Abstract
A common challenge in pathogen discovery by deep sequencing approaches is to recognize viral or subviral pathogens in samples of diseased tissue that share no significant homology with a known pathogen. Here we report a homology-independent approach for discovering viroids, a distinct class of free circular RNA subviral pathogens that encode no protein and are known to infect plants only. Our approach involves analyzing the sequences of the total small RNAs of the infected plants obtained by deep sequencing with a unique computational algorithm, progressive filtering of overlapping small RNAs (PFOR). Viroid infection triggers production of viroid-derived overlapping siRNAs that cover the entire genome with high densities. PFOR retains viroid-specific siRNAs for genome assembly by progressively eliminating nonoverlapping small RNAs and those that overlap but cannot be assembled into a direct repeat RNA, which is synthesized from circular or multimeric repeated-sequence templates during viroid replication. We show that viroids from the two known families are readily identified and their full-length sequences assembled by PFOR from small RNAs sequenced from infected plants. PFOR analysis of a grapevine library further identified a viroid-like circular RNA 375 nt long that shared no significant sequence homology with known molecules and encoded active hammerhead ribozymes in RNAs of both plus and minus polarities, which presumably self-cleave to release monomer from multimeric replicative intermediates. A potential application of the homology-independent approach for viroid discovery in plant and animal species where RNA replication triggers the biogenesis of siRNAs is discussed.
The term “viroid” was first introduced in 1971 to describe a novel class of free RNA nonviral pathogens found in plants (1–3). Viroids are single-stranded circular RNA molecules of 246–401 nt in length and do not encode any protein. Viroid RNAs display extensive intramolecular base pairing to give rod-like or quasirod-like conformations. Replication of viroids occurs via a rolling circle mechanism by host RNA polymerases to yield head-to-tail multiple-repeat replicative intermediates. Viroids identified to date infect only plants and belong to one of the two families. Viroids in the Pospiviroidae such as potato spindle tuber viroid (PSTVd) share a five-domain model including a central conserved region (CCR) and may all replicate in the nucleus. By contrast, viroids in the Avsunviroidae such as peach latent mosaic viroid (PLMVd) lack a CCR, encode a hammerhead ribozyme in both the positive and the negative strands that self-cleaves to release monomer from the multimeric intermediates, and may all replicate in the chloroplasts (1–3).
Identification of new viroids requires purification and enrichment of the naked viroid RNA by 2D gel electrophoresis before cDNA synthesis and sequencing (4, 5). However, viroids generally occur at low concentrations in the infected host, making viroid discovery a challenging task for many plant pathology laboratories. This problem is probably one of the reasons why <40 viroids from two families have so far been identified although viroids were first discovered >40 y ago (1–3).
Application of deep sequencing technologies has facilitated discovery of viruses by purification- and culture-independent approaches. In metagenomic approaches, viral sequences are enriched by partial purification of viral particles before deep sequencing (6, 7). In contrast, enrichment of viral sequences is achieved by isolating the fraction of small RNAs 20–30 nt in length for deep sequencing in virus discovery by deep sequencing and assembly of total host small RNAs (vdSAR) (8, 9). Production of virus-derived small interfering RNAs (siRNAs) is a key step in the RNA-based antiviral immunity of diverse eukaryotic hosts, in which these viral siRNAs guide specific viral RNA clearance by RNA interference (RNAi) or RNA silencing (10–12). Arabidopsis thaliana plants produce 21-, 22-, and 24-nt size classes of viral and endogenous siRNAs by Dicer-like 4 (DCL4), DCL2, and DCL3, respectively (10–12). In fruit flies and mosquitoes, viral siRNAs processed by Dicer-2 are predominantly 21 nt although recent studies also have detected virus-derived PIWI-interacting RNAs (piRNAs) that are 23–30 nt long (9, 10, 13). Notably, virus-derived siRNAs and piRNAs overlap in sequence (14) so that it is possible to assemble them into longer fragments (contigs) of the invading viral genomes (8, 9) by genome assembly algorithms developed specifically for short reads from next generation sequencing platforms such as Illumina (15).
It should be pointed out that discovery of new viruses by both vdSAR and metagenomic approaches requires presence of a detectable sequence homology with a known virus (6, 7, 9). How to recognize a novel pathogen in samples of diseased tissues that shares no statistically significant homology with a known pathogen represents a major challenge in pathogen discovery by deep sequencing approaches.
In this study, we established a homology-independent approach for the discovery of the viroid pathogens by deep sequencing. Previous studies have shown that infection of plants with viroids triggers abundant production of viroid-derived 21-, 22-, and 24-nt classes of small RNAs (sRNAs) in approximately equal plus and minus strand ratios, which are likely products of DCL4, DCL2, and DCL3, respectively (16–23). We found that these viroid sRNAs overlapped in sequence and covered the entire length of viroid RNAs with high densities. A unique computational algorithm was developed to identify viroid-specific sRNAs sequenced from a host organism and to assemble them into full-length viroid RNA sequences. We show that viroids from both the Pospiviroidae and the Avsunviroidae families could be readily identified from the published small RNA libraries sequenced from the infected plants. Analysis of a grapevine library by the algorithm led to the identification of a viroid-like circular RNA 375 nt long that shared no significant sequence homology with any known viroid but encoded an active hammerhead ribozyme in both the plus and minus strands. It is likely that our approach will facilitate discovery of unique viroids in plant and animal species in which RNA replication triggers the Dicer-mediated biogenesis of overlapping siRNAs.
