Abstract
Age is a major risk factor for many human diseases. Extremely long-lived individuals, such as centenarians, have managed to ward off age-related diseases and serve as human models to search for the genetic factors that influence longevity. The discovery of evolutionarily conserved pathways with major impact on life span in animal models has provided tantalizing opportunities to test the relevance of these pathways for human longevity. Here we specifically focus on the insulin/insulin-like growth factor-1 signaling as a prime candidate pathway. Coupled with the rapid advances in ultra high-throughput sequencing technologies, it is now feasible to comprehensively analyze all possible sequence variants in candidate genes segregating with a longevity phenotype and to investigate the functional consequences of the associated variants. A better understanding of the functional genes that affect healthy longevity in humans may lead to a rational basis for intervention strategies that can delay or prevent age-related diseases.
Keywords: Longevity, Functional genomics, IGF-1 signaling
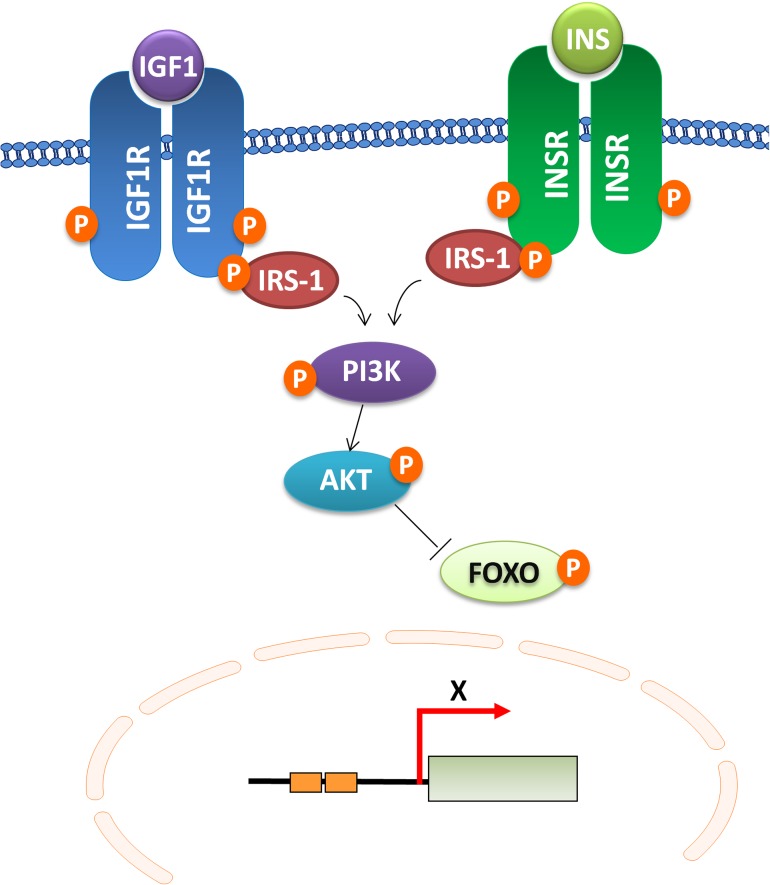
Rather than driven by stochastic events alone and being a simple entropic process of deterioration, the rate of aging and life span have been shown to be regulated by multiple pathways. Many single-gene mutations identified in invertebrates result in a reduced rate of aging and an increased life span (1,2). A large fraction of these mutations extends life span in mammals as well (3–5). These studies have provided important clues for discovering the evolutionarily conserved pathways of life span and their impact on the aging rate. Altering the pathways controlling metabolism, endocrine signaling, nutrient sensing, stress resistance, and telomere maintenance has been shown to prolong life span from yeast to mammals (6,7). Among those conserved pathways, the insulin/insulin-like growth factor-1 (IGF-1) signaling (IIS) pathway was the first one shown to regulate life span in animal models and also had the most profound effect on life-span extension (2,8). In brief, binding of IGF-1 and insulin activates an intracellular signaling cascade composed of IRS-1/PI3K/AKT that antagonizes the activity of Forkhead/FOXO transcription factors (Figure 1). Reduction-of-function mutations in IIS genes activate Forkhead/FOXO proteins that regulate expression of several hundred genes implicated in metabolism, stress resistance, and antimicrobial defense (9,10). Remarkably, these long-lived animals maintain their youthfulness (11,12) and display delayed signs of aging and a late onset of age-related diseases (12). Continued discoveries of beneficial effects of mutations on many aspects of aging in long-lived animals support the concept that aging is a major risk factor for many common human diseases and that targeting aging is a logical and economically sound strategy in developing new targets for prevention and early-stage treatment.
Figure 1.
