) (2). A pH/activity profile suggests that the pKa of the putative base is significantly elevated from the solvent-exposed pKa of aspartate (approximately 6 vs. approximately 4, respectively) (SI Appendix, Fig. S6). The pH/activity profile also shows a significant decrease in activity at high pHs, which may indicate the presence of a second ionizable group in the active site region. In addition, the TSA 5-NBT competitively inhibits the KE reaction at pH 5.0 (Ki = 330 ± 30 μM), supporting crystallographically observed TSA binding in the designed active site of HG-2 (SI Appendix, Fig. S7).
Recapitulation of Previous KE Designs.
We also tested the ability of our computational design methods to recapitulate the active sites of three functional enzymes from Röthlisberger et al. (2). KE59 was based on the Sulfolobus solfataricus indole-3-glycerolphosphate synthase scaffold (24); KE07 and KE10 were based on the Thermotoga maritima imidazoleglycerolphosphate synthase scaffold (25).
Starting with the base positions and scaffolds from the active KE07, KE10, and KE59 enzymes, TS poses and catalytic residue positions that satisfied the catalytic contacts specified in the HG-1 and HG-2 designs were retained and stabilized through packing of the surrounding amino acid side chains. We generated five designs: 1THF-1, 1THF-2, 1A53-1, 1A53-2, and 1A53-3 (SI Appendix, Table S2). Despite using the same base position as in the Röthlisberger designs, our 1THF- and 1A53-based designs differ by eight to ten mutations and give rise to active site geometries that are distinct from the Röthlisberger designs (SI Appendix, Figs. S8 and S9). These differences can be attributed to variations in the geometries used to define the active site as well as differences in the ligand pose sampling methods and force field used by Rosetta and our method. Three of the five designs showed significant activity over background (SI Appendix, Fig. S10), which indicates that multiple, geometrically unique active sites for KE catalysis can be generated from the same scaffold.
MD-Based Prediction of Enzyme Activity.
MD simulations were carried out for each of the five “recapitulation” designs with a substrate model bound in the active site. As with MD analysis of HG-2 and HG-3, analysis of the trajectories for 1A53-2, 1A53-3, 1THF-1, and 1THF-2 predicted that these four variants would be active because the structural integrity of the active site is maintained, catalytic contacts between the base and the substrate are firmly established, and the base is well-shielded from solvent. Conversely, 1A53-1 was predicted to be inactive because of reorganization of active site residues, which disrupts the catalytic contact to the substrate, as was the case for HG-1. The criteria for predicting activity were the same as those used in a recently published MD analysis of the Röthlisberger KE designs (11).
Experimental analysis showed that three out of five of these designs (1A53-2, 1A53-3, and 1THF-2) had significant catalytic activity. These results are in agreement with the MD-based predictions, which overall were correct for five of the six designs (Table 1), and suggest that MD could be a promising tool for in silico prescreening of designed enzymes.
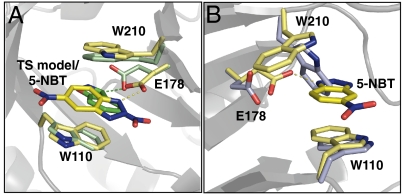
Crystallographic Analysis of 1A53-2.
X-ray crystal structures of 1A53-2 were determined in the apo and 5-NBT-bound forms to 1.6- and 1.5-Å resolution, respectively. The full protein rmsd for the ligand-bound crystal structure with the design model is 0.51 Å, which indicates that the overall fold is maintained. Active site side-chain conformations in the cocrystal structure are in general agreement with the design (Fig. 6A). As in the case of the HG-2 cocrystal structure, the position of the TSA is flipped from the designed orientation. Importantly, however, the ligand maintains a catalytically competent contact with the putative base (E178). The apo structure shows that the W210 side chain rotates from the catalytically relevant stacking position seen in the cocrystal structure to fill the substrate binding pocket (Fig. 6B). The data collection and refinement statistics for these structures are summarized in SI Appendix, Table S3.
Fig. 6.
Crystal structures of 1A53-2. (A) Overlay of 1A53-2 holostructure (yellow) and the design model (green). (B) Overlay of 1A53-2 apo crystal structure (lavender) and holostructure (yellow).