Results
Assembly of Viroid-Derived Small RNAs.
To determine whether viroid-derived small RNAs could be assembled into contigs, we analyzed two published small RNA libraries using Velvet and Vcake programs, which represent graph- and overlapping-based assembly approaches, respectively (24). The first library was from a peach tree infected with PLMVd and contained 7,862,905 reads in total, of which 423,951 and 460,283 reads showed 100% match to the plus and minus strand RNA of PLMVd, respectively (23). The second was from tendril tissues of grapevine infected with both hop stunt viroid (HSVd) and grapevine yellow speckle viroid (GYSVd) from the Pospiviroidae. The grapevine library contained a total of 4,701,135 reads, of which 95,980 and 66,933 were derived from HSVd and GYSVd, respectively (20).
We found that small RNAs from each of the three viroids in the libraries were assembled into contigs of various lengths by either Velvet or Vcake (Fig. 1), indicating that viroid sRNAs overlapped in sequence as found previously for viral siRNAs (8, 9, 14). When the minimal overlapping length (k-mer) required for joining two sRNAs into a contig was set at 17 nt, 56 of the contigs assembled by Vcake were identified as homologous to PLMVd after searching the nonredundant nucleotide sequence entries of the National Center for Biotechnology Information (NCBI) database by BLASTN (Fig. 1). Search of the NCBI database by BLASTX was not informative for viroid identification because the known viroids do not encode any proteins (1–3). Assembly of the grapevine library by Vcake (k-mer = 17) produced 12 HSVd contigs and 12 GYSVd contigs. Assembly by Velvet or a different k-mer may lead to longer viroid contigs. For example, the longest contigs assembled for PLMVd, GYSVd, and HSVd were 79 nt (Velvet; k-mer = 21), 120 nt (Velvet; k-mer = 17), and 278 nt (Velvet; k-mer = 17), respectively. However, the longest contig assembled for any of the three viroids using different parameters was much shorter than the full length of the corresponding viroid because PLMVd, GYSVd, and HSVd are 338, 363, and 293 nt in length, respectively. Importantly, we were not able to identify new viroids from either library by similarity searches of the assembled contigs to the known viroids.
Fig. 1.
Position and distribution of small RNA contigs assembled by Velvet and Vcake programs. The contigs assembled by Vcake and Velvet (k-mer = 17) are represented by black and gray arrows, respectively. The innermost line represents the viroid full-length RNA (smallest division = 20 nt).
A Computational Algorithm for Viroid Discovery.
We developed a unique computational algorithm (Dataset S1) that classifies overlapping small RNAs in a library into two groups (Fig. 2). A terminal small RNA (TSR) overlaps at least one other small RNA in the pool (default k-mer = 17) at only one end to yield a contig longer than the TSR (represented by those numbered small RNAs in each of the small RNA assembles in Fig. 2, Lower). In contrast, an internal small RNA (ISR) overlaps at least one other small RNA in the pool at both ends of the ISR to yield a longer contig (represented by those between the numbered small RNAs in each of the small RNA assemblies in Fig. 2, Lower). The algorithm then removes nonoverlapping small RNAs and all of those TSRs identified in the first step and reclassifies the small RNAs remaining in the pool into TSRs and ISRs. The algorithm, referred to as progressive filtering of overlapping small RNAs (PFOR), is repeated iteratively until no TSR is found and all of the small RNAs left in the pool are true ISRs. At the final step of PFOR, the remaining true ISRs from the small RNA library are assembled into contigs.
Fig. 2.
The key steps in progressive filtering of overlapping small RNAs (PFOR).
All of the small RNAs in a library that do not overlap or overlap <17 nt are discarded in the first round by PFOR, thereby reducing the complexity of the library. All of the siRNAs processed by a Dicer nuclease from a linear dsRNA would progressively be defined as TSRs and removed from the pool (Fig.2, Lower). However, overlapping siRNAs processed from an incomplete or complete head-to-tail direct repeat dsRNA or from a circular RNA would be classified as true ISRs because of the presence of the small RNAs derived from the junction (Fig. 2, Upper, small RNAs 9 and 10 in the small RNA assembly) and progressively enriched by PFOR. Therefore, PFOR was expected to identify viroids because current models suggest that viroid siRNAs are processed either from double-stranded head-to-tail repeats synthesized as replicative intermediates from the rolling-circle viroid replication or from the structural elements of the circular viroid RNA (3, 20–23).