Insulin/insulin-like growth factor-1 (IGF-1) signaling (IIS) is a major candidate pathway in human longevity. Binding of IGF-1 or insulin (INS) initiates the evolutionarily conserved IRS–PI3K–AKT signaling cascade to phosphorylate Forkhead transcription factor FOXO. When phosphorylated, FOXO is retained in the cytoplasm. Mutations perturbing the IIS pathway reduce phosphorylation of FOXO, enabling it to enter the nucleus and regulate transcription of diverse genes that collectively contribute to the extension of life span. The role of reduced IIS in life-span extension is well-established in model organisms from yeast to mammals, suggesting that altered IIS may also influence human life span. Human population studies established genetic associations between some of the IIS genes (19,20), for example, IGF1R, AKT, FOXO3a, and longevity, supporting the role of IIS pathway in human longevity.
IIS PATHWAY
The single-gene mutations extending life span were first discovered in nematode, Caenorhabditis elegans (13,14). Hypomorphic mutations of daf-2, encoding nematode ortholog of insulin receptor/IGF-1 receptor (IGF1R; 15), and age-1, encoding a PI3K ortholog (16), result in profound extension of life span (Figure 1). The IIS pathway negatively regulates the transcriptional activity of the Forkhead transcription factor, daf-16/FOXO, and limits longevity (17,18). Mutations that attenuate signal transduction within the IIS pathway reduce phosphorylation of DAF-16, which enables DAF-16 to translocate into the nucleus, thereby modulating transcription of several hundred target genes in distinct families including antioxidants, chaperones, and antimicrobial defenses (21–23). Inhibition of IIS also activates the heat-shock transcription factor-1 (24,25) and the Nrf-like xenobiotic-response factor, SKN-1 (26). Once activated, heat-shock transcription factor-1 induces expression of small heat-shock proteins, and SKN-1 induces genes involved in phase 2 detoxification response. Overall, IIS exerts its effects on aging in C. elegans by modulating expression of downstream genes through these transcription factors. In Drosophila melanogaster, mutations in the insulin receptor homolog and the insulin receptor substrate, CHICO, also impair IIS and prolong life span in females (27–29). Similar to C. elegans, ablation of the daf-16 homolog, dFOXO, almost completely blocks IIS-mediated life-span extension (30), whereas tissue-specific overexpression of dFOXO extends life span (31,32).
During evolution, the IIS pathway has diverged from a single receptor in invertebrates to multiple receptors and pathways forming more complicated regulatory networks in mammals with a certain degree of redundancy (33). Insulin regulates metabolic pathways, while IGF-1 controls cell growth and differentiation (33,34). Therefore, translating longevity effects of IIS gene mutations discovered in invertebrates to mammals is neither plain nor simple. However, several genetic manipulations in mice have shown that interference of this pathway prolongs murine life span. Mutations that reduce growth hormone (GH) production or GH receptor activity result in small body size (dwarfism), numerous indices of delayed aging, enhanced stress resistance, and a major increase in the life span of mice (34–41). GH is the major regulator of circulating IGF-1 suggesting that GH functions are mediated at least partially through IGF-1 action. Alterations in IIS emerged as one of the likely mechanisms linking GH with aging. The most compelling model seems to be the heterozygous IGF1R knockout mouse. Whereas mice homozygous for IGF1R knockout die at birth, female heterozygous mice live 33% longer than wild-type mice and show an increased resistance to oxidative stress (42). The same genetic alteration also protected animals from Alzheimer’s-like disease symptoms, including reduced behavioral impairment, neuroinflammation, and neuronal loss (43). Adipose tissue–specific homozygous deletion of the insulin receptor (INSR) increases life span in both sexes (44). Most recently, the homozygous deletion of IRS-1, which serves as an adapter protein that transmits signals from both IGF-1 and insulin receptors, extended life span and increased resistance to various age-related pathologies only in female mice (45). These results suggest that the GH/IGF pathway is critical in murine longevity, where the effects of reduced IIS is often more profound in females (35). Similarly, as described later, the results from human genetic studies suggest that the association of IIS gene variants with human longevity is sometimes more significant in women.
IIS PATHWAY AND HUMAN LONGEVITY
Despite the parallel between the IIS in invertebrates and mammals (3,6), the importance of this regulatory network for aging and longevity in human populations is uncertain. The question is, do candidate loci with major effects on longevity show segregating allelic variation in natural populations? Indeed, even if we adopt the premise that the basic mechanisms that control longevity and determine the rate of aging are common to all multicellular organisms, it is possible that the observed effects of candidate gene variants on longevity will prove to be exclusively associated with laboratory strains, but not in the same animals in the wild. This concept is illustrated by an elegant work by Walker and coworkers (46), who exposed mixtures of mutants and wild-type age-1 worms to more natural conditions, that is, cycles of starvation. A large reduction in the frequency of the mutant allele suggested a substantial difference in relative fitness. Furthermore, it has been demonstrated that daf-2 worms with partial loss-of-function of the IGF1R-like protein exhibit dramatically reduced fitness even under laboratory conditions (47). Thus, it remains to be demonstrated whether candidate loci with major effects on longevity in model organisms such as genes involved in insulin/IGF-1 pathway will show segregating allelic variation in human populations. Moreover, humans suffer from different age-related diseases than mice. Therefore, the only valid strategy to test the possibility that genetic variation at candidate loci contributes to phenotypic variance for longevity and aging phenotypes in natural populations would be to conduct population-based studies.
