Abstract
Immature thymocytes expressing autoreactive T-cell receptors (TCR) can adopt differing cell fates: clonal deletion by apoptosis or deviation into alternative lineages such as FoxP3+ regulatory T cells (Treg). We revisited the role of the transcription factor Nr4a1 (Nur77), an immediate-early response gene induced by TCR engagement. Nr4a1KO mice show clear quantitative defects in antigen-induced clonal deletion. The impact of the Nr4a1 deletion is not enhanced by deletion of the proapoptotic factor Bim. In addition, Nr4a1 curtails initial differentiation into the Treg lineage in TCR transgenic mice and in nontransgenic mice. Transcriptional profiling of Nr4a1KO thymocytes under selection conditions reveals that Nr4a1 activates the transcription of several targets, consistent with these diverse actions: (i) Nr4a1 partakes in the induction of Bim after TCR triggering; (ii) perhaps paradoxically, Nr4a1 positively controls several transcripts of the Treg signature, in particular Ikzf2 and Tnfrsf9; (iii) consistent with its prosurvival and metabolic role in the liver, Nr4a1 is also required for the induction by TCR of a coordinated set of enzymes of the glycolytic and Krebs cycle pathways, which we propose may antagonize Treg selection as does activation of mTOR/Akt. Thus, Nr4a1 appears to act as a balancing molecule in fate determination at a critical juncture of T-cell differentiation.
Keywords: glycolysis, immune tolerance, thymus, nuclear receptor
A hallmark of the adaptive immune system is its ability to respond to the infinite repertoire of potential pathogens to which it might someday be exposed, but the undirected process of antigen receptor rearrangement in thymocytes generates receptors that are reactive to self and hence are potentially dangerous. Fledgling T cells carrying a nascent T-cell receptor (TCR) must undergo a rigorous process of negative selection, during which cells expressing a self-reactive TCR are eliminated by induced apoptosis or deviated to alternative differentiation pathways in which self-reactivity is defused [NKT, CD8αα, or FoxP3+ regulatory T cells (Treg) (1)].
Triggering of mitochondrial apoptosis by signaling cascades downstream from the TCR is clearly established. Bim and Bax/Bak, proapoptotic members of the mitochondrial apoptosis pathway, are essential for efficient negative selection (2, 3), but this may not be the sole mechanism. Early experiments involving blockade of RNA or protein synthesis suggested that negative selection is both transcription and translation dependent (4). Indeed, the transcription factor Nr4a1 has been implicated in negative selection (5, 6). An orphan member of the nuclear receptor superfamily and the Nr4a1/Nr4a2/Nr4a3 subfamily, Nr4a1 is a transcriptional activator, but can also promote mitochondrial apoptosis. In both tumor cell lines and thymocytes undergoing negative selection, Nr4a1 undergoes phosphorylation-dependent export from the nucleus to the mitochondria, where it conformationally alters Bcl-2 to promote apoptosis (7–10). Nr4a1 is associated with apoptosis in a number of physiological and tumor systems (11–14) and induces apoptosis upon transgenic overexpression (5, 15). Paradoxically, however, it acts as a survival factor in other instances—it was originally described as a growth-factor–inducible gene—is overexpressed in several tumors, and can protect against ceramide or TNF-induced death (16, 17).
Nr4a1 is activated as part of the immediate-early response downstream of TCR signals (18), and can serve as an indicator for strength of TCR signals (19). Nr4a1 transcripts are differentially induced in double-positive (DP) and single-positive (SP) thymocytes (20) and in B6 vs. non-obese diabetic (NOD) thymocytes during negative selection, where lower levels of Nr4a1 mRNA correlate with the apparently impaired clonal deletion of NOD mice (21, 22). Nr4a1 transcripts also appear as part of the proapoptotic program in T lymphocytes during times of cellular stress such as steroid treatment or oxygen deprivation (12, 13). Winoto and colleagues first reported an association of Nr4a1 with negative selection (5): mice expressing a dominant-negative mutant of Nr4a1 in T cells showed a profound defect in thymocyte apoptosis. This hypothesis was undercut by the absence of a negative selection phenotype in a Nr4a1 knockout mouse (23). Redundancy within the Nr4a1 family may account for these disparate findings because Nr4a1, Nr4a2, and Nr4a3 have partially overlapping expression patterns and can share DNA-binding motifs within the promoter regions of target genes (24). To date, however, this compensatory activity has not been demonstrated in the context of thymocyte apoptosis, so the relative contributions of Nr4a1, Nor-1, and Nurr1 to the negative selection pathway remain undefined.
Clonal deletion is now appreciated to be closely intertwined with commitment to alternative lineages, which allow self-reactive thymocytes to escape apoptosis by adopting phenotypes in which TCR-mediated signals are mitigated (CD8αα cells) or that are more resistant to TCR-induced apoptosis (FoxP3+ Tregs, NKT cells). In this light, we revisited the fate of autoreactive T cells in relation to Nr4a1 with respect to clonal deletion and clonal deviation.
Results
Nr4a1 Affects Clonal Deletion of BDC2.5 and OTII Thymocytes.