To evaluate the performance of the algorithm (Dataset S1), we subjected the peach and grapevine small RNA libraries described above to PFOR. A single RNA species of 334, 336, and 336 nt in length was obtained from the peach library (7,862,905 reads in total) by PFOR, using k-mer = 19, 20, and 21, respectively. The computational analysis of the peach library required 3 gigabytes (GB) of random-access memory (RAM) and 2 h. The recovered RNA molecules shared ∼91–92% identities, all corresponding to PLMVd, and thus contained no host endogenous sequences. These assembled PLMVd sequences were 92%, 93%, and 95% identical, respectively to PLMVd and were only 2–4 nt shorter (Fig. S1A). PFOR analysis of the smaller grapevine small RNA library (4,701,135 reads in total) required ∼0.5 GB RAM and 2 min, which recovered both HSVd and GYSVd molecules as well as a third molecule (see below). HSVd of 294 and 296 nt in length and GYSVd of 363 and 365 nt in length were obtained by PFOR, using k-mer of 17 and 18, respectively. The sequence identity was 97% between two recovered HSVd molecules and 96% between two GYSVd molecules (Fig. S1 B and C). These findings show that PFOR analysis of the total small RNAs sequenced from the diseased tissue can readily identify and enrich siRNAs processed from viroids of both the Pospiviroidae and Avsunviroidae families and assemble them into full-length viroids.
A Hammerhead Viroid-Like RNA Identified from Grapevine.
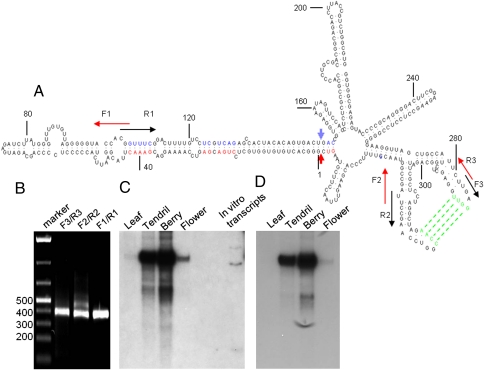
In principle, PFOR analysis of a small RNA library would identify both the known and the previously uncharacterized viroids in the disease sample. Indeed, in addition to HSVd and GYSVd, PFOR analysis of the grapevine small RNA library revealed a third RNA molecule that exhibited several key features of viroids (Fig. 3A). This RNA species, designated as grapevine hammerhead viroid-like RNA (GHVd RNA), was 372 and 375 nt in length as discovered by PFOR with k-mer of 17 and 18, respectively, and the predicted GHVd RNAs shared 96% nucleotide sequence identity (Fig. S1D). The GHVd RNA-specific contigs were also assembled by Velvet and Vcake algorithms (Fig. 1, Lower Right), but were much shorter than those identified by PFOR. Reverse transcription and PCR (RT-PCR) verified the presence of the GHVd RNA in the original “Pinot noir” grapevine in Torino, Italy (Fig. 3B). Nucleotide sequencing of the RT-PCR product revealed that GHVd RNA was 375 nt long and was 98% identical in sequence to those predicted by PFOR (Fig. S1D). Northern blot hybridizations also detected accumulation of GHVd RNA in both positive and negative polarities as a single dominant band in the grapevine sample (Fig. 3 C and D).
Fig. 3.
Predicted secondary structure and RT-PCR and Northern blot detection of the 375-nt circular GHVd RNA. (A) The minimal free-energy secondary structure of GHVd RNA designated as the plus strand. The red nucleotides represent those 13 residues in the plus-strand GHVd RNA that are conserved in all hammerhead ribozyme structures with the cleavage site indicated by a red arrowhead. The blue nucleotides represent those 13 residues conserved in the minus-strand ribozyme, with a blue arrowhead indicating the cleavage site. Positions of the plus- (F) and minus (R)-strand primers used in RT-PCR and cloning are indicated. (B–D) Detection of GHVd RNA from the tendril of the grapevine by RT-PCR (B) and in different tissues of the grapevine by Northern blotting for the plus- (C) and minus-strand (D).
We found that full-length cDNA to GHVd RNA was amplified by RT-PCR, using each of the three pairs of primers targeting distinct positions to the GHVd RNA (Fig. 3A), demonstrating a circular nature of GHVd RNA (Fig. 3B). However, no amplification product was detected when PCR was carried out without an RT step and the GHVd RNA shared no sequence similarity to the grapevine genome sequence (25, 26). These findings together indicate that the circular GHVd RNA of both polarities was exogenous in nature and was not encoded by the grapevine genome.
The nucleotide sequence of the identified GHVd RNA exhibited no significant similarity to the entries in GenBank by BLASTN. However, we found that the minimal free-energy secondary structure predicted for GHVd RNA (Fig. 3A) displayed remarkable similarity to that of PLMVd (2, 27, 28). The secondary structure of PLMVd contains a long branched region stabilized by a kissing-loop pseudoknot and an extensively base-paired arm that encodes two self-cleavable hammerhead ribozymes active in the plus and minus strands, respectively (2, 27, 28). Similarly, the predicted structure of GHVd RNA (Fig. 3A) included an extensively base-paired arm and a highly branched region with a potential kissing-loop interaction (green dashed lines) similar to those found in PLMVd and Chrysanthemum chlorotic mottle viroid (CChMVd) (2, 28).