MODELS OF HUMAN LONGEVITY: CENTENARIANS
Longevity is known to have a genetic component. Although debated, the heritability of average life expectancy has been estimated to be approximately 25% (48–50). Family studies on centenarians, those who aged 100 years or more, suggest that the relationship between genetics and longevity is stronger in the oldest-old adults. For example, the siblings of centenarians have a four times greater probability of surviving to age 90 than the siblings of people who have an average life expectancy (52). When it comes to living 100 years, the probability is 17 times greater for male siblings of centenarians and 8 times greater in fe` siblings of centenarians compared with average life span of their birth cohort (53). The immediate ancestors of Jeanne Calment, the longest ever lived human being who died at the age of 122 years and 164 days, were shown to have more than a 10-fold higher probability of living to 80 years or more compared with the control ancestors of a reference family (54). These studies support the utility of centenarians as a model system for studying the genetics of longevity, with extreme age as the phenotype toward the discovery of genetic variations predisposing people to longevity. Therefore, genetic studies of centenarians are based on the premise that they may lead to the identification of alleles that are either particularly enriched in these populations due to positive effects on extreme longevity or under represented due to a negative impact on human health. With the recognition that the IIS pathway regulates life span and healthy aging in animal models, genes involved in IIS become attractive candidates for investigating the genetic basis of human longevity using centenarians as a model (candidate gene approaches).
GENETIC ASSOCIATION STUDIES IN HUMAN LONGEVITY: IIS AS CANDIDATES
Candidate gene approaches focus on the genes or pathways that are selected based on a priori hypotheses about their role in the phenotype (55). A standard linkage approach is usually not feasible due to an almost complete absence of extensive pedigrees of long-lived individuals and the inherent complexity of longevity phenotypes, which likely involve multiple genes with small effects. Association-based studies are more effective tools than linkage studies when the focus is on complex traits such as longevity because they have greater statistical power to detect genes of small effect (56–59). By comparing the frequency of genetic variants in cases (long-lived individuals, nonagenarians, or centenarians) and unrelated controls (elderly individuals), association studies evaluate the correlation between a genetic variation and a particular trait, eg, longevity (Figure 2). Single-nucleotide polymorphisms (SNPs) are the most common type of genetic variations in the human genome and have been the workhorse of association studies. As described later, the results from genetic association studies of both common SNPs and rare variants point to a role of the IIS pathway in human longevity.
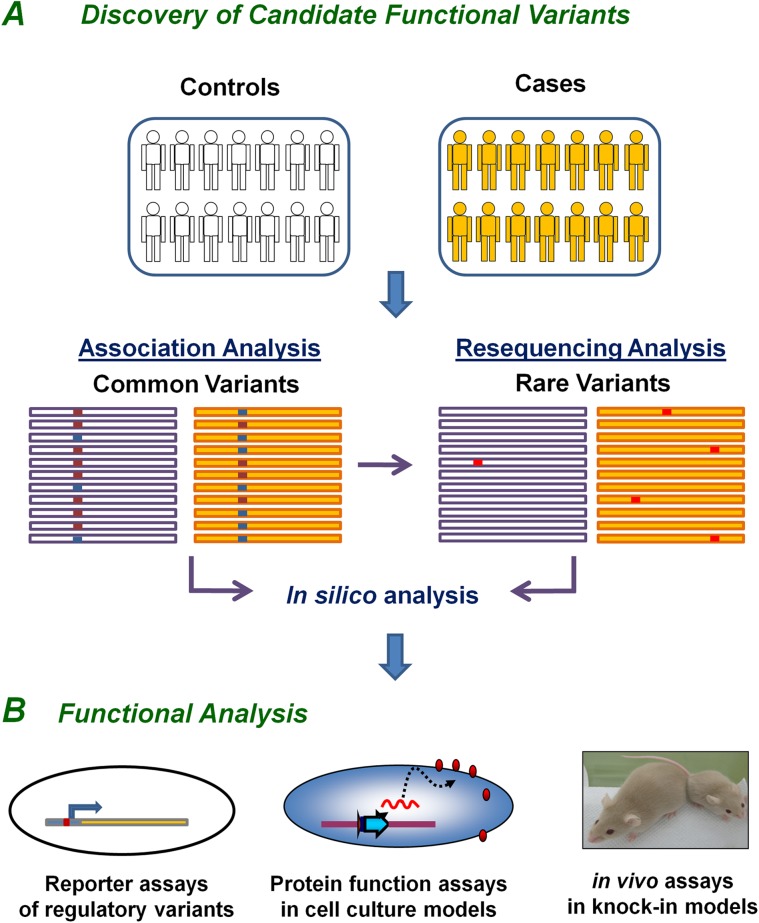
Figure 2.