Conclusions
The iterative approach to computational enzyme design described here has led to the most active computationally designed enzyme catalyst for the KE to date. Inactive designs were probed by X-ray crystallography and MD simulations to learn the likely causes of inactivity. These data informed the next round of design and led to active enzymes. In this way, computational methods and crystallography were used, rather than combinatorial experimental approaches, to create effective enzyme catalysts. We believe that this iterative approach constitutes a significant advance in enzyme design methodology that, in addition to leading to improved designs, should contribute to a more complete understanding of the mechanisms of enzymatic activity.
The relocation of the active site into the core of the HG-2 scaffold is a departure from previous enzyme design procedures, which focus designs solely in natural binding pockets of the scaffold (2–4). Although the site of the catalytic base in the HG-2 active site was manually selected, a subsequent broader computational search for possible active sites also identified D127 among a large list of potential base positions outside of the natural binding pocket. The possibility of expanded active site searches suggests an opportunity for the improvement of computational design methodology to more efficiently carry out these large searches and to rank identified active site possibilities by their likelihood of supporting catalysis.
As with previous computationally designed enzymes, the activity levels reported here are low compared to many natural enzymes. Directed evolution has been shown to be an effective strategy to increase the activity of designed enzymes (2, 21, 26) and may offer insight into the deficiencies in the design.
All-atom explicit solvent MD simulations have previously been shown to be effective at recapitulating the activity of computationally designed KE enzymes (11). Here, MD was carried out prior to experimentation for all cases except HG-1, and the integration of MD into the iterative design process proved to be useful for identifying underlying problems in the structure and dynamics of HG-1 and in guiding the improvement of HG-2. The recent design of enzymes that stereoselectively promote a Diels-Alder reaction demonstrates the applicability of MD to more complicated chemistries (4).
The discrepancy between the ligand orientation in the modeled structures and in the crystal structures of HG-2 and 1A53-2 may be due to the inaccurate modeling of the TS ligand and/or inadequate sampling of possible ligand positions within the active site. Improvements to the force field may be necessary for accurate modeling of the ligand’s nitro group in a hydrophobic environment. In addition, the utility of combining computational protein design with MD simulations suggests that future inclusion of full backbone flexibility, loop modeling, and MD move sets directly into computational design procedures may lead to more accurate predictions of ligand positions and improved de novo designed enzymes.
Materials and Methods
Full materials and methods are provided in SI Appendix.
Active Site Placement Calculations.
Possible base positions were selected on the basis of the location of wild-type carboxylate residues within the binding pocket (HG-1: E46, E131, and E237; HG-2: D127) or locations of bases in known KE enzymes (1THF-1/1THF-2: S101E; 1A53-1: L231E; 1A53-2: G178E). Design positions surrounding the natural binding pockets of the scaffolds or the base position (SI Appendix, Table S2) were allowed to sample all conformations of the catalytic amino acid types (Gly, Phe, Trp, Ser, Thr, and Tyr). In the case of 1THF-1, all conformations of Lys were also sampled at these positions to match the conditions of previously published calculations (2). Catalytic base positions were allowed to sample all conformations of Glu and Asp. A backbone-independent conformer library was used to represent side-chain flexibility (17). A library of TS poses was generated in the active site by targeted ligand placement (17). Contact geometries between the Asp/Glu and the TS model that were used to generate ligand poses are listed in SI Appendix, Table S4. During the energy calculation step, TS–side-chain interaction energies were biased to favor interactions that satisfy the geometric constraints in SI Appendix, Table S1. The energy calculation and biasing steps are discussed by Lassila et al. (17).
Active Site Repacking Calculations.