We noted that the bottom strand of the extensively base-paired arm of the secondary structure predicted for GHVd RNA contained the 13 nt conserved strictly in hammerhead ribozymes (Fig. 3A, nucleotides in red). Importantly, the region encompassing nucleotides 368–47 for the plus-strand GHVd RNA could be folded into a hammerhead structure (Fig. 4A, Left). As in the secondary structure of PLMVd, the top strand of the GHVd arm in the complementary sense could also be folded into a hammerhead structure (Fig. 4A, Center). Despite the lack of detectable similarity of GHVd RNA to known viroids at the primary nucleotide sequence level, the hammerhead structures of both the plus and the minus strands of GHVd RNA were highly similar to those encoded by members of viroids from the Avsunviroidae family (2, 27, 28). Both hammerheads of GHVd RNA contained stems I and II closed by a short loop, a stable stem III, and the 13 nucleotides conserved strictly in hammerhead ribozymes (Fig. 4A, Right) (2, 27, 28).
Fig. 4.
Predicted structures and in vitro self-cleavage of ribozymes encoded by GHVd RNA. (A) The hammerhead structures predicted for the plus- and minus-strand GHVd RNA and the consensus structure for GHVd ribozyme. The conserved 13 residues are boxed with the cleavage site indicated by an arrow. The indicated nucleotide positions correspond to those in the plus strand of GHVd RNA. (B) In vitro self-cleavage analysis of the plus- (Left) and minus (Right)-strand GHVd RNA transcripts from pGHVd and its derivatives. Transcripts transcribed in vitro from wild-type pGHVd (lanes 1 and 6), pGHVd-ΔAA (lanes 2 and 7), pGHVd-CU (lanes 3 and 8), pGHVd-ΔUU (lanes 4 and 9), and pGHVd-AG (lanes 5 and 10) were fractionated and stained with ethidium bromide. The sizes of the full-length (FL) plus- and minus-strand GHVd RNA transcripts with some linker sequence from the vector and their 5′ and 3′ cleavage fragments (5′F and 3′F) are indicated.
The Hammerhead Ribozymes of GHVd RNA Are Enzymatically Active.
The rolling-circle replication model for the hammerhead viroids from the Avsunviroidae family predicts production of unit-length viroid RNA in both polarities by autocleavages of multimeric plus- and minus-strand viroid transcripts via the hammerhead ribozyme encoded in each strand (2, 27, 29). To assay for the activity of the predicted GHVd ribozymes, the full-length cDNA of GHVd RNA linearized at nucleotide position 359 was cloned between T7 and SP6 phage promoters in the plasmid pGEM-T. The same plasmid DNA (pGHVd) was used as the template for the in vitro run-off transcription by T7 and SP6 phage RNA polymerase following digestion with restriction enzyme NotI (T7) or NcoI, yielding the full-length (FL) plus- and minus-strand GHVd transcripts, respectively (Fig. 4B). We found that in vitro transcripts of both the plus- and minus-strand GHVd autocleaved during transcription to yield the 5′ and 3′ cleavage products, which were detected in 5% denaturing polyacrylamide gel electrophoresis (Fig. 4B, lanes 1 and 6). These results indicate that both the plus- and the minus-strand GHVd RNAs encode a self-cleavable ribozyme activity.
The structural conservation found in GHVd ribozymes allowed us to predict the autocleavage sites of the plus and the minus strand of GLVd RNA (Fig. 4A). The predicted cleavage sites were consistent with the lengths of the 5′ and 3′ cleavage products (Fig. 4B, lanes 1 and 6) and were experimentally confirmed by sequencing the 3′ cleavage products obtained by rapid amplification of 5′-cDNA ends (5′-RACE)-PCR.
Four derivatives of pGHVd were constructed to either delete the AA doublet or change GA to CU in the conserved GAAAAC sequence of either plus- or minus-strand hammerhead ribozyme (Fig. 4A). As shown in Fig. 4B, the plus-strand GHVd RNA transcripts synthesized by T7 polymerase that contained either the deletion (Fig. 4B, lane 2) or the substitution (Fig. 4B, lane 3) in the hammerhead domain failed to autocleave. However, the minus-strand transcripts synthesized by SP6 polymerase from the same mutant plasmids autocleaved as efficiently as the wild-type minus-strand GHVd RNA transcripts (Fig. 4B, compare lane 6 with lanes 7 and 8). Similarly, autocleavages were not detected for the minus-strand transcripts that contained either the AA deletion or the CU substitution in the hammerhead sequence (Fig. 4B, lanes 9 and 10), but the plus-strand transcripts from the same mutant plasmids both autocleaved efficiently (Fig. 4B, lanes 4 and 5). These results therefore illustrate that self-cleavage of plus- and minus-strand GHVd RNA transcripts is mediated by the predicted hammerhead ribozymes and not altered by sequences outside the hammerhead domain.