Discovery of functional insulin/insulin-like growth factor-1 signaling (IIS) variants in human longevity. A hierarchical and multidisciplinary approach will increase the chances of identifying functional variations in the IIS genes that influence human longevity. (A) Genetic association in case/control studies establishes genetic link between IIS genes and human longevity. Identification of potentially functional variants requires either resequencing of target loci based on common single-nucleotide polymorphism association analyses or direct resequencing of candidate genes. Individually rare longevity-associated variants may be enriched in cases as a group as compared with controls. In silico analysis predicts the outcomes of potentially functional variants based on the changes in amino acid sequence, transcription factor binding, micro RNA binding, or splicing and helps prioritize candidate variants for further functional analysis. (B) Integrated multiple in vivo and in silico assays are needed to assess the functional roles of each longevity-associated IIS variants depending on the nature and location of gene variants (see text for detail).
Common IIS Variants and Human Longevity
In most candidate gene approaches, SNPs are selected based on linkage disequilibrium, which describes the nonrandom correlation between alleles at a pair of SNPs. The HapMap provides global coverage of the genome with SNP markers (60,61) to select nonredundant tagSNPs based on defined haplotype patterns. In this format, genetic association analysis is an indirect approach using a set of SNPs as surrogate markers to detect association between a candidate gene and longevity, whether or not the markers themselves have functional effects. The search for the causative variants would then be limited to the regions showing association with longevity. In addition, selected SNPs are necessarily common (minor allele frequency >5%–10%) to achieve statistical power to detect an association with a given sample size.
Six polymorphisms from IIS pathway genes (GHRHR, GH1, IGF-1, INS, and IRS1) that were previously reported to be associated with the gene expression or protein function were analyzed in Leiden 85 plus cohort by van Heemst and coworkers (62). The analyses were stratified for gender because reduced IIS extends life span of females more than in both D. melanogaster and mice. The GH1 SNP (rs2665802) was associated with reduced body height and relative mortality in women only, whereas most SNP alleles that were associated with reduced signaling showed only a trend with the same directionality. To address the combined effect of the six polymorphisms, they used an additive model (composite IIS6 score) in which each of the SNPs was weighted equally because they had no a priori knowledge regarding the relative effect of individual SNPs. They calculated composite IIS6 score by assigning a value of −1 for an allele expected to have reduced IIS activity or +1 for an allele predicted to cause increased IIS activity, therefore an individual bearing all six alleles predicted to have reduced IIS would have an IIS6 score of −6. Association of IIS6 score with height and longevity was observed only in women.
The IGF1R SNP (rs2229765) at nucleotide 3174 of messenger RNA has also been shown to be associated with lower plasma IGF-1 levels and is more frequent in long-lived individuals (63). For FOXO1A, two intronic SNPs (rs10507486 and rs4943794) were associated with age at death in a Genome-Wide Association Study (GWAS) of the Framingham study (64), and two other intronic SNPs (rs2755209 and rs2755213) showed association with the longevity of Chinese women (65). Recently, a systematic study was conducted to analyze 291 SNPs in 30 genes involved in the IIS pathway, showing the association of several genes, including IGF1R, FOXO3A, and AKT, with longevity in Caucasian women from the Study Osteoporotic Fracture cohort (19). The association of AKT and FOXO3A was replicated in other Caucasian populations, eg, Cardiovascular Health Study and Ashkenazi Jewish Centenarians. Among the associated IIS genes, FOXO3A appears to be the most robust longevity gene in humans because common variants in this gene showed consistent association with longevity in multiple populations with different ethnic backgrounds including Asians, Europeans, Caucasians, and African Americans (19,64–68). Even though each study selected different SNPs for genotyping, the fact that these SNPs show the same effect and many of them are in high linkage disequilibrium suggests that the FOXO3A gene plays a role in longevity.
Rare IIS Variants and Human Longevity
Although there is great enthusiasm for the candidate association approach using marker SNPs based on the expectation that statistically powerful genetic association studies are now feasible, a major criticism still remains. This involves the assumption that there is little allelic heterogeneity within loci and that susceptibilities for longevity are due to a small number of ancient polymorphisms that occur at high frequencies in all populations as argued in the common disease/common variant hypothesis (69,70). However, if late-onset phenotypes, such as longevity, are due to large numbers of rare variants at many loci, this strategy would fail, as no single haplotype would be strongly associated with longevity, and the contribution of most individual variants would be too small (71). These concerns turned out to be true even for common traits as evident from the GWAS. More than 1,000 published GWAS reported significant (p < 5 × 10−8) associations of approximately 4,000 SNPs for more than 200 traits/diseases (72). However, each locus has a surprisingly low to modest effect. Furthermore, there is a wide gap between the population variation in disease that is explained by the results of GWAS (usually <10%) and heritability estimates (often >50%; [73]). Much of the speculation about missing heritability from GWAS has focused on the possible contribution of rare variants (minor allele frequency < 0.5%; 74).