In the repacking calculation, the initial TS pose and catalytic residue positions were taken from the active site identified in the active site search. The TS structure was translated ± 0.4 Å in x, y, and z in 0.2-Å steps and rotated ± 10° about all three axes (origin at TS geometric center) in 5° steps. The geometric constraints from SI Appendix, Table S1 were applied to enforce the contacts between the TS and each of the three catalytic residues identified in the active site search. Residues in the immediate vicinity of the TS or catalytic residues were designated design positions (SI Appendix, Table S2) and were allowed to sample all conformations of all amino acid types except Pro and Cys. To increase the hydrophobicity of the active site, some positions were restricted to hydrophobic amino acid identities (1THF-1: 171, 201; 1THF-2: 196, 222; 1A53-2 and 1A53-3: 51, 159; HG-2: 237). A second shell of float positions was designated around the design positions; these positions were allowed to sample all conformations of the wild-type amino acid at that position (SI Appendix, Table S2). The identities of the catalytic residues were fixed and allowed to sample all conformations of that amino acid type. An occlusion-based solvation potential was applied with scale factors of 0.05 for nonpolar burial, 2.5 for nonpolar exposure, and 1.0 for polar burial (27). Other standard energy potentials and parameters were applied as by Lassila et al. (17). Side-chain–TS interaction energies were biased to favor those contacts that satisfy the geometries in SI Appendix, Table S1 as in the active site search. Sequence optimization was carried out with FASTER (28, 29), and a Monte Carlo-based algorithm (30, 31) was used to sample sequences around the minimum energy sequence.
Supplementary Material
Acknowledgments.
We thank Jens Kaiser and Pavle Nikolovski at the Caltech Molecular Observatory for assistance in crystal screening, crystallographic data collection, and structure determination. We are grateful to Daniela Röthlisberger and David Baker for providing genes for the KE positive controls and to Marie Ary and Scott A. Johnson for assistance with the manuscript. Data for the HG-2 and 1A53-2 structures were collected at beamline 12-2 at the Stanford Synchrotron Radiation Lightsource (SSRL, SLAC National Accelerator Laboratory, Menlo Park, CA). We acknowledge the Gordon and Betty Moore Foundation for support of the Molecular Observatory at Caltech and the Department of Energy and National Institutes of Health for supporting the SSRL. This work was supported by the Defense Advanced Research Projects Agency, a Department of Defense National Security Science and Engineering Faculty Fellowship (S.L.M.), and a Lawrence Livermore National Laboratory Lawrence Scholars Fellowship (G.K.). Fellowship support from the Fonds des Verbandes der chemischen Industrie and the Studienstiftung des deutschen Volkes (R.B.) is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3O2L, 3NYD, 3NYZ, and 3NZ1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118082108/-/DCSupplemental.
References
- 1.Bolon DN, Mayo SL. Enzyme-like proteins by computational protein design. Proc Natl Acad Sci USA. 2001;98:14274–14279. doi: 10.1073/pnas.251555398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Röthlisberger D, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 3.Jiang L, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel JB, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahiyat BI, Mayo SL. Protein design automation. Protein Sci. 1996;5:895–903. doi: 10.1002/pro.5560050511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Street AG, Mayo SL. Computational protein design. Structure. 1999;7:R105–R109. doi: 10.1016/s0969-2126(99)80062-8. [DOI] [PubMed] [Google Scholar]
- 7.Dahiyat BI, Mayo SL. Probing the role of packing specificity in protein design. Proc Natl Acad Sci USA. 1997;94:10172–10177. doi: 10.1073/pnas.94.19.10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahiyat BI, Gordon B, Mayo SL. Automated design of the surface positions of protein helices. Protein Sci. 1997;6:1333–1337. doi: 10.1002/pro.5560060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zollars ES, Marshall SA, Mayo SL. Simple electrostatic model improves designed protein sequences. Protein Sci. 2006;15:2014–2018. doi: 10.1110/ps.062105506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morin A, et al. Computational design of an endo-1,4-{beta}-xylanase ligand binding site. Protein Eng Des Sel. 2011;24:503–516. doi: 10.1093/protein/gzr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiss G, Röthlisberger D, Baker D, Houk KN. Evaluation and ranking of enzyme designs. Protein Sci. 2010;19:1760–1773. doi: 10.1002/pro.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frushicheva MP, Cao J, Chu ZT, Warshel A. Exploring challenges in rational enzyme design by simulating the catalysis in artificial Kemp eliminase. Proc Natl Acad Sci USA. 2010;107:16869–16874. doi: 10.1073/pnas.1010381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kemp D, Casey M. Physical organic chemistry of benzisoxazoles. II. Linearity of the Broensted free energy relation for the base-catalyzed decomposition of benzisoxazoles. J Am Chem Soc. 1973;95:6670–6680. [Google Scholar]