Detection of the Hammerhead Viroid-Like RNA in California Grapevines.
We next compared the accumulation and profile of small RNAs derived from GHVd RNA with those of HSVd and GYSVd in the same tendril, leaf, inflorescence, and berry tissues of the grapevine sequenced previously (20). Similar to HSVd and GYSVd small RNAs (20), we found that small RNAs derived from GHVd RNA were the most abundant in the tendrils and accumulated to the lowest level in the leaves (Fig. S2A). However, GHVd RNA-specific small RNAs were hardly detectable in the inflorescence, where HSVd and GYSVd small RNAs accumulated to high levels. As found for HSVd and GYSVd sRNAs (20), GHVd RNA-specific sRNAs from different size families were all divided approximately equally into the plus and minus strands and the most dominant small RNA species was the 21-nt class (Fig. S2B). However, few of the GHVd RNA-specific small RNAs were 24 nt long (Fig. S2B) whereas a notable proportion (20% and 24%) of HSVd and GYSVd sRNAs belonged to the 24-nt class (20).
Presence of GHVd RNA in the berry and inflorescent tissues of the Italian grapevine was verified by Northern blot hybridization (Fig. 3 C and D). We also detected GHVd RNA in tendrils of Pinot noir grapevine clone ENTAV115 in California by RT-PCR (Fig. S3). The GHVd RNA sequences from Italian and California grapevines exhibited ∼99% sequence identities (Fig. S1D). However, our preliminary studies indicated that transcripts corresponding to either polarity of GHVd RNA transcribed from a plasmid containing a dimeric cDNA to GHVd RNA were not infectious by slash inoculation to grapevine cuttings with a razor blade.
Discussion
In this work, we established a homology-independent approach for the discovery of one class of pathogens by deep sequencing, viroids. Our approach involves analyzing the sequences of total small RNAs sequenced from diseased tissues with a unique computational algorithm referred as PFOR. We showed that PLMVd from the Avsunviroidae and HSVd and GYSVd from the Pospiviroidae were readily identified and their full-length RNA sequences assembled by PFOR from the total small RNA libraries of the infected plants sequenced by the Illumina platform. PFOR analysis of a published Italian grapevine small RNA library further identified a previously uncharacterized viroid-like circular RNA that shares no statistically significant nucleotide sequence homology with any known viroid or sequence entry in GenBank. Notably, the identified GHVd RNA encoded an active hammerhead ribozyme in both the plus and the minus strands and was also detected in the Pinot noir variety of grapevine in California.
Deep sequencing of the abundant viroid-derived siRNAs from the infected plants facilitates the discovery of viroids, which generally accumulate to low levels during infection. Unlike PFOR results, however, we found that the available graph- and overlapping-based assembling algorithms failed to assemble the overlapping viroid-derived sRNAs into full-length viroid genomes (Fig. 1). Viroids exhibit extremely high mutation rates and the population of the viroid sequences is highly heterogeneous in an infected plant (2, 3, 23, 30). The available genome assembling programs developed for short reads such as Velvet and Vcake (24) incorporate the dominant nucleotide into a consensus sequence and discard those reads containing a nonconsensus nucleotide. Thus, the heterogeneous nature of the viroid-derived sRNAs may prevent the progressive assembly of short contigs into the full-length viroid genome by either Velvet or Vcake. Furthermore, the short contigs assembled by Velvet and Vcake are not as informative for viroid discovery as virus discovery (8, 9, 31). Unlike the noncoding viroids, use of the peptide sequences in silico translated from the contigs in database searches often leads to the identification of viruses only distantly related to a known virus. In contrast to current genome assembly programs such as Velvet and Vcake, PFOR removes nonoverlapping small RNAs and overlapping TSRs and treats each remaining overlapping ISR equally in the final genome assembly regardless of its copy number. This strategy efficiently solved the conflict caused by viroid population heterogeneity and sequencing errors and readily allowed the full genome assembly from viroid-specific siRNAs produced by the host RNA silencing machinery. As a result, PFOR with different k-mer values may retain different sets of overlapping siRNAs derived from a heterogeneous viroid population for the final assembly and the predicted molecules may not always correspond to the dominant viroid variant and must be independently verified by RT-PCR and sequencing. In our cases, we found that the longest RNA sequence assembled by PFOR differed by 2–8% from the dominant viroid sequence in the infected host.