Rare variants would not be strongly associated with any common alleles defined by the tagSNPs and are not queried in most case–control studies. Because extreme longevity is a rare phenotype (1/10,000; 75), only an exhaustive and comprehensive approach will ensure that no rare but important functional SNP escapes attention. So far, we were the first to report comprehensive resequencing analysis of IGF-1 and IGF1R genes in centenarians (n = 79) and controls (n = 161) leading to the discovery of two potentially functional missense variants in the IGF1R gene, Ala37Thr and Arg407His (76). Although rare in frequencies, these variants are significantly enriched in centenarians (n = 9) as compared with controls (n = 1) among a total of 384 centenarians and 312 controls tested (76). As increasingly recognized, most functional SNPs might be rare (77,78) and most rare variants arise as new mutations, which most likely reduce the gene function (79). These results suggest that centenarians may harbor individually rare but collectively more common genetic variations in genes encoding components of the GH/IGF signaling pathway. Additional comprehensive genetic studies in the centenarians should reveal other molecules within the IIS pathway that are important in human longevity. Unprecedented advances in sequencing technology, namely next generation sequencing, will facilitate the confirmation and expansion of the role of IIS pathway in human longevity.
FUNCTIONAL CHARACTERIZATION OF LONGEVITY-ASSOCIATED IIS GENE VARIANTS
The candidate gene approach establishes possible associations between genetic variants in the IIS pathway and longevity outcome. However, association studies typically leave open the question of whether the associated genetic variant is functionally important or serves only as a proxy, a genetic marker co-inherited with the functional allele. To complete a genetic study with solely a statistical end point is unsatisfactory in view of the uncertainties associated with the statistical interpretation. To minimize possible errors and spurious association, an integrated approach is required to assess the functional relevance of gene variations at the molecular level and to overcome the statistical limitations. Moreover, functional variants are known to be rare in frequency (80), and these rare SNPs are not queried in most case–control studies due to the insufficient statistical power (78). In this case, it is critical to directly test their impact by functional analysis. Here, we will describe later the specific approaches to test the functionality of longevity-associated IIS gene variants focusing on the IGF1R variants that we have discovered (76). In principle, these approaches can be applied to other common associated variants or potentially functional rare variants in IIS or other traits. For associated common variants of a candidate gene, a resequencing step is required to discover potentially functional variants before extensive functional analysis (Figure 2).
In Silico Analysis
Potential functionality of SNPs in the associated loci can be determined by in silico analysis. There are a number of computational tools available enabling one to predict the impact of missense SNPs on protein function and structure or regulatory SNPs on gene expression. If appropriately used, these tools greatly reduce the efforts required to test all possible missense SNPs or regulatory SNPs by allowing one to identify and prioritize candidate SNPs before an exhaustive functional assay. For coding SNPs, publicly available tools, such as SIFT (81) and PolyPhen (82), can be utilized to predict whether an amino acid substitution at a particular position in a protein will have a phenotypic effect, and SWISS-MODEL (83) can be used for automated comparative modeling of three-dimensional protein structure. For regulatory SNPs, databases such as TRANSFAC (84), Eukaryotic Promoter Database (85), or MatInspector (86) can be utilized to predict putative transcription factor binding sites and other cis-regulatory elements as well as PupaSNP (87) and ESEfinder (88) for the prediction of exonic splicing enhancers. In silico analysis of putative functionally important SNPs can pave the way for further experimental studies of the associated variants for the effects on the regulation of gene expression or protein function.
In Vitro Cell Culture Models
Cell lines established from human subjects are a useful in vitro model for studying genetic control as they may reflect the functional characteristic of the donor. Lymphoblastoid cell lines established from subjects have been used for quick readouts of the potential outcomes of particular gene variants. Caution needs to be taken as they are known to be oligoclonal in origin and thus may reflect gene actions of B cells at different stages of development from stem cells to plasma cells. The utilization and value of these cell lines for studying genotype-driven molecular endpoints have been demonstrated in functional analysis of the longevity-associated IGF1R variants (76). When treated with IGF-1, lymphoblastoid cell lines established from centenarian carriers of the rare variants, Ala37Thr or Arg407His, showed reduced activities of IGF1R compared with lymphoblastoid cell lines from centenarian noncarriers. However, because complete genotype information for the IGF1R rare variant carriers is not known, functional information obtained from lymphoblastoid cell lines of these subjects is still correlational and cannot establish a cause–effect relationship between the variants and the associated phenotypes, for example, longevity. To determine whether these human longevity-associated IGF1R variants directly affect IGF-1 signaling, mouse embryonic fibroblasts expressing the different human IGF1R variants in a mouse Igf1r null background were generated. When treated with IGF-1, mouse embryonic fibroblasts expressing the human longevity-associated IGF1R variants, Ala37Thr and Agr407His, showed attenuation of IGF-1 signaling, compared with mouse embryonic fibroblasts expressing the wild-type IGF1R. This was demonstrated by a significant reduction in the phosphorylation of both IGF1R and AKT after IGF-1 treatment, reduced expression of many IGF-1-activated genes involved in mitogenesis, apoptosis and differentiation processes, and impaired cell cycle progression (89). These results convincingly demonstrated that the human longevity-associated IGF1R variants are reduced function mutations similar to the IIS mutations causing life-span extension in model organisms (3). The mechanistic parallels between the effects of IIS mutations in model organisms and those produced by longevity-associated IGF1R mutations in humans suggest that evolutionary conservation of IIS reduction and extreme longevity includes humans.
