Abstract
Medulloblastoma is the most common malignant childhood brain tumor. The protein phosphatase and oncogene WIP1 is over-expressed or amplified in a significant number of primary human medulloblastomas and cell lines. In the present study, we examine an important mechanism by which WIP1 promotes medulloblastoma growth using in vitro and in vivo models. Human cell lines and intracerebellar xenografted animal models were used to study the role of WIP1 and the major TP53 regulator, HDM2, in medulloblastoma growth. Stable expression of WIP1 enhances growth of TP53 wild-type medulloblastoma cells, compared with cells with stable expression of an empty-vector or mutant WIP1. In an animal model, WIP1 enhances proliferation and reduces the survival of immunodeficient mice bearing intracerebellar xenografted human medulloblastoma cells. Cells with increased WIP1 expression also exhibit increased expression of HDM2. HDM2 knockdown or treatment with the HDM2 inhibitor Nutlin-3a, the active enantomer of Nutlin-3, specifically inhibits the growth of medulloblastoma cells with increased WIP1 expression. Nutlin-3a does not affect growth of medulloblastoma cells with stable expression of an empty vector or of mutant WIP1. Knockdown of WIP1 or treatment with the WIP1 inhibitor CCT007093 results in increased phosphorylation of known WIP1 targets, reduced HDM2 expression, and reduced growth specifically in WIP1 wild-type and high-expressing medulloblastoma cells. Combined WIP1 and HDM2 inhibition is more effective than WIP1 inhibition alone in blocking growth of WIP1 high-expressing medulloblastoma cells. Our preclinical study supports a role for therapies that target WIP1 and HDM2 in the treatment of medulloblastoma.
Keywords: HDM2, MDM2, medulloblastoma, PPM1D, WIP1
Patients who receive a diagnosis of medulloblastoma, the most common malignant brain tumor of childhood, are currently treated on the basis of disease stage, age at diagnosis, and extent of resection with use of a combination of surgery, chemotherapy, and ionizing radiation (IR).1 Advances in neurosurgical and medical treatments for medulloblastoma have dramatically improved the cure rate. However, disease progression or recurrence is still fatal for up to one-third of patients. Furthermore, patients who are cured often still experience chronic toxicities of treatment that can permanently inhibit appropriate growth, cognition, and motor functions, significantly detracting from their quality of life.
Initial efforts aimed at improving understanding of medulloblastoma tumor biology focused on disease classification with use of cytogenetic aberrations identified by comparative genomic hybridization (CGH).2,3 Gain of the long arm of chromosome 17 (17q) and isochromosome 17q (i17q), consisting of 17p deletion with duplication of 17q, has repeatedly been identified as the most common cytogenetic lesion affecting medulloblastoma in 30%–50% of cases.4–6 More recent attempts to categorize medulloblastoma have focused on the gene expression profile. Two recent publications have segregated medulloblastoma into 4 or 5 subtypes, based on differential expression of genes involved in Wingless (WNT) signaling (group A), Sonic-Hedgehog (SHH) signaling (group B), neuronal differentiation (groups C–D), or expression of photoreceptor genes (groups D–E). Alterations of chromosome 17 and upregulation of genes associated with tumor metastasis were strongly associated with medulloblastomas in subtypes C and D.7,8 Our analysis of this published data suggests increased expression of the TP53-induced proto-oncogene WIP1 (wild-type TP53-induced phosphatase 1 or protein phosphatase, magnesium-dependent 1, delta, PPM1D), located at chromosomal locus 17q22-23, in group C and D medulloblastomas.
TP53, on chromosome 17p13, is one of the most studied and best-characterized tumor suppressor genes. WIP1 has been implicated as an important regulator of the activity of TP53. In normal cells exposed to environmental stressors, such as ionizing radiation, WIP1 functions in a negative feedback loop with TP53. TP53 induces expression of WIP1, which in turn inactivates and limits the activity of TP53 directly by dephosphorylating serines 15 and 20 and indirectly through dephosphorylation of p38 MAPK and HDM2.9 Conversely, WIp1 has been shown to cooperate with oncogenes, including c-myc, Ras, and E1A, to transform rodent embryonic fibroblasts.10–12 Amplification of 17q22-q23 has been described in malignancies, including breast and ovarian carcinomas and in neuroblastoma, most of which are wild-type for TP53.10,12,13 We previously reported that 51% of human medulloblastomas have amplification or gain of WIP1. All primary medulloblastomas that were identified as WIP1-amplified by fluorescence in situ hybridization (FISH) were also amplified by comparative genomic hybridization and exhibited elevated WIP1 mRNA and protein expression, compared with fetal brain controls.14 Other investigators have demonstrated nuclear staining for WIP1 in 88% of human medulloblastomas.15 This suggests that WIP1 plays an important role in medulloblastoma tumorigenesis.
In the current study, we showed that stable expression of WIP1 enhances growth of TP53 wild-type medulloblastoma cells, compared with cells with stable expression of an empty vector or mutant WIP1. WIP1 enhances proliferation and reduces the survival of immunodeficient mice bearing intracerebellar xenografts of WIP1 high-expressing human medulloblastoma cells. Medulloblastoma cells with increased WIP1 expression also exhibit increased expression of HDM2. HDM2 knockdown or treatment with the HDM2 inhibitor Nutlin-3a, the active enantomer of Nutlin-3, specifically inhibits the growth of medulloblastoma cells with increased WIP1 expression. Knockdown of WIP1 or treatment with the WIP1 inhibitor CCT007093 results in increased phosphorylation of known WIP1 targets, reduced HDM2 expression, and reduced growth specifically in WIP1 wild-type, high-expressing medulloblastoma cells. Combined WIP1 and HDM2 inhibition is more effective than WIP1 inhibition alone in blocking the growth of WIP1 high-expressing medulloblastoma cells. This suggests that WIP1 and HDM2 are important targets for the treatment of medulloblastoma.
Materials and Methods
Gene Expression Analysis
WIP1 (PPM1D) expression in human medulloblastomas was determined by downloading .cel files from datasets GSE10327 and GSE21140 from the National Institutes of Health Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/). The GSE10327 dataset was generated using an Affymetrix Human Genome U33 Plus 2.0 Array. Prior to analysis, raw data were normalized using a Robust Multichip Average algorithm.16 Classification of the 62 medulloblastoma samples was maintained as previously reported.7 Partek Genomics Suite software (Partek) was used to confirm differential WIP1 expression among medulloblastoma groups A–E. Because data in the GSE21140 dataset were generated using an Affymetrix Human Exon 1.0 ST Array, raw .cel files were imported into Partek Genomics Suite and normalized using the Robust MultiChip Average algorithm.16 Classification of the 103 medulloblastoma samples was maintained according to the original published report.8 Gene expression was estimated by averaging the signals for all exons corresponding to the WIP1 (PPM1D) gene. WIP1 expression was compared among all groups (WNT, SHH, group C, and group D).
Materials
Nutlin-3a (Cayman Chemicals) was prepared at a 3.2 mM stock concentration in ethanol and diluted in culture media to 4–8 μM for use in experiments. CCT007093 (Sigma-Aldrich) was prepared at a stock concentration of 5 mM in DMSO and diluted in culture media to 0.05–5 μM for use in experiments. The maximum concentration of ethanol and of DMSO in both drug-treated and control experiments was 0.5% and 0.1%, respectively.
Cell Culture
We routinely maintained human medulloblastoma cell lines, including D556, Med8A, D425, and Daoy in DMEM or modified Eagle's medium (Invitrogen) with high glucose, 2 mM L-glutamine, and 10% (vol/vol) heat-inactivated FBS at 37°C in 5% CO2. D556 and D425 cell lines were gifts from Tobey MacDonald (Emory University); Med8A cells were obtained from John Kim (Kaiser Permanente), and Daoy parental cells were purchased from ATCC. We generated D556 and Daoy stable-expressing clones by transfecting these cells with an empty vector (pcDNA3/pcDNA), WIP1, or phosphatase-deficient, nonfunctional WIP1-D314A cDNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations and selecting with 200 μg/mL G418 (Sigma-Aldrich). After colonies were identified and growing in selection media, each colony was trypsinized in a sterile cloning cylinder (Bellco Glass) and transferred into a 24-well plate containing 200 μg/mL G418. Cell clones were allowed to grow until almost confluent and were gradually moved into larger cell culture dishes. After clones were split into 10 cm dishes, cell pellets were collected from each clone. Total RNA was extracted, and real-time reverse-transcription polymerase chain reaction (RT-PCR), described below, was conducted on each clone to determine relative WIP1 expression.
All cell lines were split twice weekly and were screened for mycoplasma contamination every 6 months (MycoAlert Detection Kit; Lonza Group). WIP1and phosphatase-deficient WIP1-D314A plasmids were gifts from Lawrence Donehower (Baylor College of Medicine). All cells were passaged for less than 6 months after receipt or resuscitation.
Effects on growth of adherent cells were assessed by plating 1 × 105 D556 or Daoy stable clones on 6-well plates in triplicate and harvesting cells with 0.25% trypsin EDTA (Invitrogen) at 24–96 h after initial plating. Cells were counted by trypan blue exclusion using standard methods.
Western Blotting
Cells were extracted from culture plates by scraping or with 0.25% trypsin EDTA (Invitrogen). Cell pellets were washed in phosphate-buffered saline (PBS), centrifuged at 200 × g, and stored at −80°C. Proteins were extracted from cell pellets using RIPA buffer (Cell Signaling), quantified using a standard Bradford assay, and electrophoretically separated on polyacrylamide denaturing gels, as previously described.17 Protein was transferred onto nitrocellulose membranes and immunoblotted with the designated antibodies as previously described.17 Antibodies used included WIP1 (Bethyl), phospho-p53 (Ser15; Cell Signaling), p53 (DO-1; Santa Cruz), HDM2 (2A10; Millipore), Phospho-p38 MAPK (Thr180/Tyr182; Cell Signaling), p38 MAPK (EMD4Biosciences), β-actin (AC-15; Sigma-Aldrich), and Vinculin (Sigma-Aldrich). Secondary antibodies Alexa Fluor 680 goat anti-mouse IgG (Invitrogen) or IRDye 800 goat anti-rabbit IgG (Rockland) were used at a dilution of 1:5000 and applied according to the manufacturer's recommendations.14 Immunoblots were imaged and the intensity of bands on Western blots was quantified using an Odyssey infrared imaging system (Li-COR Biosciences).