Identification of viroids by PFOR provides important clues about the precursor for the viroid-derived sRNAs. Asymmetric and symmetric rolling-circle replication models of viroids in the Pospiviroidae and Avsunviroidae families implicate a monomer circular viroid RNA in one and two polarities, respectively, but both predict production of plus and minus multimeric head-to-tail replicative intermediates (1–3). It has been hypothesized that the highly base-paired rod-like structures of the viroid circular RNAs have structural similarities to primary and/or precursor microRNAs (miRNAs) and serve as the substrate of Dicer, suggesting a miRNA origin for viroid sRNAs in the biogenesis (2, 3, 20). In principle, Dicer processing of such a circular RNA precursor may produce true ISRs. However, the parameters used for viroid identification and assembly by PFOR require Dicer cleavages at most nucleotide positions to generate a minimal set of small RNAs that overlap each other by ≥17 nucleotides (k-mer set for each run). This pattern of Dicer cleavages has not been observed for the stem-loop precursors of miRNAs by deep sequencing and instead has often been associated with long dsRNA substrates such as those produced during replication of a linear viral RNA genome (8, 9). Therefore, we propose that viroid sRNAs are siRNAs processed from long dsRNA precursors formed between the complementary direct-repeat viroid RNAs produced during replication by DCL2, DCL3, and DCL4, all of which are known to produce endogenous and/or exogenous siRNAs (12, 32). One reason why viroids from the Pospiviroidae induce production of a more abundant 24-nt class of siRNAs than those from the Avsunviroidae may be that the former replicates in the nucleus and DCL3 has access to the dsRNA replicative intermediates (12, 32).
We noted that only the replicating circular RNA molecules of an exogenous origin were identified by PFOR analysis of the peach and grapevine libraries. In contrast, the endogenous head-to-tail repeat elements such as the 178-bp centromeric satellite repeat, known to be targeted for DCL3-dependent siRNA biogenesis (12, 32, 33), were undetectable, suggesting that PFOR identification of these nonreplicating repeat elements may require deeper coverage of small RNA sequencing. In this regard, it should be pointed out that a viroid will not be detectable by PFOR if a minimal set of viroid-derived siRNAs that overlap each other by at least 17 nt are neither produced in the infected hosts nor obtained by deep sequencing. It is possible that PFOR can also identify those circular single-strand satellite RNAs that depend on the helper virus for their rolling-circle replication, some of which also encode the hammerhead ribozyme (34, 35). In fact, the circular RNA species identified in this study may be such a satellite RNA, which would explain why it alone was not infectious. In addition, the pattern of sequence variation among the nonconserved nucleotides in the GHVd-like RNA hammerhead ribozymes shows features associated with hammerhead ribozymes encoded by either viroids or circular satellite RNAs (27, 35, 36).
It is intriguing that viroids from two families are found in potato, horticultural species, and fruit trees, but not so far in many major crops and small fruit crops (1–3). Moreover, replication of viral RNA genomes in fruit flies, mosquitoes, and nematodes induces the biogenesis of viral siRNAs and/or piRNAs (9, 14, 37–39), which overlap in sequence and can be assembled for homology-dependent virus discovery (9). However, our analysis of these invertebrate small RNA libraries by PFOR did not reveal presence of viroids in the insects and nematodes maintained in laboratories. It is likely that PFOR will facilitate discovery of previously uncharacterized viroids in diverse plant and animal species in both agriculture and the environment.
Materials and Methods
Bioinformatics Analysis.
The small RNA library from a peach tree infected with PLMVd (accession no. GSM465746) and from a grapevine tree cultivar Pinot noir ENTAV115 (accession no. GSE18405) was downloaded from the Gene Expression Omnibus database of the NCBI (23, 40). The reference sequences of PLMVd (accession no. GQ499305), HSVd (accession no. GU825977), and GYSVd (accession no. X87909) were downloaded from the nucleotide database of the NCBI. The Velvet program was downloaded from the European Bioinformatics Institute. The Vcake1.0 program was downloaded from the web-based source code repository (htt://sourceforge.net/projects/vcake). Mapping of small RNAs was performed by Bowtie. Mapping of the assembled viroid contigs to viroid genomes was done with the BLASTN program using the standard parameters (contigs with ≥80% similarity and ≥80% coverage of contigs). Multiple sequence alignment was performed by CLUSTALW. The PFOR program was written in Perl language (Dataset S1) and installed on a Hewlett-Packard DL585 G7 server. A command line “perl PFOR.pl –f input_file –x 5” would initiate the PFOR analysis of an input small RNA library file with k-mer values from 17 to 22 (i.e., 17 + 5). Other analysis was performed by in-house scripts.
Plasmids, RT-PCR, and Northern and in Vitro Ribozyme Cleavage Analysis.