FUTURE PROSPECTS
Association studies have provided a sound-basis for the involvement of IIS in human longevity. Common variants in some IIS candidate loci, including AKT and FOXO genes, are associated with longevity. Particularly, the association of FOXO3a variants is robust and has been replicated in many diverse population-based studies. The next challenge is moving from statistical association to functional mechanism. Technological advances have made it possible to use high-throughput sequencing as a primary discovery tool in genetics (72), such as whole-genome sequencing or exom sequencing, specifically for identifying rare variants. The statistical challenge of estimating the influence of rare variants can be overcome by functional analysis of these variants to ascertain their biological significance. Despite successes in identifying IIS genes associated with longevity, the functional variants and their molecular basis have only been determined in onefigure 1 has been cited after reference (8) in the text, “(70, 89)” given in the figure caption has to be renumbered for sequential order and in turn all the reference citations also have to be renumbered. study (89). Discovery of the reduced-function variants in IGF1R that are rare but collectively enriched in centenarians as compared with controls provide the mechanistic parallel of IIS mutations between model organisms and humans. Although performed in short-term cell culture models, these results further support the role of IIS in human longevity. Taken together, a combination of common and rare SNP variants in IIS likely contributes to exceptional longevity.
Understanding the mechanisms by which longevity-associated IIS variants contribute to human longevity will be critically important to facilitate the development of strategies that can delay aging and promote health span. The complexity of aging and longevity phenotypes poses a daunting yet exciting challenge to establish functionality and causality of longevity-associated IIS variants. A variety of experimental approaches in multiple model systems are needed to elucidate the functional consequences of gene variants. For regulatory variants, in vitro reporter assays can be utilized to characterize the variants in a promoter or enhancer region (Figure 2B). Genotype–phenotype relationship can be directly assessed in patient-derived cells as described for lymphoblastoid cells. Recently, induced pluripotent stem cells generated from patient-derived primary cells such as fibroblasts present potential opportunities for human gene variant modeling to interrogate the cellular and biochemical consequences of gene variants (73). The field is poised to make important advances.
Eventually, the in vivo roles of longevity-associated IIS variants will have to be determined especially for their impact on life span. Classically, the generation of knock-in models expressing human gene variants in model organisms has been explored and mice have been the model system par excellence for human genetic variants. However, caution needs to be taken in interpreting the results because human phenotypes may not be successfully replicated in mice owing to fundamental biological differences between the two species. Hopefully, identification and elucidation of molecular pathways leading to exceptional longevity in humans could provide a rational basis for intervention strategies that slow aging and delay the onset of age-related diseases.
Funding
National Institutes of Health (AG024391, AG027734, and AG17242).
Acknowledgments
C.T. is a recipient of Ellison/American Federation for Aging Research postdoctoral fellowship. Y.S. is the recipient of a Glenn Award for Research in Biological Mechanisms of Aging.
References
- 1.Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 2007;3:1565–1571. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 4.Bartke A. Insulin and aging. Cell Cycle. 2008;7:3338–3343. doi: 10.4161/cc.7.21.7012. [DOI] [PubMed] [Google Scholar]
- 5.Barbieri M, Bonafe M, Franceschi C, Paolisso G. Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am J Physiol Endocrinol Metab. 2003;285:E1064–E1071. doi: 10.1152/ajpendo.00296.2003. [DOI] [PubMed] [Google Scholar]
- 6.Vijg J, Campisi J. Puzzles, promises and a cure for ageing. Nature. 2008;454:1065–1071. doi: 10.1038/nature07216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontana L, Partridge L, Longo VD. Extending healthy life span—from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen E, Dillin A. The insulin paradox: aging, proteotoxicity and neurodegeneration. Nat Rev Neurosci. 2008;9:759–767. doi: 10.1038/nrn2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy CT. The search for DAF-16/FOXO transcriptional targets: approaches and discoveries. Exp Gerontol. 2006;41:910–921. doi: 10.1016/j.exger.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 10.Greer EL, Brunet A. FOXO transcription factors in ageing and cancer. Acta Physiol (Oxf) 2008;192:19–28. doi: 10.1111/j.1748-1716.2007.01780.x. [DOI] [PubMed] [Google Scholar]
- 11.Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herndon LA, Schmeissner PJ, Dudaronek JM, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 13.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman DB, Johnson TE. Three mutants that extend both mean and maximum life span of the nematode, Caenorhabditis elegans, define the age-1 gene. J Gerontol. 1988;43:B102–B109. doi: 10.1093/geronj/43.4.b102. [DOI] [PubMed] [Google Scholar]