We first re-explored the involvement of Nr4a1 in negative selection in the context of the BDC2.5 TCR transgenic model, in which our previous analyses had shown Nr4a1 to be differentially induced in the NOD and B6.H2g7 (B6g7) backgrounds in response to negative selection cues (22). This system also allows an analysis of apoptosis induction in response to graded signals, perhaps less stringent than the H-Y and AND systems used initially (23). Clonal deletion was examined in fetal thymic organ culture (FTOC), where embryonic day 15 (E15) thymi were cultured in vitro in the presence of a mimotope peptide [BDCmi (25)] that triggers clonal deletion of BDC2.5+ thymocytes at the immature CD4+CD8+ DP stage. We crossed the Nr4a1 KO allele onto BDC2.5/B6g7 mice and tested for clonal deletion after 3 d in FTOC with agonist BDCmi peptide. More BDC2.5 DP thymocytes survived in Nr4a1KO FTOC over a range of BDCmi doses (Fig. 1), although the protective effect of the knockout was overcome at high peptide doses. This was true whether DP cells were estimated as a proportion or as absolute numbers per FTOC (Fig. 1B). FTOCs from the Nr4a1KO background always behaved as intermediate between FTOCs on the B6g7 and NOD backgrounds.
Fig. 1.
Impaired clonal deletion of BDC2.5 TCR transgenic thymocytes in Nr4a1KO. E15.5 BDC2.5-tg FTOCs from B6g7 or B6g7.Nr4a1KO mice were cultured for 3 d to promote CD4+CD8+ differentiation and then incubated for 16 h with BDC mimotope peptide (BDCmi). (A) Representative cytometry plots after 16 h with 10 ng/mL BDCmi. Percentage of surviving DPs is indicated in upper right of each panel. (B) Differences in percentage (Upper) and absolute cell count (Lower) of DPs surviving after 16 h with titrated BDCmi; each point represents a single FTOC culture (representative experiment). (C) Compilation of five experiments comparing percentage of surviving DPs after 16 h FTOC with no or 3 ng/mL BDCmi; each point represents a single FTOC.
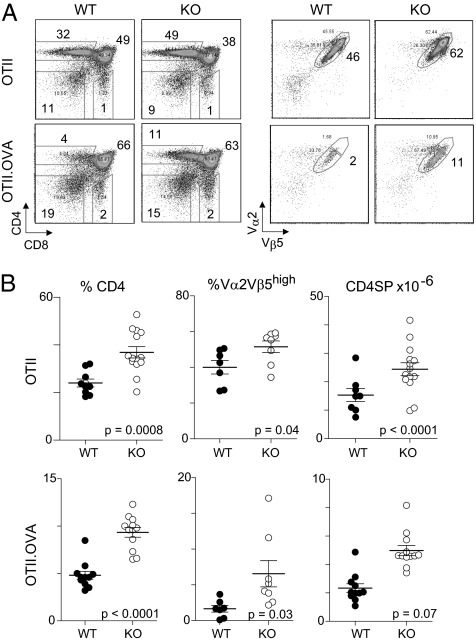
To determine whether the reduced negative selection of BDC2.5 Nr4a1KO DP thymocytes represents a general phenomenon, we crossed the Nr4a1KO allele to B6.OTII/RIP-mOVA double-transgenic mice. The OTII transgene encodes an MHC-II–restricted TCR (Vα2Vβ5) reactive to ovalbumin (OVA); the RIP-mOVA transgene encodes OVA under the dictates of the rat insulin promoter, which primarily directs expression to the pancreas but also causes low-level ectopic expression in medullary epithelial cells of the thymus (26, 27). Perhaps because RIP-mOVA is principally active in the medulla, it primarily results in a dearth of OTII CD4+CD8− SPs. Thymi from OTII and OTII/RIP-mOVA were analyzed at 6 wk of age (Fig. 2). As expected, in the presence of RIP-mOVA, there was a marked decrease in the frequency and numbers of CD4SPs (fourfold on average) with a shift in the expression of the Vα2Vβ5 clonotype, such that very few Vα2Vβ5hi cells remained in the presence of antigen. These negative selection effects were partially reversed by the Nr4a1 deficiency (a twofold recovery in numbers), with a partial restoration of the Vα2Vβ5hi fraction. A more modest yet significant effect of the Nr4a1 deficiency was also observed in the absence of RIP-mOVA, perhaps resulting from impairment of the deletion that may occur in the OTII thymus in response to endogenous superantigen or from weak cross-reactivity to Ab complexed with self-peptides. Thus, Nr4a1 makes a contribution to negative selection in response to antigen in SPs as well as in DPs. Notably, Nr4a1KO mice devoid of TCR transgenes showed no changes in proportion or numbers of DPs or SPs, suggesting that shifts in the window of TCR/peptide/MHC affinity of the polyclonal repertoire mask the effects on single specificities.
Fig. 2.