Anchorage-Independent Growth Assays
A total of 8 × 104 cells per well were plated in triplicate in 6-well dishes in 1 mL of matrigel (BD Biosciences) diluted 3:1 (media to matrigel). The matrigel cell suspension was allowed to solidify for at least 2 h at 37°C. For Nutlin experiments, after the matrigel solidified, 1 mL of media containing ethanol control or Nutlin-3a (final concentration, 8 μM) was added to each well. Photographs were taken with an Olympus IX50 inverted microscope at 4–40× magnification. To quantify the cell number, matrigel was digested using dispase (BD Biosciences) 0–96 h after initial plating. Media were removed from each well, and 2 mL of dispase was added per well and incubated at 37°C for 1 h. Each 3-mL sample was then transferred to a centrifuge tube, and 10 mM EDTA was added to stop the enzymatic activity of dispase. Each sample was centrifuged at 1000 rpm for 5 min and washed 3 times with PBS. Cells were then counted by trypan blue exclusion.
Real-Time Cell Analysis
Cell growth was monitored in real time using a Real-Time Cell Analyzer Dual-Plate instrument (Roche Applied Science) placed in a humidified incubator maintained with 5% CO2 at 37°C. This system records cell index (CI) as a unitless number that is a function of electrical impedance of cells attached to interdigitated electrodes built into the bottom of wells of a 16-well E-plate (Roche Applied Science). Recording of CI and subsequent data analysis were performed using the RTCA Software 1.2 (Roche Applied Science). Background impedance in each well in the presence of media alone was measured before cell seeding and automatically subtracted by the RTCA software. For proliferation assays, 5000–20,000 cells/well in 10% FBS containing DMEM or MEM media (Invitrogen) were seeded in E-plates. CI was recorded every 15 min for the next 96 h. Assays were performed at least twice with reproducible results.
Cell Cycle Analysis
Cells were plated at a density of 2.5 × 105 cells per well in 6-well plates in media containing 0.1% FBS. Cells were serum starved for 72 h to synchronize cells in G0. At 72 h, cells were harvested using trypsin, washed twice with DPBS + 10%FBS, fixed in ice-cold 80% ethanol, and stored at −20°C for at least 24 h. Remaining cells were treated with either ethanol or Nutlin-3a (final concentration, 8 µM) in media with 10% FBS. Treated cells were harvested and fixed at 24 h after the start of treatment. Fixed cells were incubated in 50 µL of PI buffer (20 µg/mL PI, 0.1% Triton-X 100, 200 µg/mL RNaseA in DPBS) for 30 min in the dark. Cells were then resuspended in 400 µL of DPBS for flow analysis. The samples were analyzed using a BD FACS Canto II cytometer (BD Biosciences) with BD FACS Diva software. All experiments were performed in triplicate and repeated at least twice.
Short Hairpin RNA Lentivirus Production and Infection
EGFP-tagged negative control and shWIP1 lentiviral expression constructs have been previously described.18 The HIV-EF-1-EGFP,19 psPAX2, and pVSVG plasmids were gifts from H. Trent Spencer (Emory University); 2 × 106 293T cells were plated on collagen-1–coated 100-mm dishes (BIOCOAT; Becton Dickinson) and, 24 h later, were transfected with 8 μg EGFP-tagged empty vector (HIV-EF-1-EGFP) or the FG12-hv6-1-shWIP1 lentiviral vector along with 4 μg packaging construct, psPAX2, and 4 μg vesicular stomatitis virus G expression plasmid (pVSVG), using Lipofectamine 2000 (Invitrogen). Supernatant from transfected cells was collected at 48 and 72 h following transfection and stored at −80°C. Virus production was verified using the green fluorescent protein expression marker. Cell supernatant was pooled and centrifuged overnight at 10,000 × g at 4°C. Virus was resuspended in 1:200 × volume serum-free DMEM media and stored at −80°C; 1 × 106 medulloblastoma cells were plated in 6-well plates and, 24 h later, were transduced with virus particles at a multiplicity of infection (MOI) of 2 with 8 µg/mL of polybrene. WIP1 knockdown following lentivirus infection was confirmed in cells by immunoblotting.
siRNA Transfection
For siRNA experiments, 1 × 106 medulloblastoma cells were plated in 6-well plates and, 24 h later, were transfected with 50 nM siWIP1, siHDM2, or negative control (siNC) siRNA oligomers (Dharmacon), using HiPerfect (Qiagen) reagent, according to the manufacturer's recommendations. Gene knockdown following siRNA transfection was confirmed at various time points by immunoblotting.
Dose-Response Viability Assays
D556 clones with stable expression of empty vectors (D556-pcDNA3), of WIP1 (D556-WIP1), and of a phosphatase-dead WIP1 (D556-WIP1 D314A) were either plated as adherent cell cultures or in matrigel and treated with 8 μM Nutlin-3a for 24–72 h, and then cell survival was determined by trypan blue exclusion. D556-pcDNA3 and D556-WIP1 cells were also plated as adherent cultures and treated with DMSO, 0.5 μM, or 5 μM CCT007093 (Sigma-Aldrich) for 6 days, and then cell survival was determined by trypan blue exclusion. Finally, D556-pcDNA3, D556-WIP1, and D425 cells were plated as adherent cultures and treated with ethanol, 8 μM Nutlin-3a, and shWIP1 lentivirus (MOI = 2), alone or in combination for 24–72 h, and then cell survival was determined by trypan blue exclusion. Experiments were done in triplicate and repeated at least twice.
Quantitative Real-Time RT-PCR
Total cellular RNA was extracted using the RNeasy Mini Kit (Qiagen). Total RNA (1 µg) was reverse transcribed into cDNA with Superscript III (Invitrogen) using random hexamers, as previously described.17 RNA integrity was verified using a Nanodrop spectrophotometer (ThermoScientific); only samples with an A260/A280 ratio more than 1.9 and less than 2.1 were used in quantitative real-time RT-PCR reactions. Quantitative real-time RT-PCR reactions containing cDNA, Syber Green PCR Master Mix (Applied Biosystems), and primers for human WIP1, HDM2, TATA-box Binding Protein (TBP), and/or Glyceraldehyde-3 Phosphate Dehydrogenase (GAPDH) in 25 µL final volume were performed for 45 cycles in triplicate on an ABI 7500 Real-Time PCR Cycler (Applied Biosystems) using absolute quantification with a standard curve. Serial dilutions of cDNA were used to determine a standard curve for each primer. Primer sequences are available upon request. Amplification products were verified by agarose gel electrophoresis and analysis of melting curves. WIP1 and HDM2 expression was normalized internally to TBP20 or GAPDH expression, accounting for differences in primer efficiencies. RNA expression was determined by the ΔΔCt method.21 Results from at least 3 separate experiments were analyzed.
Stereotactic Implantation of Tumor Cells
SCID/Beige mice (Charles River Laboratories) were anesthetized using 100 mg/kg ketamine (Pfizer) and 9 mg/kg xylazine (Ben Venue Laboratories) and were positioned in a stereotactic frame with a mouse adaptor (David Kopf Instruments). An incision was made in the midline of the scalp over the cerebellum, and a small hole was made in the skull (1 mm lateral to midline) using a beveled (sharp point) 18G needle. A 24G Hamilton syringe loaded with 5 × 105 cells was mounted on a micromanipulator and introduced through the hole at a 30° angle to the surface of the cerebellum, at a depth of 1 mm. Cells in tissue culture media were injected over the course of 2 min, and the needle was left in place for another 2 min to avoid reflux. After removing the mouse from the frame, 1–2 drops of 0.25% (2.5 mg/mL) bupivicaine (Hospira) were applied along the incision for postoperative analgesia, and the skin was closed with 6-0 fast-absorbing plain gut suture using a 3/8 PC-1 cutting needle (Ethicon).
Mouse Handling
All mice were housed in an American Association of Laboratory Animal Care–accredited facility and were maintained in accordance with the National Institutes of Health guidelines. Mice were followed and sacrificed by CO2 inhalation after they developed symptoms of medulloblastoma, which included head doming, hunched posture, preferential turning to one side, lethargy, and/or more than 15% weight loss. All animal care and experiments were approved by the Institutional Animal Care and Use Committee of Emory University (Protocol # 145-2009). Survival was analyzed using GraphPad Prism 4 software (GraphPad Software).
Mouse Necropsy and Tissue Handling
After euthanasia, the cranium and skull were removed from symptomatic mice using scissors and forceps. After severing the cranial nerves, we sagittally sectioned each mouse brain with a sterile scalpel and fixed them in 4% formalin for pathological examination. Tissue blocks were paraffin embedded, cut into 4-μm sections, and then stained with hematoxylin and eosin (H&E).22 Slides were processed in Emory's Winship Cancer Institute Histology Core and were visualized with an Olympus Provis AX-70 microscope (Olympus America). Images were captured with an Olympus DP70 digital camera and were analyzed using the DP Controller software package. Images were processed for publication using Adobe Photoshop Elements 5.0 (Adobe Systems).
H&E stained slides and immunohistochemistry for PCNA (Cell Signaling) of mid-sagittal sections from each mouse brain were examined by a pathologist (M.S.) using an Olympus CH-series light microscope. Three randomly selected, high-magnification fields (400×; field diameter, 0.45 mm) were assessed for numbers of mitotic figures on H&E slides and averaged for each specimen. Percent positivity of tumor cell nuclei for PCNA was determined by examination of a portion of one randomly selected 400× field and counting positive and negative nuclei until a total of 200 cells were counted.
Statistical Analysis
Results were analyzed using a 2-tailed Student's t test or 1-way analysis of variance (ANOVA) in Microsoft Excel or Graphpad Prism 4 software to assess statistical significance. Values of P < .05 were considered to be statistically significant.
Results
Group C and D Human Medulloblastomas Express WIP1 at High Levels
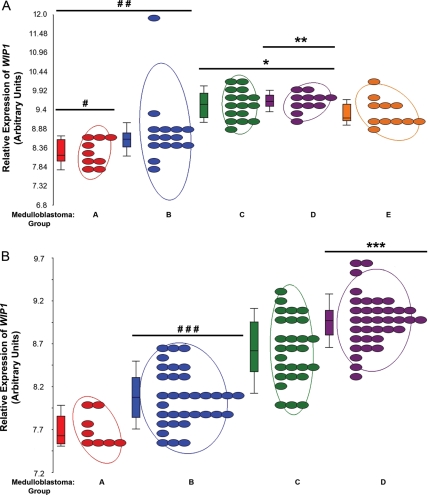
We have previously reported amplification or over-expression of WIP1 in 51% of human medulloblastomas.14 Another study has reported strong nuclear expression of WIP1 by immunohistochemistry in 148 (88%) of 168 human medulloblastomas on a tissue microarray.15 To place these prior findings in the context of recent reports that have categorized medulloblastoma into 3–5 distinct groups based on gene expression, we confirmed WIP1 expression in 2 independent medulloblastoma datasets.7,8 In the 62 human medulloblastomas profiled by Kool et al., WIP1 expression was 2.1-fold lower in group A's WNT pathway-activated medulloblastomas, compared with the medulloblastomas in groups B–E (Fig. 1A). WIP1 expression was also 2-fold lower in combined groups A and Sonic-hedgehog (SHH) pathway-activated, group B, compared with medulloblastoma groups C–E (Fig. 1A). Conversely, WIP1 expression was 1.8-fold higher in the medulloblastomas in groups C and D combined than those in groups A, B, and E combined (Fig. 1A). WIP1 expression was also 1.6-fold higher in group D than other medulloblastoma groups (Fig. 1A).