- 15.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 16.Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 17.Ogg S, Paradis S, Gottlieb S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 18.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 19.Pawlikowska L, Hu D, Huntsman S, et al. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8:460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slagboom PE, Beekman M, Passtoors WM, et al. Genomics of human longevity. Philos Trans R Soc Lond B Biol Sci. 2011;366:35–42. doi: 10.1098/rstb.2010.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 22.Ayyadevara S, Tazearslan C, Bharill P, Alla R, Siegel E, Shmookler Reis RJ. Caenorhabditis elegans PI3K mutants reveal novel genes underlying exceptional stress resistance and lifespan. Aging Cell. 2009;8:706–725. doi: 10.1111/j.1474-9726.2009.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy CT, McCarroll SA, Bargmann CI, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 24.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 25.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tullet JM, Hertweck M, An JH, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Junger MA, Rintelen F, Stocker H, et al. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. 2003;2:20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 29.Clancy DJ, Gems D, Harshman LG, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 30.Slack C, Giannakou ME, Foley A, Goss M, Partridge L. dFOXO-independent effects of reduced insulin-like signaling in Drosophila. Aging Cell. 2011;10:735–748. doi: 10.1111/j.1474-9726.2011.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 32.Giannakou ME, Goss M, Junger MA, Hafen E, Leevers SJ, Partridge L. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- 33.Werner H, Weinstein D, Bentov I. Similarities and differences between insulin and IGF-I: structures, receptors, and signalling pathways. Arch Physiol Biochem. 2008;114:17–22. doi: 10.1080/13813450801900694. [DOI] [PubMed] [Google Scholar]
- 34.Bartke A. Minireview: role of the growth hormone/insulin-like growth factor system in mammalian aging. Endocrinology. 2005;146:3718–3723. doi: 10.1210/en.2005-0411. [DOI] [PubMed] [Google Scholar]
- 35.Bartke A. Impact of reduced insulin-like growth factor-1/insulin signaling on aging in mammals: novel findings. Aging Cell. 2008;7:285–290. doi: 10.1111/j.1474-9726.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- 36.Ladiges W, Van Remmen H, Strong R, et al. Lifespan extension in genetically modified mice. Aging Cell. 2009;8:346–352. doi: 10.1111/j.1474-9726.2009.00491.x. [DOI] [PubMed] [Google Scholar]
- 37.Berryman DE, Christiansen JS, Johannsson G, Thorner MO, Kopchick JJ. Role of the GH/IGF-1 axis in lifespan and healthspan: lessons from animal models. Growth Horm IGF Res. 2008;18:455–471. doi: 10.1016/j.ghir.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonkowski MS, Pamenter RW, Rocha JS, Masternak MM, Panici JA, Bartke A. Long-lived growth hormone receptor knockout mice show a delay in age-related changes of body composition and bone characteristics. J Gerontol A Biol Sci Med Sci. 2006;61:562–567. doi: 10.1093/gerona/61.6.562. [DOI] [PubMed] [Google Scholar]
- 39.Coschigano KT, Clemmons D, Bellush LL, Kopchick JJ. Assessment of growth parameters and life span of GHR/BP gene-disrupted mice. Endocrinology. 2000;141:2608–2613. doi: 10.1210/endo.141.7.7586. [DOI] [PubMed] [Google Scholar]
- 40.Coschigano KT, Holland AN, Riders ME, List EO, Flyvbjerg A, Kopchick JJ. Deletion, but not antagonism, of the mouse growth hormone receptor results in severely decreased body weights, insulin, and insulin-like growth factor I levels and increased life span. Endocrinology. 2003;144:3799–3810. doi: 10.1210/en.2003-0374. [DOI] [PubMed] [Google Scholar]
- 41.Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci USA. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holzenberger M, Dupont J, Ducos B, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 43.Cohen E, Paulsson JF, Blinder P, et al. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139:1157–1169. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 45.Selman C, Lingard S, Choudhury AI, et al. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J. 2008;22:807–818. doi: 10.1096/fj.07-9261com. [DOI] [PubMed] [Google Scholar]
- 46.Walker DW, McColl G, Jenkins NL, Harris J, Lithgow GJ. Evolution of lifespan in C. elegans. Nature. 2000;405:296–297. doi: 10.1038/35012693. [DOI] [PubMed] [Google Scholar]
- 47.Jenkins NL, McColl G, Lithgow GJ. Fitness cost of extended lifespan in Caenorhabditis elegans. Proc Biol Sci. 2004;271:2523–2526. doi: 10.1098/rspb.2004.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gudmundsson H, Gudbjartsson DF, Frigge M, Gulcher JR, Stefansson K. Inheritance of human longevity in Iceland. Eur J Hum Genet. 2000;8:743–749. doi: 10.1038/sj.ejhg.5200527. [DOI] [PubMed] [Google Scholar]
- 49.Perls T, Shea-Drinkwater M, Bowen-Flynn J, et al. Exceptional familial clustering for extreme longevity in humans. J Am Geriatr Soc. 2000;48:1483–1485. [PubMed] [Google Scholar]
- 50.Herskind AM, McGue M, Holm NV, Sorensen TI, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97:319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- 51.McGue M, Vaupel JW, Holm N, Harvald B. Longevity is moderately heritable in a sample of Danish twins born 1870-1880. J Gerontol. 1993;48:B237–B244. doi: 10.1093/geronj/48.6.b237. [DOI] [PubMed] [Google Scholar]