The sequence of GHVd RNA shown in Fig. 3A (see Fig. S4 for the sequence of GHVd RNA) was designated as the plus strand and the nucleotide (G) to the left of the ribozyme cleavage site as the first nucleotide of GHVd RNA. Primers F1, F2, and F3 corresponded to nucleotides 43–80, 352–15, and 293–328 of RNA whereas primers R1, R2, and R3 were complementary to nucleotides 8–45, 321–359, and 259–297 of GHVd, respectively. F1, R2, and R3 were each used to prime cDNA synthesis using total RNAs from the Italian grapevine tree and combined, respectively, with R1, F2, and F3 in RT-PCR to verify the circular nature of both the plus and the minus strands of GHVd RNA. Total RNA from grapevine cultivars Thompson Seedless and Pinot noir clone ENTAV115 obtained from California Grapevine Nursery, Inc. (St. Helena, CA) were also analyzed by RT-PCR. The full-length GHVd cDNA obtained by RT-PCR using F2 and R2 was cloned into pGEM-T (Promega) to generate pGHVd and multiple clones were sequenced as described in ref. 41. Four derivatives of pGHVd were constructed by site-directed mutagenesis and contained either substitution at nucleotides 38–39 (GA →CU) and 110–111 (UC →AG) or deletion of nucleotides 39–40 (AA) and 109–110 (UU), yielding pGHVd-CU, pGHVd-AG, pGHVd-ΔAA, and pGHVd-ΔUU, respectively.
pGHVd and its derivatives linearized by NotI or NcoI were used as the template for in vitro transcription of the full-length GHVd RNA in the plus and minus polarities, respectively. The conditions used allowed self-cleavages during transcription and the cleavage products were fractionated by 5% denaturing polyacrylamide gel electrophoresis and visualized by ethidium bromide staining as described in ref. 42. The 5′-terminal sequence of the 3`-cleavage products was determined by the 5′-RACE system (Invitrogen). Full-length plus- and minus-strand transcripts of GHVd RNA labeled with [α-32P]UTP were used as strand-specific riboprobes in Northern blot analysis to detect the plus and minus strands of GHVd RNA in the leaf, tendril, berry, and inflorescence tissues of the Italian grapevine as described in ref. 43. Plasmids containing a head-to-tail dimeric cDNA to GHVd RNA were also generated and plus- and minus-strand transcripts were used for slash inoculation of the grapevine cultivar Thompson Seedless with a razor blade (44).
Supplementary Material
Acknowledgments
This project was supported by National Institutes of Health Grants RC1 GM091896 and R01 GM094396, by grants from the California Citrus Research Board and University of California Discovery (to S.-W.D.), in part by the State Key Development Program for Basic Research of China Grant 2011CBA01103 (to Q.W.), and by the European Union-funded FP6 Integrated Project SIROCCO (Silencing RNAs: organisers and coordinators of complexity in eukaryotic organisms) LSHG-CT-2006-037900 (to J.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117815109/-/DCSupplemental.
References
- 1.Diener TO. Discovering viroids—a personal perspective. Nat Rev Microbiol. 2003;1:75–80. doi: 10.1038/nrmicro736. [DOI] [PubMed] [Google Scholar]
- 2.Flores R, Hernández C, Martínez de Alba AE, Daròs JA, Di Serio F. Viroids and viroid-host interactions. Annu Rev Phytopathol. 2005;43:117–139. doi: 10.1146/annurev.phyto.43.040204.140243. [DOI] [PubMed] [Google Scholar]
- 3.Ding B. The biology of viroid-host interactions. Annu Rev Phytopathol. 2009;47:105–131. doi: 10.1146/annurev-phyto-080508-081927. [DOI] [PubMed] [Google Scholar]
- 4.Owens RA. Curr Protoc Microbiol. 2008. Identification of viroids by gel electrophoresis. Chap 16, Unit 16G.1.1–16G.1.9. [DOI] [PubMed] [Google Scholar]
- 5.Schumacher J, Randles JW, Riesner D. A two-dimensional electrophoretic technique for the detection of circular viroids and virusoids. Anal Biochem. 1983;135:288–295. doi: 10.1016/0003-2697(83)90685-1. [DOI] [PubMed] [Google Scholar]
- 6.Lipkin WI. Microbe hunting. Microbiol Mol Biol Rev. 2010;74:363–377. doi: 10.1128/MMBR.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Victoria JG, Kapoor A, Dupuis K, Schnurr DP, Delwart EL. Rapid identification of known and new RNA viruses from animal tissues. PLoS Pathog. 2008;4:e1000163. doi: 10.1371/journal.ppat.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreuze JF, et al. Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: A generic method for diagnosis, discovery and sequencing of viruses. Virology. 2009;388:1–7. doi: 10.1016/j.virol.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q, et al. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc Natl Acad Sci USA. 2010;107:1606–1611. doi: 10.1073/pnas.0911353107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 11.Llave C. Virus-derived small interfering RNAs at the core of plant-virus interactions. Trends Plant Sci. 2010;15:701–707. doi: 10.1016/j.tplants.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 13.Scott JC, et al. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and -incompetent mosquito cells. PLoS Negl Trop Dis. 2010;4:e848. doi: 10.1371/journal.pntd.0000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aliyari R, et al. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papaefthimiou I, et al. Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 2001;29:2395–2400. doi: 10.1093/nar/29.11.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itaya A, Folimonov A, Matsuda Y, Nelson RS, Ding B. Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol Plant Microbe Interact. 2001;14:1332–1334. doi: 10.1094/MPMI.2001.14.11.1332. [DOI] [PubMed] [Google Scholar]