Nr4a1KO impairs clonal deletion of OTII TCR transgenic thymocytes. (A) Representative cytometry profiles of OTII TCR transgenic thymi on WT or Nr4a1KO backgrounds, without (Upper panels) or with (Lower panels) the RIP-mOVA transgene. (Left panels) CD4 vs. CD8. (Right panels) Anti-clonotypic antibodies. The gates drawn outline the OTIIhi and OTIIlow populations, and numbers refer to OTIIhi cells. (B) Compilation of seven experiments comparing percentage of CD4SP (Left panels), Vα2Vβ5hi of gated CD4SP (Center panels), and absolute cell count of CD4SP (Right panels).
Relationship Between Nr4a1 and Bim in Negative Selection.
These results indicate that, in two different TCR models of negative selection, one can detect a clear impact of Nr4a1, results that contrast with a previous report (23). Thus, it became of interest to analyze the relationship between Nr4a1 and Bim because the latter is considered the dominant player in apoptosis induced by negative selection. We performed an intercross between the Nr4a1 and Bim knockouts; if these factors have an independent impact on apoptosis, one would expect additive or synergistic effects; if they belong to the same pathway, one might expect no additional effect beyond that of each individual mutation. Both mutations were intercrossed together with OTII and OTII/RIP-mOVA transgenes (Fig. 3; for unknown reasons, the single BimKO could not be obtained together with OTII/RIP-mOVA in these complex intercrosses). Unexpectedly, there was little impact of the Bim mutation beyond the increased cell yield already achieved by the Nr4a1 deletion alone [which was of a magnitude similar to the reported effects of Bim in BimKO/OTII.RIP-mOVA mice (2)]; this was true with or without the strong selecting pressure from RIP-mOVA. This absence of additional effect from the Bim deficiency may mean that Bim and Nr4a1 act in sequence or through distinct and noncomplementing pathways (with Nr4a1 being rate limiting) and/or that Bim plays a relatively minor role compared with Nr4a1.
Fig. 3.
No additional clonal deletion defect in Bim/Nr4a1 double KO. (A) Representative CD4xCD8 plots of OTII/B6 thymi from 6-wk-old male mice of WT (Left panels), Nr4a1KO (Center panels), and Nr4a1/Bim double KO (Right panels) genotypes, transgenic for OTII (Upper panels) or OTII/RIP-mOVA (Lower panels). (B) Compilation of three experiments comparing the percentage of CD4SP.
Nr4a1 Influences Selection in the FoxP3+ Treg Lineage.
Several models of negative selection have linked clonal deletion of autoreactive thymocytes with the genesis of thymus-derived Treg cells. Engagement by agonist ligands favors the selection of Treg cells either by induced differentiation (28–31) or because FoxP3+ cells are more resistant to antigen-induced apoptosis (32–34). Given the differences in the efficiency of clonal deletion in Nr4a1KO mice, we wondered whether Nr4a1KO might affect selection of FoxP3+ cells as well. In nontransgenic thymi, the absence of Nr4a1 led to a twofold increase in the proportion and absolute numbers of FoxP3+ thymocytes (Fig. 4 A and B; similar results were obtained in the NOD background). These overrepresented Treg cells had normal levels of CD25 and FoxP3 (Fig. 4A). We also analyzed Nr4a1KO thymi with OTII and OTII/RIP-mOVA transgenes (Fig. 4 C and D). As in other TCR/Ag systems, exposure to antigen boosted the proportion and absolute numbers of FoxP3+ cells among wild-type (WT) CD4+ SPs, and the absence of Nr4a1 further increased this number in OTII/RIP-mOVA/Nr4a1KO mice (Fig. 4D, Lower Right); the proportion of FoxP3+ cells among CD4+ SPs decreased slightly (Fig. 4D, Upper Right), owing to the large number of conventional cells spared from deletion. This increase in Treg numbers was also observed in the absence of RIP-mOVA antigen.
Fig. 4.
Fop3+ thymic Tregs populations increased in Nr4a1KO. (A) Representative plots showing increased FoxP3+ population in gated CD4SPs from 6-wk-old WT and Nr4a1KO mice. (B) Compilation of three experiments comparing percentage (Left) and number (Right) FoxP3+ cells within gated CD4SPs. (C) Representative FoxP3xCD25 plot of gated CD4SPs from OTII or OTII.RIP-mOVA on WT and Nr4a1KO background. (D) Compilation of eight experiments comparing FoxP3+ frequency within CD4SPs and absolute counts in WT and Nr4a1KO. (E) Representative CD25xFoxP3 staining of CD4SPs, with FoxP3-CD25+ “Treg precursor” population frequencies indicated (Left). (Right) Compilation from individual mice. (F) FC/FC plot depicting the ratio of expression in Treg vs. Tconv CD4SPs from WT (x axis) and Nr4a1KO (y axis) thymi. Treg up (red) and down (blue) signatures are highlighted. (G) Representative suppression assay at varying Treg:Teffector ratios, comparing Tregs from WT or Nr4a1KO.