Fig. 1.
Group C and D human medulloblastomas express WIP1 at high levels. (A) WIP1 expression in medulloblastomas from group A versus groups B-E (#P = 3.8 × 10−5) in the Kool et al. dataset (GEO GSE10327). WIP1 expression was also lower in groups A and B than in groups C–E (##P = 3.7 × 10−7). Conversely, WIP1 expression was higher in groups C and D than in other groups (*P = 1.7 × 10−5). WIP1 expression was also higher in group D than in group C (**P = .012). (B) In the Northcott et al. dataset (GEO GSE21140), WIP1 expression was lower in the SHH-pathway activated group than in other groups (###P = 2.9 × 10−5). Conversely, WIP1 expression was higher in group D than in other groups (***P = 4.7 × 10−18). Each colored oval denotes expression in an individual tumor sample. Adjacent boxes represent first, second, and third quantiles of data. The middle line in each box represents the median value for each group. Whiskers represent the bottom 10th and top 90th percentiles for each group. Values on the Y-axis denote relative expression after raw expression values were quantile-normalized and log2 transformed. All P values were false discovery rate corrected.
Of the 103 independent medulloblastoma specimens profiled by Northcott et al., WIP1 expression was 1.3-fold lower in those activated by the SHH-pathway than in the 3 other groups (Fig. 1B). Conversely, WIP1 expression was 1.8-fold higher in the medulloblastomas in group D than in the other groups (Fig. 1B). Northcott et al. also re-analyzed the Kool et al. dataset, separating those tumors into 4 rather than 5 categories. They report increased significance of the reduced expression of WIP1 in the medulloblastomas of the WNT-activated (P = 3 × 10−6) group and increased WIP1 expression in those of group D (P = 7.9 × 10−6) in the Kool et al. dataset. Because of the chromosomal locus of WIP1on 17q22-23, these data are consistent with prior reports that have suggested that WNT- and SHH-activated medulloblastomas are biologically distinct from medulloblastomas with gain of chromosome 17q or i17q.
WIP1 Promotes Growth of TP53 Wild-Type Human Medulloblastoma Cell Lines
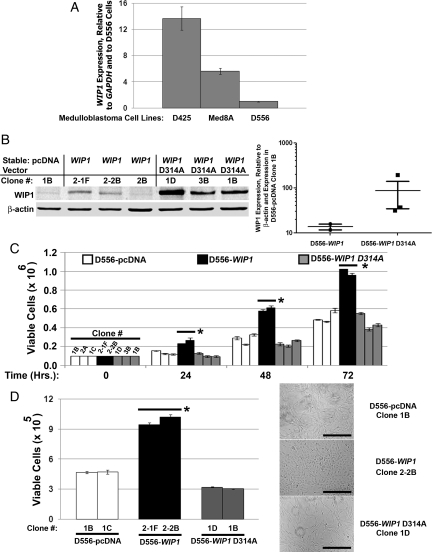
Because WIP1 is expressed at increased levels in group D and, possibly, in group C medulloblastomas, we next sought to understand the mechanisms by which WIP1 promotes medulloblastoma tumor growth. We first quantified WIP1 mRNA expression in human medulloblastoma cell lines. Because WIP1 over-expression or gene amplification has previously been described in human breast cancers that are wild-type for the TP53 gene, we examined WIP1 expression in the TP53 wild-type human medulloblastoma cell lines D425, Med8A, and D556 (http://www.sanger.ac.uk/genetics/CGP/CellLines/)23,24 by quantitative real-time RT-PCR. We found that D556 cells express relatively low levels of WIP1 mRNA, comparable to levels previously described in the TP53 mutant Daoy cell line.14 D425 and Med8A cells expressed endogenous WIP1 mRNA at 13.7 and 5.6 times that in D556 cells, respectively (Fig. 2A). Next, we generated stable, empty vector (pcDNA3), WIP1-expressing, and mutant, phosphatase-deficient WIP1-D314A25 clones of D556 cells and screened for expression of WIP1 transcript using quantitative real-time RT-PCR. D556 cells stably transfected with WIP1 (D556-WIP1) expressed up to 2.5 times more WIP1 mRNA than D556 cells with stable expression of an empty vector (D556-pcDNA; data not shown). D556 cells stably transfected with a mutant WIP expression vector (D556-WIP1 D314A) expressed WIP1 transcript by quantitative real-time RT-PCR that was 5–16 times higher than in D556-pcDNA stable cells (data not shown). Western blotting (Fig. 2B) revealed up to 15.5-fold higher WIP1 expression in D556-WIP1 clones and up to 194-fold higher WIP1 expression in D556-WIP1 D314A stable clones than in the D556-pcDNA clone 1B (Fig. 2B).
Fig. 2.
Stable increased WIP1 expression enhances growth of human medulloblastoma cells. (A) WIP1 mRNA expression by quantitative real-time reverse-transcription polymerase chain reaction (qRT RT-PCR) in medulloblastoma cell lines, relative to expression of the internal gene control, GAPDH, and to expression in parental D556 human medulloblastoma cells. Error bars represent standard deviation among triplicates. (B) Western blotting of whole cell lysates from D556 clones with stable expression of an empty vector (pcDNA), of WIP1, or of a mutated, phosphatase-deficient WIP1 (WIP1 D314A). Right panel, Scatter plot of quantified WIP1 expression, relative to β-actin and to expression in the D556 empty-vector (D556-pcDNA) clone 1B. Error bars represent standard error of the mean. (C) Cell viability assayed by trypan blue exclusion in adherent cultures of D556 clones 24–72 h after initial plating (*P < .0001 at all time points). Error bars represent standard deviation among triplicates. (D) Viability of D556 clones grown in matrigel, harvested, and counted by trypan blue exclusion 72 h after initial plating. Error bars represent standard deviation among triplicates (*P < .0001). Right panel, Representative photomicrographs of D556 clones grown in matrigel, 72 h after initial plating (Mag. 10×; Bars measure 400 μm). All experiments other than matrigel assays were repeated at least three times. Matrigel assays were repeated at least twice.
To assess the effect of increased WIP1 expression on medulloblastoma cell growth, D556-pcDNA, D556-WIP1, and D556-WIP1 D314A clones were plated at a density of 1 × 104 cells/cm2 and harvested at 0, 24, 48, and 72 h after initial plating. D556-WIP1 stable clones exhibited increased viability at all time points, compared with either D556-pcDNA or D556-WIP1 D314A stable clones (P < .0001, 1-way ANOVA) (Fig. 2C). We confirmed these findings by examining anchorage-independent growth, a hallmark of transformed cells that refers to the ability of cells to proliferate in the absence of adhesion to a surface, which has been characterized as one of the best in vitro correlates of tumorgenicity.26 Seventy-two hours after initial plating in matrigel, cells were counted by trypan blue exclusion assays. D556-WIP1 cells demonstrated significantly increased viability, compared with D556-pcDNA or D556-WIP1 D314A stable-expressing clones (P < .0001, 1-way ANOVA) (Fig. 2D). We also plated 5 000 D556-pcDNA, D556-WIP1, or D556-WIP1 D314A clones per well on E-plates and simultaneously monitored real-time changes in CI, which can be used as an indicator of cell proliferation, over 96 h.27,28 Log-phase growth of D556-WIP1 (clone 2-1F) cells was apparent as early as 12 h after plating and was apparent much earlier than in the D556-pcDNA or D556-WIP1 D314A clones (Supplementary material, Fig. S1A). This difference was quantified by measuring the slope of the CI and doubling times of all 3 cell lines 24–48 h after plating. The slope of the cell index of D556-WIP1 (clone 2-1F) cells was more than twice that of D556-pcDNA (clone 1B) and more than 12 times that of D556-WIP1 D314A (clone 1D) cells (P < .0001) (Supplementary material, Fig. S1B). The doubling time of D556-WIP1 (clone 2-1F) cells was 1.7 times faster than that of D556-pcDNA (clone 1B) cells and 5.3 times faster than that of D556-WIP1 D314A (clone 1D) cells (P = .0022) (Supplementary material, Fig. S1C).
WIP1 Does Not Alter the Growth of TP53 Mutant Daoy Medulloblastoma Cells
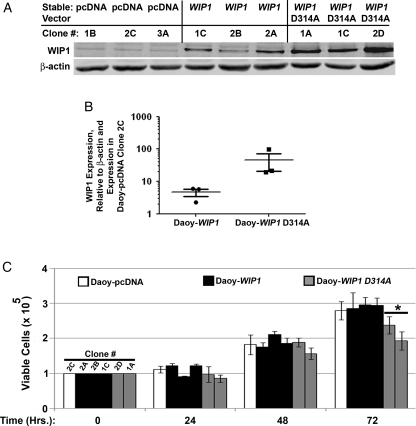
Having demonstrated the growth-promoting effects of over-expressed WIP1 in TP53 wild-type medulloblastoma cells, we hypothesized that mutation of TP53 would inhibit the proliferative effects of WIP1. Daoy medulloblastoma cells are known to contain a mutant TP53 gene (http://www.sanger.ac.uk/genetics/CGP/CellLines/). We have also previously shown that Daoy cells express low levels of WIP1 mRNA and protein.14 To assess the effect of increased WIP1 expression in TP53 mutant medulloblastoma cells, we generated clones of Daoy cells that stably expressed empty vectors (pcDNA3), WIP1, and mutant, phosphatase-deficient WIP1-D314A clones of Daoy cells and then screened these clones for expression of the WIP1 protein. Western blotting (Fig. 3A and B) revealed up to a 7.1-fold higher WIP1 expression in Daoy-WIP1 clones and up to a 119-fold higher WIP1 expression in Daoy-WIP1 D314A stable clones than in the Daoy-pcDNA clone 2C. To assess effects on proliferation, 1 Daoy-pcDNA, 3 Daoy-WIP1, and 2 Daoy-WIP1 D314A clones were plated at a density of 1 × 104 cells/cm2 and harvested at 0, 24, 48, and 72 h after initial plating (Fig. 3C). At 24 and 48 h, there were no significant differences in viability among Daoy clones. At 72 h, there were fewer of both Daoy-WIP1 D314 clones 2D and 1A (P = .0037). However, there was no difference in viability between Daoy-pcDNA and Daoy-WIP1 stable clones.
Fig. 3.