- 18.Martínez de Alba AE, Flores R, Hernández C. Two chloroplastic viroids induce the accumulation of small RNAs associated with posttranscriptional gene silencing. J Virol. 2002;76:13094–13096. doi: 10.1128/JVI.76.24.13094-13096.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markarian N, Li HW, Ding SW, Semancik JS. RNA silencing as related to viroid induced symptom expression. Arch Virol. 2004;149:397–406. doi: 10.1007/s00705-003-0215-5. [DOI] [PubMed] [Google Scholar]
- 20.Navarro B, et al. Deep sequencing of viroid-derived small RNAs from grapevine provides new insights on the role of RNA silencing in plant-viroid interaction. PLoS ONE. 2009;4:e7686. doi: 10.1371/journal.pone.0007686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Serio F, et al. Deep sequencing of the small RNAs derived from two symptomatic variants of a chloroplastic viroid: Implications for their genesis and for pathogenesis. PLoS ONE. 2009;4:e7539. doi: 10.1371/journal.pone.0007539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, et al. Accumulation of Potato spindle tuber viroid-specific small RNAs is accompanied by specific changes in gene expression in two tomato cultivars. Virology. 2011;413:72–83. doi: 10.1016/j.virol.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 23.Bolduc F, Hoareau C, St-Pierre P, Perreault JP. In-depth sequencing of the siRNAs associated with peach latent mosaic viroid infection. BMC Mol Biol. 2010;11:16. doi: 10.1186/1471-2199-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Velasco R, et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaillon O, et al. French-Italian Public Consortium for Grapevine Genome Characterization The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 27.Symons RH. Small catalytic RNAs. Annu Rev Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- 28.Gago S, De la Peña M, Flores R. A kissing-loop interaction in a hammerhead viroid RNA critical for its in vitro folding and in vivo viability. RNA. 2005;11:1073–1083. doi: 10.1261/rna.2230605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Branch AD, Robertson HD. A replication cycle for viroids and other small infectious RNA's. Science. 1984;223:450–455. doi: 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- 30.Gago S, Elena SF, Flores R, Sanjuán R. Extremely high mutation rate of a hammerhead viroid. Science. 2009;323:1308. doi: 10.1126/science.1169202. [DOI] [PubMed] [Google Scholar]
- 31.van Mierlo JT, van Cleef KW, van Rij RP. Small silencing RNAs: Piecing together a viral genome. Cell Host Microbe. 2010;7:87–89. doi: 10.1016/j.chom.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, Lee HR, Koo DH, Jiang J. Epigenetic modification of centromeric chromatin: Hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell. 2008;20:25–34. doi: 10.1105/tpc.107.057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayo MA, et al. Satellites. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy – Eighth Report of the International Committee on Taxonomy of Viruses. San Diego: Academic; 2005. pp. 1163–1169. [Google Scholar]
- 35.Symons RH, Randles JW. Encapsidated circular viroid-like satellite RNAs (virusoids) of plants. Curr Top Microbiol Immunol. 1999;239:81–105. doi: 10.1007/978-3-662-09796-0_5. [DOI] [PubMed] [Google Scholar]
- 36.Fadda Z, Daròs JA, Fagoaga C, Flores R, Duran-Vila N. Eggplant latent viroid, the candidate type species for a new genus within the family Avsunviroidae (hammerhead viroids) J Virol. 2003;77:6528–6532. doi: 10.1128/JVI.77.11.6528-6532.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flynt A, Liu N, Martin R, Lai EC. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc Natl Acad Sci USA. 2009;106:5270–5275. doi: 10.1073/pnas.0813412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myles KM, Wiley MR, Morazzani EM, Adelman ZN. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc Natl Acad Sci USA. 2008;105:19938–19943. doi: 10.1073/pnas.0803408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brackney DE, Beane JE, Ebel GD. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS Pathog. 2009;5:e1000502. doi: 10.1371/journal.ppat.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pantaleo V, et al. ) Identification of grapevine microRNAs and their targets using high throughput sequencing and degradome analysis. Plant J. 2010;62:960–976. doi: 10.1111/j.0960-7412.2010.04208.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang XB, et al. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell. 2011;23:1625–1638. doi: 10.1105/tpc.110.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hernández C, Flores R. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc Natl Acad Sci USA. 1992;89:3711–3715. doi: 10.1073/pnas.89.9.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daròs JA, Marcos JF, Hernández C, Flores R. Replication of avocado sunblotch viroid: Evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc Natl Acad Sci USA. 1994;91:12813–12817. doi: 10.1073/pnas.91.26.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ambrós S, Hernández C, Desvignes JC, Flores R. Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: Implications of the existence of constraints limiting the heterogeneity of viroid quasispecies. J Virol. 1998;72:7397–7406. doi: 10.1128/jvi.72.9.7397-7406.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.