Thus, Nr4a1 influences the efficiency of selection and the size of the thymic Treg pool. In theory, the increase observed in the absence of Nr4a1 could be achieved by an enhanced rate of differentiation or by increased survival or retention. Hsieh and colleagues have described a two-step model of Treg differentiation in which immature thymocytes destined to the Treg lineage first appear as CD25hiFoxP3−, a transient state during which signals via the IL2R-STAT5 axis finally induce FoxP3 and establish the stable Treg phenotype (35). The results depicted in Fig. 4E show that the absence of Nr4a1 led to an increase in the precursor population commensurate with that of the FoxP3+ Treg pool. These data confirmed the notion that Nr4a1 dampens selection into the Treg lineage and does so at the earliest stage when TCR signals lead to the CD25hi phenotype.
FoxP3+ cells can also be generated by “conversion” from naive CD4+ T cells by stimulation in culture in the presence of IL-2 and TGFβ (36). In such cultures, FoxP3 was induced as robustly in Nr4a1-deficient cells as in WT counterparts (Fig. S1). Furthermore, Nr4a1KO mice showed normal numbers and proportions of peripheral Treg populations, suggesting that Nr4a1 is not involved in peripheral Treg differentiation or in niche size control.
Do the FoxP3+ cells elicited in higher numbers by Nr4a1 deficiency have the normal phenotype of Treg cells? FoxP3+ Treg cells have a gene expression signature that distinguishes them from conventional CD4+ cells (37); Tregs from Nr4a1KO thymi appeared very similar to their WT counterparts in this respect, with the expected distribution of signature genes on expression profiles (Fig. 4F). In addition, Treg cells from Nr4a1KO mice were as effective as WT in the classic in vitro suppression assay (Fig. 4G). Thus, within the informativity of these assays, the enhanced clonal deviation to the Treg lineage in Nr4a1KO mice generates phenotypically and functionally normal Treg cells.
Nr4a1 Transcriptional Footprint in Developing Thymocytes.
We turned to transcriptional profiling for clues about Nr4a1's mechanism of action in both apoptosis and Treg differentiation. Profiles from WT or Nr4a1-deficient thymocytes were compared primarily in two different settings: (i) DPs from age/sex-matched WT or Nr4a1KO stimulated in vitro for 3 h with plate-bound anti-TCRβ/CD28/CD2, conditions used by others to mimic negative selection and under which Nr4a1 and Bim are rapidly induced (20); this short response time should bring forth Nr4a1's direct transcriptional footprint; (ii) DP and CD4+CD25−Vα2hiVβ5hi SP thymocytes from adult OTII and OTII/RIP-mOVA transgenic mice for a footprint of Nr4a1 in vivo under steady-state conditions of antigen-induced clonal deletion. The latter in vivo datasets should show, under more physiological conditions, the more integrated (direct and indirect) influence of Nr4a1 in cells that survive apoptosis.
We focused first on the Nr4a1 target genes differentially induced in DP thymocytes by short-term stimulation. In unstimulated cells, the absence of Nr4a1 had very little effect (Fig. 5A, Left, in the same range as the experimental background estimated by Monte Carlo randomization). However, a clear difference between WT and Nr4a1KO emerged after 3 h (Fig. 5A, Right). These Nr4a1-dependent transcripts (listed in Table S1) were not affected by the Nr4a1 deficiency before stimulation. They included genes belonging to several different pathways, but the most notable candidates in this context were Bcl2l11 (Bim) and the Treg signature genes Tnfrsf9 (4.1BB) and Ikzf2 (Helios).
Fig. 5.
Nr4a1-dependent transcripts identified by microarray profiling. (A) FoldChange vs. expression plots for WT vs. Nr4a1KO in resting DP thymocytes (Left) or DP thymocytes stimulated in vitro for 3 h with plate-bound anti-CD2/CD28/TCRβ. Transcripts highlighted indicate ratio of expression >2× (orange) or <0.5× (aqua) in WT/Nr4a1KO. (B) Volcano plots comparing WT and KO thymocytes (FoldChange: x axis; t test P value: y axis) for DPs stimulated 3 h with plate-bound anti-CD2/CD28/TCRβ (Left), DPs from OTII.OVA (Center), or CD4SPs from OTII.OVA (Right). (C) FC/FC plot showing the ratio of expression in CD4 SPs from OTII vs. OTII.OVA thymi from WT or Nr4a1KO mice. Treg up (red) and down (blue) signatures are highlighted. (D) FC/FC plot depicting the WT vs. KO ratio of expression in OTII.OVA CD4SPs (x axis) or in 3-h anti-TCR–stimulated DPs (y axis). Highlights as in C.
We analyzed more directly the effect of Nr4a1 on the expression of Bim in the three conditions analyzed (red highlight in Fig. 5B). Bim was represented in all contexts, particularly in DPs, suggesting that Nr4a1 does have a transcriptional impact on Bim. It should be pointed out, however, that the impact of Nr4a1 is only partial (1.5- to 2-fold) relative to the strong induction of Bim by TCR ligands (8.7-fold in in vitro DPs, 4.7-fold in OTII SPs, 2.3-fold in OTII DPs).