Stable increased WIP1 expression failed to affect growth of TP53 mutant medulloblastoma cells. (A) Western blotting of whole cell lysates from TP53 mutant Daoy clones with stable expression of an empty vector (pcDNA), of WIP1, or of a mutated, phosphatase-deficient WIP1 (WIP1 D314A). (B) Scatter plot of quantified WIP1 expression, relative to β-actin and to expression in the Daoy empty-vector (Daoy-pcDNA) clone 2C. Error bars represent standard error of the mean. (C) Cell viability assayed by trypan blue exclusion in adherent cultures of Daoy clones 24–72 h following initial plating of cells (*P = .004 at 72 h). Error bars represent standard deviation among triplicates.
WIP1 Promotes Medulloblastoma Growth and Inhibits Mouse Survival In Vivo
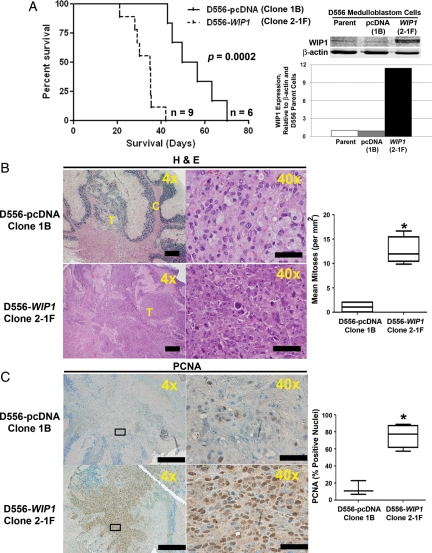
To validate the effects of WIP1 in vivo, we injected 5 × 105 D556-pcDNA (clone 1B) or D556-WIP1 (clone 2-1F) cells into the cerebellum of 3–6-month-old immunodeficient, SCID/Beige mice. Mice were observed daily and sacrificed after they exhibited symptoms of medulloblastoma, such as head doming, hunched posture, preferential turning to one side, lethargy, and/or significant weight loss. Mice bearing D556-WIP1 orthotopic xenografts (n = 9) exhibited inferior survival (median survival, 35 days following xenografting), compared with mice xenografted with D556-pcDNA cells (n = 6; median survival, 53 days; P = .0002) (Fig. 4A). The number of mice in each group differed because of problems with anesthesia (one death in each group of xenografts) or significant intracranial hemorrhage within a week of xenografting.
Fig. 4.
High WIP1 expression promotes growth of intracerebellar xenografts of human medulloblastoma cells in vivo; 5 × 105 D556-pcDNA (Clone 1B) or D556-WIP1 (Cone 2-1F) cells were injected into the cerebellum of 3–6-month-old SCID/Beige mice. (A) Kaplan-Meier plot comparing survival among mice bearing D556-WIP1 and D556-pcDNA xenografts (P = .0002). Right panel, WIP1 expression by Western blotting in parental D556, D556-pcDNA (Clone 1B), and D556-WIP1 (Cone 2-1F) cells with β-actin as a loading control. Below Western blot, quantification of WIP1 expression relative to β-actin and expression in parental D556 cells. (B) Representative hematoxalin and eosin (H&E)-stained saggital sections of tumors from symptomatic SCID/Beige mice xenografted with D556-pcDNA (Clone 1B) or D556-WIP1 (Clone 2-1F) cells. Right panel, quantification of mitotic figures in H&E-stained sections of D556-pcDNA (Clone 1B, n = 4) or D556-WIP1 (Clone 2-1F, n = 4) xenografted tumors (*P = .0003). (C) Representative immunohistochemistry for the marker of proliferation, PCNA (n = 4 for each group). Right panel, Quantification of nuclear staining for PCNA in D556-pcDNA (Clone 1B) or D556-WIP1 (Clone 2-1F) xenografted tumors (*P = .001). The middle line in each box represents the median value for each group. Whiskers represent the minimum and maximum values in each group. Bars in photomicrographs measure 500 μm (4×) or 100 μm (40×).
Nevertheless, a clear difference in both histology and proliferation could be seen in the D556-WIP1 tumors. Compared with the D556-pcDNA xenografts, the D556-WIP1 xenografts were overall larger in size, had larger cells with more open chromatin and prominent nucleoli, and had more mitotic figures (P = .0003) (Fig. 4B). One of the D556-WIP1 xenografted animals even had distant intracranial metastases on the cerebral surface, detected by H&E staining. The D556-WIP1 xenografts had frequent pyknotic nuclear debris, which indicates a higher rate of apoptosis, although this debris was not noted in the D556-pcDNA xenografts. These findings are similar to the differences between classic and large-cell/anaplastic medulloblastoma in human clinical specimens. However, not all histological features of large-cell/anaplastic histology were noted in the D556-WIP1 xenografts. We identified no unequivocal cell-cell wrapping and only modest nuclear molding. D556-WIP1 xenografted tumors did exhibit increased staining for the marker of proliferation, PCNA,29 compared with tumors in mice injected with the D556-pcDNA stable clone 1B (P = .001) (Fig. 4C), which likely explains the increased mortality of mice xenografted with D556-WIP1 cells.
WIP1-Driven Medulloblastoma Growth is Mediated by HDM2
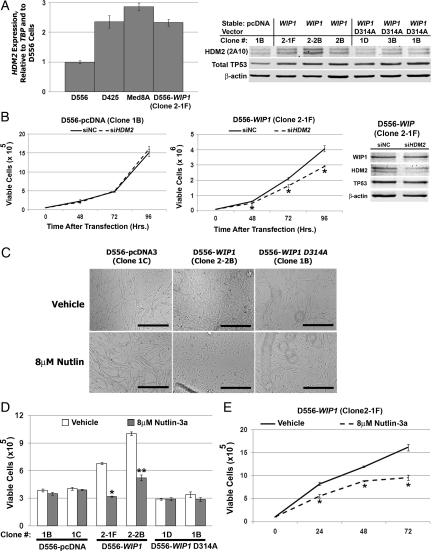
HDM2 has previously been defined as an important target of WIP1's phosphatase activity. WIP has been shown to dephosphorylate and, thus, stabilize HDM2, preventing it from ubiquitination and degredation through the 26S proteosome.9 We examined the expression of HDM2 in medulloblastoma cells by quantitative real-time RT-PCR and Western blotting. HDM2 mRNA expression was more than 2-fold higher in medulloblastoma cells with increased WIP1 expression, compared with the D556-pcDNA clone 1B (Fig. 5A). D556-WIP1 stable clones exhibited a slight increase in HDM2 protein expression, compared with clones containing empty vectors or mutant WIP1 D556 (Fig. 5A, right panel). Of interest, there was no clear difference in TP53 expression between WIP1 wild-type and mutant D556 clones.
Fig. 5.
WIP1 effects on medulloblastoma growth are mediated by HDM2. (A) Left panel, HDM2 expression by real-time reverse-transcription polymerase chain reaction (RT-PCR) in medulloblastoma cell lines, relative to expression of TBP and expression in D556-pcDNA clone 1B. Right panel, Western blotting of whole cell lysates from D556-pcDNA, D556-WIP1, or D556-WIP1 D314A clones for HDM2 and TP53. (B) Viability of D556-pcDNA (Clone 1B) and D556-WIP1 (Clone 2-1F) cells determined by trypan blue exclusion 48–96 h following transfection with either scrambled negative-control (siNC) or HDM2 siRNA (*P < .003). Far right panel: HDM2 expression by Western blotting 48 h after transfection with siNC or siMDM2 oligomers. Error bars represent standard deviation among triplicates. (C) Representative photomicrographs of D556-pcDNA (Clone 1C), D556-WIP1 (Clone 2-2B), and D556-WIP1 D314A (Clone 1B) cells grown in matrigel 72 h following treatment with vehicle or 8 μM Nutlin-3a (Mag. 10×). Bars measure 400 μm. (D) Viability of D556 clones grown in matrigel and counted by trypan blue exclusion 72 h following treatment with vehicle or 8 μM Nutlin-3a (*P < .00003; **P < .0005). Error bars represent standard deviation among triplicates. (E) Viability of adherent D556-WIP1 (Clone 2-1F) cells treated with vehicle or 8 μM Nutlin-3a from 0–72 h and counted by trypan blue exclusion (*P < .004). Error bars represent standard deviation among triplicates. All experiments other than matrigel assays were repeated at least three times. Matrigel assays were repeated at least twice.
To examine whether or not the effects of WIP1 on medulloblastoma growth were mediated by HDM2, we transfected medulloblastoma cells with either scrambled siRNA (siNC) or siRNA against HDM2 (siHDM2). The D556-WIP1 clone 2-1F exhibited reduced viability in response to HDM2 knockdown at all time points examined (Fig. 5B). HDM2 knockdown did not affect the growth of D556-pcDNA cells. We next examined the effect of the HDM2 small molecule antagonist, Nutlin-3a, on medulloblastoma cell growth. Nutlin-3a prevents HDM2 from interacting with p53 and has been used to inhibit the growth of a variety of cancer cells in vitro, including Hodgkin's lymphoma,30 acute lymphocytic leukemia,31 and neuroblastoma.32 Although Nutlin-3a inhibited anchorage independent growth of D556-WIP1 stable cells by more than 50% (P < .0003), it did not affect the growth of D556-pcDNA or of mutant D556-WIP1 D314A clones (Fig. 5C and D). Similarly, while Nutlin-3a did not significantly alter the growth of D556-pcDNA cells grown as a monolayer (data not shown), it significantly inhibited the growth of adherent D556-WIP1 cells (P < .004) (Fig. 5E). We have shown that high WIP1 expression is associated with increased HDM2 expression. However, there are likely additional genetic differences among medulloblastoma models that affect HDM2 protein expression. Of importance, we showed that only D556 cells with increased expression of WIP1, compared with cells with stable expression of an empty vector or of mutant WIP1, exhibit sensitivity to genetic and pharmacologic inhibition of HDM2. This suggests that Nutlin treatment may be most effective in suppressing the growth of medulloblastomas that exhibit high WIP1 expression.