- 14.Thorn SN, Daniels RG, Auditor MM, Hilvert D. Large rate accelerations in antibody catalysis by strategic use of haptenic charge. Nature. 1995;373:228–230. doi: 10.1038/373228a0. [DOI] [PubMed] [Google Scholar]
- 15.Hollfelder F, Kirby AJ, Tawfik DS. Off-the-shelf proteins that rival tailor-made antibodies as catalysts. J Org Chem. 2001;66:5866–5874. doi: 10.1038/383060a0. [DOI] [PubMed] [Google Scholar]
- 16.Korendovych IV, et al. Design of a switchable eliminase. Proc Natl Acad Sci USA. 2011;108:6823–6827. doi: 10.1073/pnas.1018191108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lassila JK, Privett HK, Allen BD, Mayo SL. Combinatorial methods for small-molecule placement in computational enzyme design. Proc Natl Acad Sci USA. 2006;103:16710–16715. doi: 10.1073/pnas.0607691103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Debler EW, et al. Structural origins of efficient proton abstraction from carbon by a catalytic antibody. Proc Natl Acad Sci USA. 2005;102:4984–4989. doi: 10.1073/pnas.0409207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo Leggio L, et al. Substrate specificity and subsite mobility in T aurantiacus xylanase 10A. FEBS Lett. 2001;509:303–308. doi: 10.1016/s0014-5793(01)03177-5. [DOI] [PubMed] [Google Scholar]
- 20.Casey M, Kemp D, Paul K, Cox D. Physical organic chemistry of benzisoxazoles. I. Mechanism of the base-catalyzed decomposition of benzisoxazoles. J Org Chem. 1973;38:2294–2301. [Google Scholar]
- 21.Khersonsky O, et al. Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution. J Mol Biol. 2011;407:391–412. doi: 10.1016/j.jmb.2011.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steiner T. The hydrogen bond in the solid state. Angew Chem Int Ed. 2002;41:48–76. doi: 10.1002/1521-3773(20020104)41:1<48::aid-anie48>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 23.Ruscio J, Kohn J, Ball K, Head-Gordon T. The influence of protein dynamics on the success of computational enzyme design. J Am Chem Soc. 2009;131:14111–14115. doi: 10.1021/ja905396s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hennig M, Darimont BD, Jansonius JN, Kirschner K. The catalytic mechanism of indole-3-glycoerol phosphate synthase: crystal structures of complexes of the enzyme from Sulfolobus solfataricus with substrate analogue, substrate, and product. J Mol Biol. 2002;319:757–766. doi: 10.1016/S0022-2836(02)00378-9. [DOI] [PubMed] [Google Scholar]
- 25.Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M. Structural evidence for evolution of the β/α barrel scaffold by gene duplication and fusion. Science. 2000;289:1546–1550. doi: 10.1126/science.289.5484.1546. [DOI] [PubMed] [Google Scholar]
- 26.Khersonsky O, et al. Evolutionary optimization of computationally designed enzymes: Kemp eliminases of the KE07 series. J Mol Biol. 2010;396:1025–1042. doi: 10.1016/j.jmb.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 27.Chica RA, Moore MM, Allen BD, Mayo SL. Generation of longer emission wavelength red fluorescent proteins using computationally designed libraries. Proc Natl Acad Sci USA. 2010;107:20257–20262. doi: 10.1073/pnas.1013910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desmet J, Spriet J, Lasters I. Fast and accurate side-chain topology and energy refinement (FASTER) as a new method for protein structure optimization. Proteins. 2002;48:31–43. doi: 10.1002/prot.10131. [DOI] [PubMed] [Google Scholar]
- 29.Allen BD, Mayo SL. Dramatic performance enhancements for the FASTER optimization algorithm. J Comput Chem. 2006;27:1071–1075. doi: 10.1002/jcc.20420. [DOI] [PubMed] [Google Scholar]
- 30.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH. Equation of state calculations by fast computing machines. J Chem Phys. 1953;21:1087–1092. [Google Scholar]
- 31.Voigt CA, Gordon DB, Mayo SL. Trading accuracy for speed: A quantitative comparison of search algorithms in protein sequence design. J Mol Biol. 2000;299:789–803. doi: 10.1006/jmbi.2000.3758. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.