We also noted, among the genes most clearly influenced by Nr4a1, the glycolytic enzyme Enolase 3, which was reported to be directly regulated by Nr4a1 in an analysis of metabolic control in the liver (38). Enolase 3 catalyzes an important step in the glycolytic pathway (39), so we asked whether Eno3 was part of a broader set of metabolic-control transcripts influenced by Nr4a1. Indeed, transcripts encoding glycolytic enzymes were essentially all under-represented in Nr4a1-deficient cells, whether in short-term activated DPs or in long-term antigen-exposed OTII DPs and SPs (Fig. 5B and Table S2). Nr4a1 similarly impacted transcripts encoding enzymes of the Krebs cycle (blue). Thus, Nr4a1 effects a global activation of the energy-producing metabolic pathways in thymocytes, which can have a profound effect on cell survival and proliferation (40), consistent with effects in skeletal muscle and adipose tissue in addition to liver (reviewed in ref. 41).
The absence of Nr4a1 leads to an enhanced efficiency of Treg selection. We thus asked whether we could distinguish, among Nr4a1-controlled genes, transcripts that might reflect this effect on selection (e.g., a repression of key controlling genes in the Treg signature. We examined the relative expression of Treg signature genes in the CD4SP OTII samples, which were generated from CD25− thymocytes, thereby excluding most thymic Tregs and Treg precursor cells. The effect of Nr4a1 on the set of genes induced by the presence of OVA in WT and KO thymi is displayed on the FoldChange/FoldChange (FC/FC) plots of Fig. 5C. Transcripts along the diagonal represent genes equally up- or down-regulated by OVA in WT and Nr4a1KO, whereas transcripts biased toward the horizontal are those whose induction requires Nr4a1 (listed in Table S3). A general transcriptional activation and repression of “Treg up” and “Treg down” signature genes were induced by OVA, likely because some bona fide FoxP3+ Treg cells are CD25− and thus partake in this bias. A number of transcripts of the Treg up signature were less induced (Ikzf2 and Tnfrsf9) or even uninduced (Itgae, Cst7, Elk3) in the absence of Nr4a1. On the other hand, there was no influence of Nr4a1 on the Treg down signature. Thus, a marked effect of Nr4a1 was observed, but an unexpected one: instead of mediating a generalized repression, Nr4a1 turned out to activate a subset of Treg signature transcripts.
The FC/FC plot of Fig. 5D addresses the relation between Nr4a1's immediate target genes relative to its longer-term impact in CD4SPs by representing the variation between WT and Nr4a1KO cells for both antigen-stimulated OTII CD4SPs and in vitro-stimulated DPs. Here, transcripts lining the diagonal would reflect target genes that Nurr77 affects immediately and persistently, whereas transcripts along the x axis reflect delayed effects (because they are active only in SPs, because they are Nr4a1-dependent but late responders, or because they are indirectly affected). A few transcripts showed the constant imprint of Nr4a1 (notably Ikzf2 and Tnfrsf9, previously noted), but the plot also delineated a set of Treg transcripts uniquely influenced in the steady-state population (Nd5e, Pdcd1, Itgae). As expected, no Nr4a1-related differences were observed in OTII SPs from OVA-negative mice. Nr4a1's footprint seemed mainly inductive, with little or no repressive signature, suggesting that it comes into play as a positive transcriptional inducer, of both the proapoptotic Bim and, paradoxically, some Treg signature genes.
Discussion
We have observed an important role for the transcription factor Nr4a1 in modulating two key aspects of tolerance to self during differentiation of immature T cells in the thymus: clonal deletion of self-reactive thymocytes and commitment to the Treg lineage. Nr4a1 expression in immature thymocytes, at a stage where self-recognition can elicit either death by clonal deletion or deviation into the Treg lineage, is at the crux of this important cell-lineage determination. The early literature on Nr4a1 and T cells hinges on its molecular description as a transcription factor, with defined transcriptional activation domains and DNA-binding motif, and on its functional description as a proapoptotic factor, whose overexpression in cell lines and in transgenic mice promotes T-cell apoptosis. In subsequent reviews, this description has been abbreviated to “proapoptotic transcription factor” Nr4a1. However, a growing literature also describes Nr4a1 as a survival factor and activator of metabolic pathways in a variety of cell types (12, 41, 42), and one should consider both facets in attempting to integrate Nr4a1's role in T-cell differentiation.
Clonal deletion results, obtained in two TCR transgenic mouse systems, are conceptually concordant with the effect of dominant-negative Nr4a1 transgenic mice, which showed defects in negative selection of TCR transgenic thymocytes (5, 6). On the other hand, they disagree with a previous report that found no detectable phenotype in Nr4a1KO thymus (23). Why did this original study not find any defect in negative selection? Different TCR transgenes were used (H-Y and AND), both of which lead to strong negative selection at the DP stage, whereas the OT-II and BDC2.5 systems show only partial and/or late negative selection. The BDC2.5 TCR stems from a diabetogenic T-cell clone that had, by definition, escaped clonal deletion in NOD mice; the OT-II TCR responds to OVA peptides presented by medullary epithelial cells in RIP-mOVA transgenic mice with a late (medullary SP stage only) and incomplete negative selection. Whatever the explanation, it is clear that Nr4a1 can have a dominant impact, which is not redundant with its Nor-1 and Nurr1 cousins.