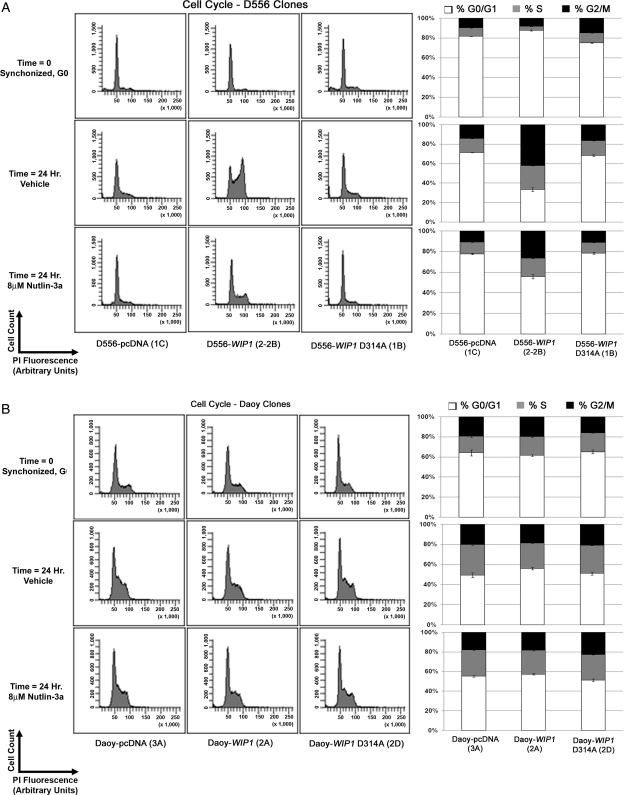
To confirm these findings, we synchronized D556 and Daoy stable clones in the G0 phase of the cell cycle by serum starvation for 72 h and then treated cells with either vehicle or Nutlin-3a for 24 h. Once released from G0 arrest, there was a significant shift of vehicle-treated D556-WIP1 (Clone 2-2B) cells into both the S and the G2/M phases of the cell cycle, consistent with the increased proliferative capacity of the D556-WIP1 stable clones (Fig. 6A; Table 1). Twenty-two percent of vehicle-treated D556-WIP1 cells were found in the S phase 24 h after being released from the G0 phase of the cell cycle. By comparison, 4.5%–6.7% of D556-pcDNA or mutant D556-WIP1 D314A cells were found in the S phase 24 h after being released from the G0 phase. There were no significant differences in the cell-cycle profile or in the percent of cells in the S phase among vehicle-treated Daoy pcDNA, WIP1, or mutant WIP1 stable clones 24 h after release from the G0 phase (Fig. 6B; Table 1). The only cells significantly affected by treatment with Nutlin-3a were the D556-WIP1 clones. Although Nutlin-3a treatment did not alter the percentage of cells in the S phase, it did significantly reduce the percentage of D556-WIP1 (clone 2-2B) cells in the G2/M phase (from 36% to 25%; P = .008) and increase the percentage of cells in the G0/G1 phase from 42% to 51% (P = .007). Nutlin-3a did not affect the cell-cycle profile of the TP53-mutant Daoy clones.
Fig. 6.
Nutlin-3 promotes the G0/G1 arrest of TP53 wild-type but not mutant TP53 medulloblastoma cells. Cell-cycle profiles of (A) TP53 wild-type D556 or (B) mutant TP53 Daoy medulloblastoma cell clones stably transfected with an empty vector (pcDNA), WIP1, or mutant WIP1 (D314A) arrested (time = 0) in G0 phase of the cell cycle by serum starvation. Cells were subsequently treated with serum-containing media and vehicle or Nultin-3a for 24 h. Right panels, Mean percentages of cells in the G0/G1, the S, and the G2/M phases of the cell cycle. Error bars represent standard error among triplicates. Experiments were repeated at least twice.
Table 1.
Nutlin-3 promotes the G0/G1 arrest of TP53 wild-type D556-WIP1 but not TP53 mutant Daoy-WIP1 medulloblastoma cells
| Variable | % G0/G1 | % S | % G2/M |
|---|---|---|---|
| D556-pcDNA (Clone 1C) | |||
| Vehicle | 90 ± 0.34 | 4.5 ± 0.18 | 5.7 ± 0.18 |
| Nutlin-3a | 91 ± 0.54 | 4.1 ± 0.28 | 5 ± 0.32 |
| D556-WIP1 (Clone 2-2B) | |||
| Vehicle | 42 ± 1.4 | 22 ± 0.16 | 36 ± 1.4 |
| Nutlin-3a | 51 ± 0.87 | 24 ± 0.27 | 25 ± 0.61 |
| D556-WIP1 D314A (Clone 1B) | |||
| Vehicle | 83 ± 0.51 | 6.7 ± 0.35 | 5 ± 0.17 |
| Nutlin-3a | 86 ± 0.37 | 5.5 ± 0.17 | 8.4 ± 0.32 |
| Daoy-pcDNA (Clone 3A) | |||
| Vehicle | 49 ± 2.5 | 31 ± 0.62 | 20 ± 1.9 |
| Nutlin-3a | 55 ± 0.6 | 27 ± 0.53 | 18 ± 1.1 |
| Daoy-WIP1 (Clone 2A) | |||
| Vehicle | 56 ± 1.4 | 25 ± 0.41 | 19 ± 1.7 |
| Nutlin-3a | 57 ± 0.69 | 24 ± 0.42 | 18 ± 0.37 |
| Daoy-WIP1 D314A (Clone 2D) | |||
| Vehicle | 51 ± 1.4 | 28 ± 0.17 | 21 ± 1.5 |
| Nutlin-3a | 51 ± 1.2 | 26 ± 0.32 | 23 ± 0.84 |
Quantification of mean percentages ± standard error among triplicates of TP53 wild-type D556 or mutant TP53 Daoy medulloblastoma cell clones stably transfected with an empty vector (pcDNA), of WIP1, or of mutant WIP1 (D314A) arrested (time = 0) in the G0 phase of the cell cycle by serum starvation. Cells were subsequently treated with serum-containing media and vehicle or Nultin-3a for 24 h. Experiments were repeated at least twice.
WIP1 Inhibition Blocks Medulloblastoma Cell Growth
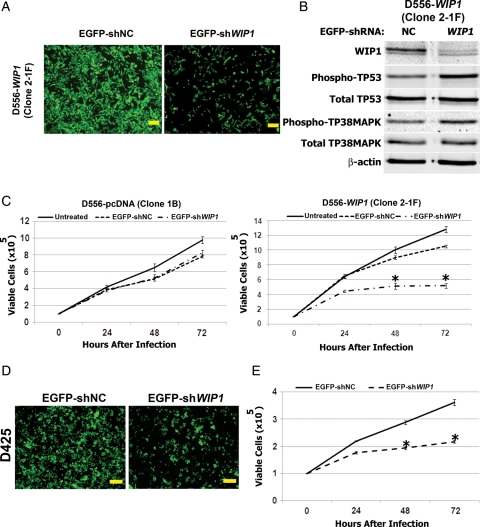
Human malignancies often evolve as a consequence of progressive accumulation of mutations and epigenetic changes in multiple genes. The term “oncogene addiction” has been coined to describe the phenomenon by which reversal of only one or a few of these abnormalities can profoundly inhibit tumor growth and affect patient survival.33 To demonstrate addiction to the effects of WIP1, we infected not only D556-pcDNA and D556-WIP1 cells (Fig. 7A) but also endogenous, WIP1 high-expressing D425 medulloblastoma cells (Fig. 7D) with EGFP-tagged lentiviral particles containing an empty vector or shWIP1. Western blotting of whole cell lysates from the negative control (NC) or from shWIP1-infected D556-WIP1 or D425 cells 48 h after infection revealed 80%–90% knockdown of WIP1 protein with increased phosphorylation and, thus, activation of p38 MAPK and p53, which are both known targets of WIP1's phosphatase activity (Fig. 7B).9 Quantification by trypan blue exclusion revealed a significant reduction in the viability of D556-WIP1 and D425 cells 48 and 72 h after infection with shWIP1, compared with shNC-infected controls (P < .0001) (Fig. 7C and E). Although lentivirus appeared to inhibit the growth of both D556-WIP1 and D556-pcDNA cells at 48 and 72 h, compared with uninfected controls, WIP1 knockdown did not affect the viability of D556-pcDNA stable cells, compared with EGFP-shNC infected D556-pcDNA controls (Fig. 7C).
Fig. 7.
WIP1 knockdown inhibits growth of WIP1 high-expressing medulloblastoma cells. Representative photomicrographs of (A) D556-WIP1 (Clone 2-1F) and (D) D425 medulloblastoma cells 72 h following infection with enhanced green fluorescent protein (EGFP)-tagged shWIP1 or with empty vector (shNC)-containing lentiviral particles at a multiplicity of infection (MOI) of 2 (4×). Bars measure 200 μm. (B) Western blotting of whole cell lysates of shWIP1- and negative-control (NC)-infected D556-WIP1 (Clone 2-1F) cells. (C) Viability of D556-pcDNA (Clone 1B) and D556-WIP1 (Clone 2-1F) cells 0–72 h following infection with EGFP-empty vector or shWIP1 lentiviral particles at an MOI of 2, determined by trypan blue exclusion (*P < .0001). (D-E) Representative photomicrographs (magnification 4×, bars measure 200 μm) and viability of D425 medulloblastoma cells 0 to 72 h following infection with EGFP-shNC or EGFP-shWIP1 lentiviral particles at an MOI of 2, determined by trypan blue exclusion (*P < .0001). Error bars represent standard deviation among triplicates. All experiments were repeated at least three times.
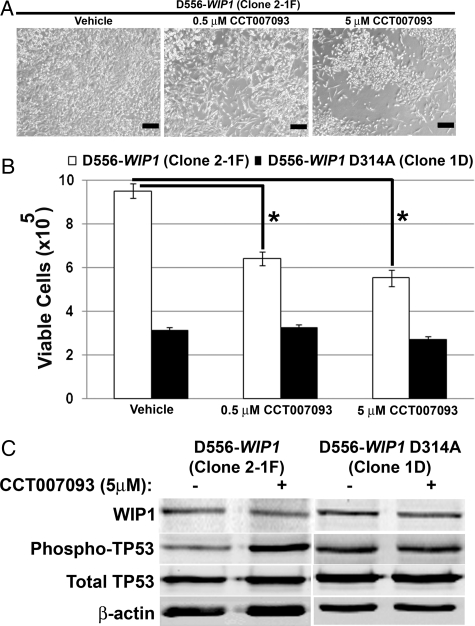
To validate our findings from the WIP1 knockdown in medulloblastoma cell lines and to examine the role of a WIP1 inhibitor as a targeted therapy for medulloblastoma, we treated D556-WIP1 cells with the small molecule inhibitor, CCT007093. Rayter et al. previously demonstrated that CCT007093 inhibits WIP1's ability to dephosphorylate p38 MAPK and specifically inhibits proliferation of WIP1-expressing cells.34 We found that treatment of D556-WIP1 cells with CCT007093 inhibited proliferation in a dose-dependent manner (P < .0005) (Fig. 8A and B). CCT007093 treatment did not have any significant effect on the growth of D556 cells that contain a mutant WIP1 (D556-WIP1 D314A). Growth inhibition of D556-WIP1 cells was associated with increased phosphorylation of the WIP1 target p53 (Fig. 8C). Together, our data suggests that WIP1 targeting may be most useful for treatment of medulloblastomas with high WIP1 expression.
Fig. 8.