- 52.Perls TT, Bubrick E, Wager CG, Vijg J, Kruglyak L. Siblings of centenarians live longer. Lancet. 1998;351:1560. doi: 10.1016/S0140-6736(05)61126-9. [DOI] [PubMed] [Google Scholar]
- 53.Perls TT, Wilmoth J, Levenson R, et al. Life-long sustained mortality advantage of siblings of centenarians. Proc Natl Acad Sci USA. 2002;99:8442–8447. doi: 10.1073/pnas.122587599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robine JM, Allard M. The oldest human. Science. 1998;279:1834–1835. doi: 10.1126/science.279.5358.1831h. [DOI] [PubMed] [Google Scholar]
- 55.Tabor H, Risch NJ, Myers RM. Opinion: candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3:391–397. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- 56.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 57.Long AD, Langley CH. The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits. Genome Res. 1999;9:720–731. [PMC free article] [PubMed] [Google Scholar]
- 58.Halushka MK, Fan J-B, Bentley K, et al. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- 59.Martin ER, Lai EH, Gilbert JR, et al. SNPing away at complex diseases: analysis of single-nucleotide polymorphisms around APOE in Alzheimer disease. Am J Hum Genet. 2000;67:383–394. doi: 10.1086/303003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Consortium TIH. The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 61.Belmont JW, Gibbs RA. Genome-wide linkage disequilibrium and haplotype maps. Am J Pharmacogenomics. 2004;4:253–262. doi: 10.2165/00129785-200404040-00005. [DOI] [PubMed] [Google Scholar]
- 62.van Heemst D, Beekman M, Mooijaart SP, et al. Reduced insulin/IGF-1 signalling and human longevity. Aging Cell. 2005;4:79–85. doi: 10.1111/j.1474-9728.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- 63.Bonafe M, Barbieri M, Marchegiani F, et al. Polymorphic variants of insulin-like growth factor I (IGF-I) receptor and phosphoinositide 3-kinase genes affect IGF-I plasma levels and human longevity: cues for an evolutionarily conserved mechanism of life span control. J Clin Endocrinol Metab. 2003;88:3299–3304. doi: 10.1210/jc.2002-021810. [DOI] [PubMed] [Google Scholar]
- 64.Lunetta KL, D’Agostino RB, Karasik D, et al. Genetic correlates of longevity and selected age-related phenotypes: a genome-wide association study in the Framingham study. BMC Med Genet. 2007;8(suppl 1):S13. doi: 10.1186/1471-2350-8-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Y, Wang WJ, Cao H, et al. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18:4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Willcox BJ, Donlon TA, He Q, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flachsbart F, Caliebe A, Kleindorp R, et al. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci USA. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anselmi CV, Malovini A, Roncarati R, et al. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- 69.Lander ES. The new genomics: global views of biology. [Comment] Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 70.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 71.Slager SL, Huang J, Vieland VJ. Effect of allelic heterogeneity on the power of the transmission disequilibrium test. Genet Epidemiol. 2000;18:143–156. doi: 10.1002/(SICI)1098-2272(200002)18:2<143::AID-GEPI4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 72.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 73.Zhu H, Lensch MW, Cahan P, Daley GQ. Investigating monogenic and complex diseases with pluripotent stem cells. Nat Rev Genet. 2011;12:266–275. doi: 10.1038/nrg2951. [DOI] [PubMed] [Google Scholar]
- 74.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perls TT, Bochen K, Freeman M, Alpert L, Silver MH. Validity of reported age and centenarian prevalence in New England. Age Ageing. 1999;28:193–197. doi: 10.1093/ageing/28.2.193. [DOI] [PubMed] [Google Scholar]
- 76.Suh Y, Atzmon G, Cho MO, et al. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci USA. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 78.Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am J Hum Genet. 2008;82:100–112. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Crawford DC, Akey DT, Nickerson DA. The patterns of natural variation in human genes. Annu Rev Genomics Hum Genet. 2005;6:287–312. doi: 10.1146/annurev.genom.6.080604.162309. [DOI] [PubMed] [Google Scholar]
- 81.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sunyaev S, Ramensky V, Koch I, Lathe W, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 83.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wingender E, Chen X, Fricke E, et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schmid CD, Praz V, Delorenzi M, Perier R, Bucher P. The Eukaryotic Promoter Database EPD: the impact of in silico primer extension. Nucleic Acids Res. 2004;32:D82–D85. doi: 10.1093/nar/gkh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cartharius K, Frech K, Grote K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 87.Conde L, Vaquerizas JM, Santoyo J, et al. PupaSNP Finder: a web tool for finding SNPs with putative effect at transcriptional level. Nucleic Acids Res. 2004;32:W242–W248. doi: 10.1093/nar/gkh438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tazearslan C, Huang J, Barzilai N, Suh Y. Impaired IGF1R signaling in cells expressing longevity-associated human IGF1R alleles. Aging Cell. 2012;10:551–554. doi: 10.1111/j.1474-9726.2011.00697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]