How does Nr4a1 effect clonal deletion? An open question is whether Nr4a1 induces deletion through transcriptional means by inducing proapoptotic molecules or through its direct interactions with Bcl-2 (7–10). Initially thought to be transcriptional (15, 24), the phenotype of the early Nr4a1 mutants that led to this conclusion may have reflected perturbed nuclear export signals contained within the transcriptional activation domain (8). We find that Nr4a1 is necessary for the full induction of Bim transcripts in all three conditions tested, including a very early time point, which suggests direct transcriptional regulation (consistent with the double-KO phenotype). However, the share of Bim induction that can be imparted to Nr4a1 matches only a fraction of the Bim induction during negative selection. Nr4a1 cannot be the sole controller of Bim. No other clear proapoptotic candidate was readily identified among Nr4a1-controlled genes.
In addition to the negative-selection defect, we find that Nr4a1 expression negatively impacts differentiation of thymic Treg cells. There are no apparent differences in Treg phenotypes between WT and Nr4a1KO mice, suggesting that Nr4a1's role in Tregs is a matter of differentiation and not of function. Indeed, peripheral homeostasic mechanisms restore the peripheral Treg population to normal numbers, as Treg over-representation in Nr4a1KO is seen only in the thymus, not in secondary lymphoid organs. Mechanisms that promote Treg differentiation are incompletely understood, but likely are controlled by the balance of signals along the NF-κB vs. Akt-mTOR pathways. We did test the hypothesis that Akt/mTOR might act directly through Nr4a1, which proved false as the inhibitory effects of activated Akt were still observed in Nr4a1KO cells.
Importantly and quite paradoxically, Nr4a1 had a positive effect on a fraction of the Treg signature (but not on FoxP3 itself, consistent with ref. 43). Thus, Nr4a1 biases Treg transcriptional differentiation even while restricting their numbers. One might speculate that Nr4a1, by promoting survival, boosts negative feedback mechanisms that control the efficacy of Treg selection and/or thymic Treg niche size. Or perhaps, in a cell-autonomous fashion, the transcripts that it induces are actually negative feedback controls on Tregs themselves.
In addition, Sekiya et al. (43) recently reported that transduction of Nr4a2 induces FoxP3 in vitro; Nr4a1 and Nr4a3 did not, the difference mapping to the N-terminal transactivation domain. In Nr4a2-deficient mice, thymic selection of Treg cells was normal, but there was reduced FoxP3 induction in response to IL-2/TGFβ and differential Treg cell stability (43). Because Nr4a family members have conserved DNA-binding motifs and can heterodimerize, it is possible that some of the present results reflect competition or cooperation of Nr4a1 with Nr4a2. We note, however, that Nurr1 is expressed at low levels in both Treg and conventional thymocyte populations relative to Nr4a1 and Nr4a3 (Fig. S2).
Within this context, can the Nr4a1-regulated transcripts identified here be interpreted for their potential involvement at the clonal deletion vs. clonal deviation branch point? Genes of the costimulatory family, including PD-1 and 4.1BB, are differentially expressed and relevant to both fates. All of these are differentially expressed in Treg cells, and several studies have associated PD-1 with negative selection (44). A previous study also identified both Pdcd1 and Ctla4 as genes induced by Nr4a1 upon transgenic over-expression in thymocytes (15). One might speculate that, by dampening the signals associated with the TCR (Ctla4) or by modulating its downstream footprint (PD-1, 4.1BB), the costimulatory molecules controlled by Nr4a1 may affect the balance of survival and lineage commitment.
In addition, the transcriptional profiles clearly denote a prosurvival effect of Nr4a1: the whole group of glycolytic and TCA enzymes behave as early response genes and Nr4a1 participates in their activation, thus boosting the whole cascade of energy production in the cell. This is most obvious for Eno3, which Kasler et al. identified as a Nr4a1-transcriptional target in D011.10 cells overexpressing Nr4a1 (45) and in analyses of hepatic glucose metabolism (38). Nr4a1 binds directly to a response element in the Eno3 promoter (38). Enhanced activation of metabolism in Nr4a1-expressing thymocytes may blunt differentiation into the Treg lineage, similar to that recently described in peripheral T cells in which transcriptional activation of glycolysis preferentially promotes Th17 rather than Treg differentiation via a HIF1α-dependent mechanism (46). In that study, as here, genetic inhibition of glycolysis increased Treg cell numbers. (We attempted to directly test the role of glycolysis in Treg differentiation by inhibition with 2-DG during the FTOC, but the results were confounded by toxicity of 2-DG on immature thymocytes). We note that activation of the Akt/mTOR axis, which boosts glucose metabolism, also results in lower Treg differentiation.
One might propose, then, that the induction of Nr4a1, through its different biochemical activities, forces decisions in relation to thymocyte fate: promoting clonal deletion via Nr4a1's proapoptotic activity at the mitochondrial or transcriptional levels and facilitating cell survival by boosting ATP generation, a condition that may promote commitment away from clonal deviation.
Materials and Methods
Mice.