CCT007093 inhibits growth of WIP1 high-expressing medulloblastoma cells. (A) Representative photomicrographs of D556-WIP1 (Clone 2-1F) cells 6 days after treatment with vehicle or with the WIP1 small molecule inhibitor CCT007093 (Mag. 4×). Bars measure 500 μm. (B) Viability of D556-WIP1 (Clone 2-1F) and D556-WIP1 D314A (Clone 1D) cells, determined by trypan blue exclusion, 6 days following treatment with vehicle or with 0.5–5 μM CCT007093 (*P < .0005 at both doses). Error bars represent standard deviation among triplicates. (C) Western blotting of whole cell lysates of D556-WIP1 (Clone 2-1F) and D556-WIP1 D314A (Clone 1D) cells 72 h following treatment with vehicle or with 5 μM CCT007093. All experiments were repeated at least 3 times.
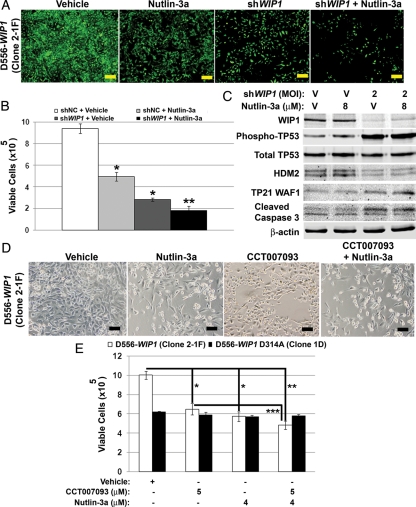
Nutlin Potentiates the Effect of WIP1 Inhibition Against WIP1 High-Expressing Medulloblastoma Cells
Because inhibition of HDM2 and WIP1 independently suppressed medulloblastoma growth, we also examined the effect of combined inhibition of both targets. D556-WIP1 cells were treated with Nutlin-3a, shWIP1 lentivirus, or both (Fig. 9A). Seventy-two hours after the start of treatment with Nutlin or shWIP1, the viability of D556-WIP1 cells was reduced by 47.5% and 69.9%, respectively, relative to the vehicle-treated controls (P < .0007). Combined treatment with Nutlin-3a and shWIP1 resulted in an 80.9% reduction in viability determined by trypan blue exclusion, relative to the vehicle-treated controls (P < .00005) (Fig. 9B). The effect of combined HDM2 and WIP1 inhibition on medulloblastoma growth was also significant when compared with Nutlin-3a or shWIP1 treatment alone (P < .05). Western blotting confirmed significant WIP1 knockdown in shWIP1 lentivirus-infected cells. Cells infected with shWIP1 also exhibited increased phosphorylation of TP53 on serine 15, reduced expression of HDM2, increased expression of the TP53 target TP21 WAF1, and increased cleavage of caspase 3. Treatment with Nutlin-3a resulted in increased expression of HDM2 (Fig. 9C).
Fig. 9.
Combined inhibition of WIP1 and HDM2 enhances growth suppression of WIP1 high-expressing medulloblastoma cells. (A) Representative photomicrographs of EGFP-expressing D556-WIP1 (Clone 2-1F) cells 72 h after infection with enhanced green fluorescent protein (EGFP)-tagged shWIP1 or empty vector (shNC)-containing lentiviral particles at a multiplicity of infection (MOI) of 2 and treatment with either vehicle or 8 μM Nutlin-3a (Mag. 4×). Bars measure 500 μm. (B) Viability of D556-WIP1 (Clone 2-1F) cells treated as described in (A), assessed by trypan blue exclusion at 72 h (*P < .0007, both relative to EGFP-shNC infected and vehicle-treated cells; **P < .04 relative to either shWIP1-infected or Nutlin-3a-treated cells). Error bars represent standard deviation among triplicates. (C) Western blotting of whole cell lysates of D556-WIP1 (Clone 2-1F) cells treated as described in (A). (D) Representative photomicrographs of D556-WIP1 (Clone 2-1F) cells 6 days after treatment with vehicle, 4 μM Nutlin-3a, 5 μM CCT007093, or 4 μM Nutlin-3a along with 5 μM CCT007093 (Mag. 10×). Bars measure 500 μm. (E) Growth of D556-WIP1 (Clone 2-1F) and WIP1-mutated D556-WIP1 D314A (Clone 1D) cells treated as described in (D), assessed by trypan blue exclusion 6 days following treatment (*P < .005 relative to vehicle-treated controls; **P < .0002 relative to vehicle-treated controls; ***P < .03 relative to CCT007093-treated cells). Error bars represent standard deviation among triplicates. All experiments were repeated at least twice.
Similar experiments in endogenous high WIP1-expressing D425 cells showed a 43.7% and 45.6% reduction in viability, compared with vehicle-treated controls, 72 h after treatment with Nutlin-3a or shWIP1, respectively (P < .00005). Combined Nutlin-3a and shWIP1 treatment suppressed medulloblastoma growth by 56%, compared with vehicle-treated controls (P < .000004). Combined HDM2 and WIP1 inhibition on medulloblastoma growth was also significant when compared with Nutlin-3a or shWIP1 treatment alone (P < .005) (Supplementary material, Fig. S2).
We also treated wild-type WIP1, D556-WIP, and WIP1-mutant D556-WIP1 D314A cells with Nutlin-3a, CCT007093, or both (Fig. 9D). Treatment of D556-WIP1 cells with nutlin-3a or CCT007093-treated reduced viability by 42.9% and 35.2%, respectively, relative to the vehicle-treated controls (P < .005). Combined Nutlin-3a and CCT007093 treatment resulted in a 51.8% reduction in viability determined by trypan blue exclusion, relative to the vehicle-treated controls (P < .0002) (Fig. 9E). The effect of combined HDM2 and WIP1 inhibition on medulloblastoma growth was significant when compared with CCT007093 treatment alone (P < .03). This suggests a role for combined HDM2 and WIP1 inhibition in the treatment of medulloblastomas with high WIP1 expression.
Discussion
Multiple studies have reported that gain of the long arm of chromosome 17 (17q) and isochromosome 17q (i17q), consisting of 17p deletion with duplication of 17q, are present in 30%–50% of medulloblastomas.4–6 Either loss of 17p (17p-) and/or gain of 17q have been associated with poor survival in patients who have received a diagnosis of medulloblastoma. Loss of the TP53 tumor suppressor gene, located on 17p13.1 and frequently deleted or mutated in 50% of adult cancers, has been considered a possible mechanism of tumorigenesis in 17p- medulloblastomas. However, prior studies have found that at least 50% of medulloblastomas with loss of heterozygosity of 17p maintain the TP53 gene locus.35 In addition, less than10% of medulloblastomas have been reported to contain a TP53 mutation.36,37
The prevalence of the TP53 mutation and its significance in medulloblastoma biology have re-emerged as an area of controversy. A recent retrospective study reported 8 (16%) of 49 medulloblastomas had mutations in the DNA binding domain of TP53, and 19 (18%) of 108 showed strong staining for TP53 by immunohistochemistry. Strikingly, all TP53 mutated tumors were initially characterized as average-risk but exhibited local recurrence within 2 years after diagnosis and 0% 5-year survival, compared with 74% 5-year survival among patients bearing average-risk, TP53 wild-type medulloblastomas.38 Another study reports a 75% overall survival among patients whose medulloblastomas stain negatively, for TP53 compared with 35% overall survival among patients whose medulloblastomas stain positively. Three of 10 tumors that stained positively were found to harbor mutations of TP53.39 Another recent study identified TP53 mutations in 21 (6.8%) of 310 medulloblastomas but no association with chromosome 17p loss or patient survival.40 Thus, the role that TP53 mutations play in medulloblastoma tumorigenesis awaits investigation in larger, prospective randomized clinical trials.
Nevertheless, evidence suggests that signaling through TP53 pathways is abnormal in medulloblastomas. Frank et al.41 described activation of the TP53-P14ARF pathway as a consequence of TP53 mutation, or homozygous deletion or methylation of the P14ARF promoter, particularly in large-cell/anaplastic medulloblastomas. Multiple transgenic mouse models have used Trp53 deletion to accelerate medulloblastoma tumorigenesis.42 We and others have described a role for splice variants of the TP53 family member TP73 in medulloblastoma formation.17,43 Amplification of the TP53 regulator HDM2 is another potential mechanism of medulloblastoma formation. Prior studies have reported that HDM2 amplification is present in 7% of human malignancies that lack mutation of p53.44 HDM2 over-expression has been associated with poor survival among adult patients with medulloblastoma; however, other groups have failed to document HDM2 amplification in pediatric medulloblastomas.37,45,46 We have described amplification and over-expression of the WIP1 oncogene in primary medulloblastomas and cell lines and have implicated high WIP1 expression in chemo-resistance.14 We have verified the results of previous studies that identified increased expression of PPM1D (i.e., WIP1) transcripts in a subset of medulloblastomas (Fig. 1).7,8 The infrequent nature of TP53 mutations in WIP1-amplified malignancies47 and the absence of TP53 mutation in most medulloblastomas suggest that increased WIP1 expression may be one mechanism by which gain of chromosome 17q promotes medulloblastoma growth.
In this study, we investigated the role of the WIP1 oncogene on medulloblastoma growth in TP53 wild-type human medulloblastoma cell lines D556, D425, and Med8A. At baseline, D425 and Med8A cells exhibited relative high levels of WIP1 transcript and protein, compared with that expressed in parental D556 or Daoy14 medulloblastoma cells. To examine the ability of WIP1 to promote medulloblastoma cell growth, we stably transfected either an empty vector, WIP1, or a phosphatase-dead WIP1 (WIP1 D314A)25 into D556 cells. Of interest, expression of WIP1 RNA and protein was significantly higher in D556 cells with stable expression of a mutant, phosphatase-dead WIP1 than in D556-WIP1 stable clones and was similar to expression in endogenous WIP1 high-expressing D425 and Med8A cells. This is likely because TP53 and WIP1 normally function in a negative-feedback control loop. TP53 induces expression of WIP1, which, in turn, dephosphorylates and inactivates TP53.48 Stable transfection of mutant WIP1-D314A is likely to be acting as a dominant negative, reducing endogenous WIP1 function, resulting in reduced viability, compared with wild-type WIP1-expressing or empty vector–containing control cells. This is likely to be the reason that D556-WIP1 D314A stable clones grew at a significantly slower rate as adherent cultures and in anchorage independence than D556 cell clones containing the WIP1 oncogene (Fig. 2C and D; Supplementary material, Fig. S1).