C57BL/6.Nr4a1−/− [Nr4a1KO (23)] mice, a gift from J. Milbrandt (Washington University, St Louis, MO), were crossed with the transgenic lines BDC2.5/B6g7 (47), OT-II (26), and RIP-mOVA (27). C57BL/6.Bim−/− [BimKO (48)] mice were from The Jackson Laboratory. Mice were bred in an specific-pathogen-free barrier facility.
Antigen Exposure.
Fetal thymus lobes from E15.5 embryos were cultured as described (22). On culture day 3, BDC-specific mimotope peptide (BDCmi, peptides 1,040–1,063) (25) was added to culture media, and lobes were analyzed 16 h later. For in vitro thymocyte stimulation, single-cell thymocyte suspensions were incubated at 5 × 106 cells/mL for 3 h at 37 °C with plates precoated with anti-TCRβ (10 μg/mL), anti-CD2 (10 μg/mL), and anti-CD28 (50 μg/mL) as described (20). Cell sorting for microarray RNA preparation and hybridization to Affymetrix MoGene 1.0ST and RMA normalization were performed as described (49).
Supplementary Material
Acknowledgments
We thank K. Hattori, J. LaVecchio, G. Buruzula, and S. Davis for help with mice, cytometry, and computational analysis. This work was supported by a grant from the Juvenile Diabetes Research Foundation (4-2007-1057) and by Grant AI051530 from the National Institute of Allergy and Infectious Diseases (to C.B. and D.M.). M.S.F. was supported by a National Institutes of Health Training Grant (T32 DK7260) and the Harvard-MIT Medical Scientist Training Program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200090109/-/DCSupplemental.
References
- 1.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: Learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 2.Bouillet P, et al. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature. 2002;415:922–926. doi: 10.1038/415922a. [DOI] [PubMed] [Google Scholar]
- 3.Rathmell JC, Lindsten T, Zong WX, Cinalli RM, Thompson CB. Deficiency in Bak and Bax perturbs thymic selection and lymphoid homeostasis. Nat Immunol. 2002;3:932–939. doi: 10.1038/ni834. [DOI] [PubMed] [Google Scholar]
- 4.D'Adamio L, Clayton LK, Awad KM, Reinherz EL. Negative selection of thymocytes. A novel polymerase chain reaction-based molecular analysis detects requirements for macromolecular synthesis. J Immunol. 1992;149:3550–3553. [PubMed] [Google Scholar]
- 5.Calnan BJ, Szychowski S, Chan FK, Cado D, Winoto A. A role for the orphan steroid receptor Nur77 in apoptosis accompanying antigen-induced negative selection. Immunity. 1995;3:273–282. doi: 10.1016/1074-7613(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 6.Zhou T, et al. Inhibition of Nur77/Nurr1 leads to inefficient clonal deletion of self-reactive T cells. J Exp Med. 1996;183:1879–1892. doi: 10.1084/jem.183.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin B, et al. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116:527–540. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- 8.Thompson J, Winoto A. During negative selection, Nur77 family proteins translocate to mitochondria where they associate with Bcl-2 and expose its proapoptotic BH3 domain. J Exp Med. 2008;205:1029–1036. doi: 10.1084/jem.20080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang A, Rud J, Olson CM, Jr, Anguita J, Osborne BA. Phosphorylation of Nur77 by the MEK-ERK-RSK cascade induces mitochondrial translocation and apoptosis in T cells. J Immunol. 2009;183:3268–3277. doi: 10.4049/jimmunol.0900894. [DOI] [PubMed] [Google Scholar]
- 10.Li H, et al. Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science. 2000;289:1159–1164. doi: 10.1126/science.289.5482.1159. [DOI] [PubMed] [Google Scholar]
- 11.Liu ZG, Smith SW, McLaughlin KA, Schwartz LM, Osborne BA. Apoptotic signals delivered through the T-cell receptor of a T-cell hybrid require the immediate-early gene nur77. Nature. 1994;367:281–284. doi: 10.1038/367281a0. [DOI] [PubMed] [Google Scholar]
- 12.Moll UM, Marchenko N, Zhang XK. p53 and Nur77/TR3: Transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25:4725–4743. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- 13.Winoto A, Littman DR. Nuclear hormone receptors in T lymphocytes. Cell. 2002;109(Suppl):S57–S66. doi: 10.1016/s0092-8674(02)00710-9. [DOI] [PubMed] [Google Scholar]
- 14.Woronicz JD, Calnan B, Ngo V, Winoto A. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature. 1994;367:277–281. doi: 10.1038/367277a0. [DOI] [PubMed] [Google Scholar]
- 15.Rajpal A, et al. Transcriptional activation of known and novel apoptotic pathways by Nur77 orphan steroid receptor. EMBO J. 2003;22:6526–6536. doi: 10.1093/emboj/cdg620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brás A, Albar JP, Leonardo E, de Buitrago GG, Martínez-A C. Ceramide-induced cell death is independent of the Fas/Fas ligand pathway and is prevented by Nur77 overexpression in A20 B cells. Cell Death Differ. 2000;7:262–271. doi: 10.1038/sj.cdd.4400653. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki S, et al. Nur77 as a survival factor in tumor necrosis factor signaling. Proc Natl Acad Sci USA. 2003;100:8276–8280. doi: 10.1073/pnas.0932598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osborne BA, et al. Identification of genes induced during apoptosis in T lymphocytes. Immunol Rev. 1994;142:301–320. doi: 10.1111/j.1600-065x.1994.tb00894.x. [DOI] [PubMed] [Google Scholar]