Of interest, stable transfection of WIP1 into TP53-mutant Daoy medulloblastoma cells failed to affect proliferation, compared with Daoy cells stably transfected with an empty vector. Similar to D556 cell clones, Daoy-WIP1 stable clones expressed almost 10-fold more WIP1 protein, compared with expression in Daoy-pcDNA stable clones. However, unlike the D556-WIP1 stable clones, proliferation and the cell-cycle profile were similar between Daoy-WIP1 and Daoy-pcDNA stable clones at all time points (Figs 3C and 6C). This is consistent with prior findings of WIP1 amplification primarily in cancers that retain wild-type TP53 and suggests that TP53 status is important for the effects of WIP1 on medulloblastoma growth.49 However, although the effects were not as striking in TP53 mutated Daoy cells as in TP53 wild-type D556-WIP1 D314A clones, stable expression of a phosphatase-dead WIP1 construct in Daoy cells (Daoy-WIP1 D314A) also inhibited proliferation. This suggests a p53-independent function for WIP1. In fact, another group has previously reported antiproliferative effects of WIP1 knockdown in Daoy medulloblastoma cells.50
We verified the tumorigenic effects of WIP1 in vivo using human medulloblastoma cells and found that the level of expression of WIP1 significantly altered the survival of orthotopic, xenografted immunodeficient mice (Fig. 4). D556-WIP1 stable cells injected into the cerebellum of SCID/Beige mice reduced mouse survival by almost one-half to a median of 35 days, compared with survival of mice injected with the identical number of empty vector-transfected D556 cells. D556-WIP1 xenografted tumors exhibited some histological features of large-cell/anaplastic medulloblastoma, with increased atypia and mitotic figures and increased staining for a marker of cell proliferation, PCNA. Human large-cell medulloblastomas are a highly malignant variant that contain large, round, or pleomorphic nuclei with prominent nucleoli and more abundant cytoplasm than is found in non-large-cell medulloblastomas. Large-cell medulloblastomas also exhibit cell-cell wrapping, high mitotic activity, and a high apoptotic rate.51 Although D556-WIP1 xenografted murine medulloblastomas did not exhibit all the findings of human large-cell tumors, they were histologically distinct from tumors in mice orthotopically xenografted with empty vector D556 cells and were more aggressive with at least one D556-WIP1 xenograft that displayed evidence of leptomeningeal metastasis. Together, our in vitro and in vivo data suggest that WIP1 is an important tumor-promoting gene in medulloblastoma.
We further demonstrate that WIP1 knockdown is associated with increased phosphorylation of WIP1 targets and reduced growth of human medulloblastoma cells with stable (D556-WIP1) and endogenous high expression of WIP1 (D425) (Fig. 7). Treatment of WIP1-expressing medulloblastoma cells with the small-molecule WIP1 inhibitor CCT007093 similarly results in increased phosphorylation of the WIP1 target TP53 and dose-dependent growth inhibition of D556-WIP1 cells (Fig. 8). WIP1 knockdown had no effect on the growth of D556 empty vector-transfected cells. In addition, treatment with CCT007093 similarly had no significant effect on the growth of D556 cells containing a phosphatase-dead WIP1 gene. Our results suggest that medulloblastoma cells become addicted to the growth-promoting properties of the WIP1 gene34 and validate the potential use of a small molecule WIP1 antagonist as a targeted therapeutic agent in malignancies with high WIP1 expression.
The mechanism by which WIP1 promotes medulloblastoma growth is not entirely clear but is mediated in part by HDM2. D556-WIP1 and D556-WIP1 D314A cells both exhibited similar levels of expression and phosphorylation of p53. However, HDM2 levels were slightly higher in D556-WIP1 stable clones (Fig. 5A). HDM2 knockdown inhibited growth of D556-WIP1 stable cells but did not affect the growth of D556 empty vector stable clones. Nutlin-3a, the HDM2 small molecule inhibitor, similarly and most significantly suppressed the growth of D556-WIP1 stable clones grown as adherent cells and in anchorage independence. We also provide evidence for increased growth suppression when both HDM2 and WIP1 are targeted in WIP1 high-expressing medulloblastoma cells (Fig. 9).
Recently published work suggests that aberrant HDM2 expression plays an important role in brain development and in medulloblastoma tumor formation. Mdm2, the mouse homologue of HDM2, is an E3 ubiquitin ligase that has been extensively characterized as an important inhibitor of the tumor suppressive function of p53.52 Absence of Mdm2 expression is known to result in death of mice in utero because of Trp53-mediated apoptosis; lethality can be fully rescued by deletion of the Trp53 gene.53,54 A recent study using mice that express a hypomorphic allele of Mdm2, Mdm2puro/Δ7-9 found that a 70% reduction in expression of Mdm2 protein results in hypoplasia and abnormal foliation of the developing cerebellum in mice. These findings were most pronounced in granule neuron precursor cells, a well-characterized cell of origin of de novo medulloblastoma tumors in mice. Mdm2puro/Δ7-9, Mdm2-deficient mouse granule neuron precursors exhibited increased expression of p53 and its downstream target genes, reduced proliferation, and increased apoptosis. The effects of Mdm2 expression on development of the cerebellum were highly dose-dependent, because mice with 50% Mdm2 expression did not exhibit noticeable cerebellar abnormalities. Furthermore, Mdm2 deficiency inhibited formation of preneoplastic lesions in medulloblastoma-prone Patched1-deficient (Ptch1 +/−) mice.55 Mdm2 plays an important role not only in the appropriate development of the cerebellum but also through its effects on p53 function and therefore may be an important therapeutic target for the treatment of medulloblastoma.
Recent evidence also suggests that HDM2 does not function on its own in regulating the activity of TP53. A recent report suggests an important role for the U-box E3/E4 ligase ubiquitin factor E4B (UBE4B) in regulation of TP53 and in medulloblastoma tumorigenesis. U-box ubiquitin ligases feature a U-box domain, which is structurally related to the RING finger domain required for function of other ubiquitin ligases, such as HDM2.56 Mice harboring homozygous loss-of-function Ube4b mutations die in utero, whereas heterozygotes show multiple nervous system anomalies that worsen with age. UBE4B has recently been shown to be essential for HDM2-mediated polyubiquitination and degredation of TP53. The same study showed an inverse relation between TP53 expression and UBE4B amplification and over-expression in mouse medulloblastomas and in human cell lines and primary medulloblastomas, suggesting a novel target for treatment of pediatric brain tumors.57 It has yet to be determined whether UBE4B can be regulated by phosphorylation events and whether it may be a target of the WIP1 phosphatase.
In conclusion, we have demonstrated that the WIP1 phosphatase is important for medulloblastoma tumor growth in vitro and in vivo. Evidence in cell lines shows that high WIP1 expression is associated with increased expression of HDM2. Targeting HDM2 with Nutlin-3a phenocopies the effects of WIP knock-down or targeting with the small molecule drug CCT007093 in suppression of medulloblastoma growth. This suggests an important role for WIP1 and HDM2 inhibition, either alone or together, especially for the treatment of childhood medulloblastomas that exhibit high expression of the WIP1 oncogene.
Supplementary Material
Funding
This work was supported by St. Baldrick's Foundation Scholar Award (R.C.C.), CURE Childhood Cancer Foundation (R.C.C.), Southeastern Brain Tumor Foundation (R.C.C.), American Association for Cancer Research-Aflac, Inc. Career Development Award (R.C.C.), Emory Egleston Children's Research Center Seed Grant (R.C.C.), Winship Cancer Institute's Molecular Pathways and Biomarker Program Seed Grant (R.C.C.), American Brain Tumor Association (R.C.C.), and Alex's Lemonade Stand-Young Investigator Award (R.C.C.).
Supplementary Material
Acknowledgments
We thank Jennifer McAnally, Rosemary Maxwell, Dianne Alexis, and Dr. Jeanne Kowalski for technical assistance, and Dr. Rita Nahta and Dr. Tobey MacDonald for editorial assistance.
Conflict of interest statement. None declared.
References
- 1.Rood BR, Macdonald TJ, Packer RJ. Current treatment of medulloblastoma: recent advances and future challenges. Semin Oncol. 2004;31:666–675. doi: 10.1053/j.seminoncol.2004.07.009. doi:10.1053/j.seminoncol.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Packer RJ, Rood BR, MacDonald TJ. Medulloblastoma: present concepts of stratification into risk groups. Pediatr Neurosurg. 2003;39:60–67. doi: 10.1159/000071316. doi:10.1159/000071316. [DOI] [PubMed] [Google Scholar]
- 3.Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. doi:10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 4.Biegel JA. Cytogenetics and molecular genetics of childhood brain tumors. Neuro Oncol. 1999;1:139–151. doi: 10.1215/15228517-1-2-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan E, Pellarin M, Holmes E, et al. Isochromosome 17q is a negative prognostic factor in poor-risk childhood medulloblastoma patients. Clin Cancer Res. 2005;11:4733–4740. doi: 10.1158/1078-0432.CCR-04-0465. doi:10.1158/1078-0432.CCR-04-0465. [DOI] [PubMed] [Google Scholar]
- 6.Rossi MR, Conroy J, McQuaid D, Nowak NJ, Rutka JT, Cowell JK. Array CGH analysis of pediatric medulloblastomas. Genes Chromosomes Cancer. 2006;45:290–303. doi: 10.1002/gcc.20292. doi:10.1002/gcc.20292. [DOI] [PubMed] [Google Scholar]
- 7.Kool M, Koster J, Bunt J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS ONE. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. doi:10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Northcott PA, Korshunov A, Witt H, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. doi:10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu X, Nguyen TA, Zhang X, Donehower LA. The Wip1 phosphatase and Mdm2: cracking the “Wip” on p53 stability. Cell Cycle. 2008;7:164–168. doi: 10.4161/cc.7.2.5299. doi:10.4161/cc.7.2.5299. [DOI] [PubMed] [Google Scholar]
- 10.Bulavin DV, Demidov ON, Saito S, et al. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat Genet. 2002;31:210–215. doi: 10.1038/ng894. doi:10.1038/ng894. [DOI] [PubMed] [Google Scholar]
- 11.Nannenga B, Lu X, Dumble M, et al. Augmented cancer resistance and DNA damage response phenotypes in PPM1D null mice. Mol Carcinog. 2006;45:594–604. doi: 10.1002/mc.20195. doi:10.1002/mc.20195. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Yang Y, Peng Y, et al. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31:133–134. doi: 10.1038/ng888. doi:10.1038/ng888. [DOI] [PubMed] [Google Scholar]