- 19.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham NR, et al. Immature CD4+CD8+ thymocytes and mature T cells regulate Nur77 distinctly in response to TCR stimulation. J Immunol. 2006;177:6660–6666. doi: 10.4049/jimmunol.177.10.6660. [DOI] [PubMed] [Google Scholar]
- 21.Liston A, et al. Generalized resistance to thymic deletion in the NOD mouse; a polygenic trait characterized by defective induction of Bim. Immunity. 2004;21:817–830. doi: 10.1016/j.immuni.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 22.Zucchelli S, et al. Defective central tolerance induction in NOD mice: genomics and genetics. Immunity. 2005;22:385–396. doi: 10.1016/j.immuni.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Lee SL, et al. Unimpaired thymic and peripheral T cell death in mice lacking the nuclear receptor NGFI-B (Nur77) Science. 1995;269:532–535. doi: 10.1126/science.7624775. [DOI] [PubMed] [Google Scholar]
- 24.Cheng LE, Chan FK, Cado D, Winoto A. Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis. EMBO J. 1997;16:1865–1875. doi: 10.1093/emboj/16.8.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Judkowski V, et al. Identification of MHC class II-restricted peptide ligands, including a glutamic acid decarboxylase 65 sequence, that stimulate diabetogenic T cells from transgenic BDC2.5 nonobese diabetic mice. J Immunol. 2001;166:908–917. doi: 10.4049/jimmunol.166.2.908. [DOI] [PubMed] [Google Scholar]
- 26.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 27.Kurts C, et al. Constitutive class I-restricted exogenous presentation of self antigens in vivo. J Exp Med. 1996;184:923–930. doi: 10.1084/jem.184.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat Immunol. 2002;3:756–763. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 29.Jordan MS, et al. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2:283–284. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 30.Kawahata K, et al. Generation of CD4(+)CD25(+) regulatory T cells from autoreactive T cells simultaneously with their negative selection in the thymus and from nonautoreactive T cells by endogenous TCR expression. J Immunol. 2002;168:4399–4405. doi: 10.4049/jimmunol.168.9.4399. [DOI] [PubMed] [Google Scholar]
- 31.Walker LS, Chodos A, Eggena M, Dooms H, Abbas AK. Antigen-dependent proliferation of CD4+ CD25+ regulatory T cells in vivo. J Exp Med. 2003;198:249–258. doi: 10.1084/jem.20030315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonasio R, et al. Clonal deletion of thymocytes by circulating dendritic cells homing to the thymus. Nat Immunol. 2006;7:1092–1100. doi: 10.1038/ni1385. [DOI] [PubMed] [Google Scholar]
- 33.Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ-specific T cells. Nat Immunol. 2003;4:350–354. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 34.Smith KA. The quantal theory of how the immune system discriminates between “self and non-self.”. Med Immunol. 2004;3:3. doi: 10.1186/1476-9433-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burchill MA, et al. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–121. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen W, et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill JA, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Pei L, et al. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med. 2006;12:1048–1055. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- 39.Warburg O, Christian W. Isolation and crystallization of fermentation enzyme enolase. Biochem Z. 1941;310:384–421. [Google Scholar]
- 40.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearen MA, Muscat GE. Minireview: Nuclear hormone receptor 4A signaling: Implications for metabolic disease. Mol Endocrinol. 2010;24:1891–1903. doi: 10.1210/me.2010-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pols TW, Bonta PI, de Vries CJ. NR4A nuclear orphan receptors: Protective in vascular disease? Curr Opin Lipidol. 2007;18:515–520. doi: 10.1097/MOL.0b013e3282ef77d1. [DOI] [PubMed] [Google Scholar]
- 43.Sekiya T, et al. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat Commun. 2011;2:269. doi: 10.1038/ncomms1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baldwin TA, Hogquist KA. Transcriptional analysis of clonal deletion in vivo. J Immunol. 2007;179:837–844. doi: 10.4049/jimmunol.179.2.837. [DOI] [PubMed] [Google Scholar]
- 45.Kasler HG, Verdin E. Histone deacetylase 7 functions as a key regulator of genes involved in both positive and negative selection of thymocytes. Mol Cell Biol. 2007;27:5184–5200. doi: 10.1128/MCB.02091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katz JD, Wang B, Haskins K, Benoist C, Mathis D. Following a diabetogenic T cell from genesis through pathogenesis. Cell. 1993;74:1089–1100. doi: 10.1016/0092-8674(93)90730-e. [DOI] [PubMed] [Google Scholar]
- 48.Bouillet P, et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 49.Yamagata T, Mathis D, Benoist C. Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha alpha lineage with characteristics of innate immune cells. Nat Immunol. 2004;5:597–605. doi: 10.1038/ni1070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.