- 13.Saito-Ohara F, Imoto I, Inoue J, et al. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res. 2003;63:1876–1883. [PubMed] [Google Scholar]
- 14.Castellino RC, De Bortoli M, Lu X, et al. Medulloblastomas overexpress the p53-inactivating oncogene WIP1/PPM1D. J Neurooncol. 2008;86:245–256. doi: 10.1007/s11060-007-9470-8. doi:10.1007/s11060-007-9470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendrzyk F, Radlwimmer B, Joos S, et al. Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J Clin Oncol. 2005;23:8853–8862. doi: 10.1200/JCO.2005.02.8589. doi:10.1200/JCO.2005.02.8589. [DOI] [PubMed] [Google Scholar]
- 16.Qu Y, He F, Chen Y. Different effects of the probe summarization algorithms PLIER and RMA on high-level analysis of Affymetrix exon arrays. BMC Bioinformatics. 2010;11:211. doi: 10.1186/1471-2105-11-211. doi:10.1186/1471-2105-11-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castellino RC, De Bortoli M, Lin LL, et al. Overexpressed TP73 induces apoptosis in medulloblastoma. BMC Cancer. 2007;7:127. doi: 10.1186/1471-2407-7-127. doi:10.1186/1471-2407-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moon SH, Lin L, Zhang X, et al. Wild-type p53-induced phosphatase 1 dephosphorylates histone variant gamma-H2AX and suppresses DNA double strand break repair. J Biol Chem. 2010;285:12935–12947. doi: 10.1074/jbc.M109.071696. doi:10.1074/jbc.M109.071696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, Kim GJ, Miyoshi H, et al. Efficiency of the elongation factor-1alpha promoter in mammalian embryonic stem cells using lentiviral gene delivery systems. Stem Cells Dev. 2007;16:537–545. doi: 10.1089/scd.2006.0088. doi:10.1089/scd.2006.0088. [DOI] [PubMed] [Google Scholar]
- 20.Kreth S, Heyn J, Grau S, Kretzschmar HA, Egensperger R, Kreth FW. Identification of valid endogenous control genes for determining gene expression in human glioma. Neuro Oncol. 2010;12:570–579. doi: 10.1093/neuonc/nop072. doi:10.1093/neuonc/nop072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. doi:10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Kim JY, Nelson AL, Algon SA, et al. Medulloblastoma tumorigenesis diverges from cerebellar granule cell differentiation in patched heterozygous mice. Dev Biol. 2003;263:50–66. doi: 10.1016/s0012-1606(03)00434-2. doi:10.1016/S0012-1606(03)00434-2. [DOI] [PubMed] [Google Scholar]
- 23.von Bueren AO, Shalaby T, Rajtarova J, et al. Anti-proliferative activity of the quassinoid NBT-272 in childhood medulloblastoma cells. BMC Cancer. 2007;7:19. doi: 10.1186/1471-2407-7-19. doi:10.1186/1471-2407-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bunt J, de Haas TG, Hasselt NE, et al. Regulation of cell cycle genes and induction of senescence by overexpression of OTX2 in medulloblastoma cell lines. Mol Cancer Res. 2010;8:1344–1357. doi: 10.1158/1541-7786.MCR-09-0546. doi:10.1158/1541-7786.MCR-09-0546. [DOI] [PubMed] [Google Scholar]
- 25.Fujimoto H, Onishi N, Kato N, et al. Regulation of the antioncogenic Chk2 kinase by the oncogenic Wip1 phosphatase. Cell Death Differ. 2006;13:1170–1180. doi: 10.1038/sj.cdd.4401801. doi:10.1038/sj.cdd.4401801. [DOI] [PubMed] [Google Scholar]
- 26.Thullberg M, Stromblad S. Anchorage-independent cytokinesis as part of oncogenic transformation? Cell Cycle. 2008;7:984–988. doi: 10.4161/cc.7.8.5674. doi:10.4161/cc.7.8.5674. [DOI] [PubMed] [Google Scholar]
- 27.Atienza JM, Zhu J, Wang X, Xu X, Abassi Y. Dynamic monitoring of cell adhesion and spreading on microelectronic sensor arrays. J Biomol Screen. 2005;10:795–805. doi: 10.1177/1087057105279635. doi:10.1177/1087057105279635. [DOI] [PubMed] [Google Scholar]
- 28.Mosse YP, Laudenslager M, Longo L, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. doi:10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castellino RC, Barwick BG, Schniederjan M, et al. Heterozygosity for Pten promotes tumorigenesis in a mouse model of medulloblastoma. PLoS ONE. 2010;5:e10849. doi: 10.1371/journal.pone.0010849. doi:10.1371/journal.pone.0010849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drakos E, Thomaides A, Medeiros LJ, et al. Inhibition of p53-murine double minute 2 interaction by nutlin-3A stabilizes p53 and induces cell cycle arrest and apoptosis in Hodgkin lymphoma. Clin Cancer Res. 2007;13:3380–3387. doi: 10.1158/1078-0432.CCR-06-2581. doi:10.1158/1078-0432.CCR-06-2581. [DOI] [PubMed] [Google Scholar]
- 31.Gu L, Zhu N, Findley HW, Zhou M. MDM2 antagonist nutlin-3 is a potent inducer of apoptosis in pediatric acute lymphoblastic leukemia cells with wild-type p53 and overexpression of MDM2. Leukemia. 2008;22:730–739. doi: 10.1038/leu.2008.11. doi:10.1038/leu.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Maerken T, Speleman F, Vermeulen J, et al. Small-molecule MDM2 antagonists as a new therapy concept for neuroblastoma. Cancer Res. 2006;66:9646–9655. doi: 10.1158/0008-5472.CAN-06-0792. doi:10.1158/0008-5472.CAN-06-0792. [DOI] [PubMed] [Google Scholar]
- 33.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. discussion 80 doi:10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 34.Rayter S, Elliott R, Travers J, et al. A chemical inhibitor of PPM1D that selectively kills cells overexpressing PPM1D. Oncogene. 2008;27:1036–1044. doi: 10.1038/sj.onc.1210729. doi:10.1038/sj.onc.1210729. [DOI] [PubMed] [Google Scholar]
- 35.Burnett ME, White EC, Sih S, von Haken MS, Cogen PH. Chromosome arm 17p deletion analysis reveals molecular genetic heterogeneity in supratentorial and infratentorial primitive neuroectodermal tumors of the central nervous system. Cancer Genet Cytogenet. 1997;97:25–31. doi: 10.1016/s0165-4608(96)00319-6. doi:10.1016/S0165-4608(96)00319-6. [DOI] [PubMed] [Google Scholar]
- 36.Saylors RL, 3rd, Sidransky D, Friedman HS, et al. Infrequent p53 gene mutations in medulloblastomas. Cancer Res. 1991;51:4721–4723. [PubMed] [Google Scholar]
- 37.Adesina AM, Nalbantoglu J, Cavenee WK. p53 gene mutation and mdm2 gene amplification are uncommon in medulloblastoma. Cancer Res. 1994;54:5649–5651. [PubMed] [Google Scholar]
- 38.Tabori U, Baskin B, Shago M, et al. Universal poor survival in children with medulloblastoma harboring somatic TP53 mutations. J Clin Oncol. 2010;28:1345–1350. doi: 10.1200/JCO.2009.23.5952. doi:10.1200/JCO.2009.23.5952. [DOI] [PubMed] [Google Scholar]
- 39.Gessi M, von Bueren AO, Rutkowski S, Pietsch T. p53 expression predicts dismal outcome for medulloblastoma patients with metastatic disease. J Neurooncol. 2011;106:135–141. doi: 10.1007/s11060-011-0648-8. [DOI] [PubMed] [Google Scholar]
- 40.Pfaff E, Remke M, Sturm D, et al. TP53 Mutation Is Frequently Associated With CTNNB1 Mutation or MYCN Amplification and Is Compatible With Long-Term Survival in Medulloblastoma. J Clin Oncol. 2010;28:5188–5196. doi: 10.1200/JCO.2010.31.1670. doi:10.1200/JCO.2010.31.1670. [DOI] [PubMed] [Google Scholar]
- 41.Frank AJ, Hernan R, Hollander A, et al. The TP53-ARF tumor suppressor pathway is frequently disrupted in large/cell anaplastic medulloblastoma. Brain Res Mol Brain Res. 2004;121:137–140. doi: 10.1016/j.molbrainres.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 42.Huse JT, Holland EC. Genetically engineered mouse models of brain cancer and the promise of preclinical testing. Brain Pathol. 2009;19:132–143. doi: 10.1111/j.1750-3639.2008.00234.x. doi:10.1111/j.1750-3639.2008.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zitterbart K, Zavrelova I, Kadlecova J, et al. p73 expression in medulloblastoma: TAp73/DeltaNp73 transcript detection and possible association of p73alpha/DeltaNp73 immunoreactivity with survival. Acta Neuropathol. 2007;114:641–650. doi: 10.1007/s00401-007-0298-2. doi:10.1007/s00401-007-0298-2. [DOI] [PubMed] [Google Scholar]
- 44.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. doi:10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 45.Giordana MT, Duo D, Gasverde S, et al. MDM2 overexpression is associated with short survival in adults with medulloblastoma. Neuro Oncol. 2002;4:115–122. doi: 10.1093/neuonc/4.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Batra SK, McLendon RE, Koo JS, et al. Prognostic implications of chromosome 17p deletions in human medulloblastomas. J Neurooncol. 1995;24:39–45. doi: 10.1007/BF01052657. doi:10.1007/BF01052657. [DOI] [PubMed] [Google Scholar]
- 47.Lu X, Nguyen TA, Moon SH, Darlington Y, Sommer M, Donehower LA. The type 2C phosphatase Wip1: an oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metastasis Rev. 2008;27:123–135. doi: 10.1007/s10555-008-9127-x. doi:10.1007/s10555-008-9127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007;12:342–354. doi: 10.1016/j.ccr.2007.08.033. doi:10.1016/j.ccr.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 49.Rauta J, Alarmo EL, Kauraniemi P, Karhu R, Kuukasjarvi T, Kallioniemi A. The serine-threonine protein phosphatase PPM1D is frequently activated through amplification in aggressive primary breast tumours. Breast Cancer Res Treat. 2006;95:257–263. doi: 10.1007/s10549-005-9017-7. doi:10.1007/s10549-005-9017-7. [DOI] [PubMed] [Google Scholar]
- 50.Baxter EW, Milner J. p53 Regulates LIF expression in human medulloblastoma cells. J Neurooncol. 2010;97:373–382. doi: 10.1007/s11060-009-0043-x. doi:10.1007/s11060-009-0043-x. [DOI] [PubMed] [Google Scholar]
- 51.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. doi:10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bond GL, Hu W, Levine AJ. MDM2 is a central node in the p53 pathway: 12 years and counting. Curr Cancer Drug Targets. 2005;5:3–8. doi: 10.2174/1568009053332627. doi:10.2174/1568009053332627. [DOI] [PubMed] [Google Scholar]
- 53.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. doi:10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 54.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. doi:10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 55.Malek R, Matta J, Taylor N, Perry ME, Mendrysa SM. The p53 inhibitor MDM2 facilitates Sonic Hedgehog-mediated tumorigenesis and influences cerebellar foliation. PLoS ONE. 2011;6:e17884. doi: 10.1371/journal.pone.0017884. doi:10.1371/journal.pone.0017884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marin I. Ancient origin of animal U-box ubiquitin ligases. BMC Evol Biol. 2010;10:331. doi: 10.1186/1471-2148-10-331. doi:10.1186/1471-2148-10-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu H, Pomeroy SL, Ferreira M, et al. UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat Med. 2011;17:347–355. doi: 10.1038/nm.2283. doi:10.1038/nm.2283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.