SUMMARY
Many larval sponges possess pigment ring eyes that apparently mediate phototactic swimming. Yet sponges are not known to possess nervous systems or opsin genes, so the unknown molecular components of sponge phototaxis must differ fundamentally from those in other animals, inspiring questions about how this sensory system functions. Here we present molecular and biochemical data on cryptochrome, a candidate gene for functional involvement in sponge pigment ring eyes. We report that Amphimedon queenslandica, a demosponge, possesses two cryptochrome/photolyase genes, Aq-Cry1 and Aq-Cry2. The mRNA of one gene (Aq-Cry2) is expressed in situ at the pigment ring eye. Additionally, we report that Aq-Cry2 lacks photolyase activity and contains a flavin-based co-factor that is responsive to wavelengths of light that also mediate larval photic behavior. These results suggest that Aq-Cry2 may act in the aneural, opsin-less phototaxic behavior of a sponge.
KEY WORDS: Porifera, cryptochrome, evolution, eye, vision
INTRODUCTION
Like many sponge larvae, Amphimedon queenslandica parenchymellae (flagellated free-living larvae) are capable of detecting differing light intensities for use as settlement cues (Leys et al., 2002). This phototactic ability is conferred by posterior, concentric rings of cells that are pigmented and/or possess long cilia that act as light-responsive ‘rudders’ to steer the animal, with maximal sensitivity near 440 nm (blue) (Leys et al., 2002). With pigment adjacent to photoreceptors, the sponge ring structure meets a minimal definition of an eye (Arendt, 2001). In all non-poriferan animals examined to date, including cnidarians (Koyanagi et al., 2008; Kozmik et al., 2008), eyes utilize photo-sensitive opsin proteins expressed in neurons that communicate information about the light environment to the nervous system. However, despite possessing many other G-protein coupled receptors (GPCRs), the fully sequenced genome of A. queenslandica lacks opsin, defined as opsin-clade GPCRs that possess a conserved Shiff base lysine that is central to opsin function (Plachetzki et al., 2007; Srivastava et al., 2010). Like other sponges, A. queenslandica lacks a nervous system. Therefore, A. queenslandica pigment ring eyes likely evolved convergently in the absence of opsins and nervous systems, and probably use as-yet-unknown molecular mechanisms that are fundamentally different from those employed by other animal eyes. One step in understanding the function of these convergently evolved sponge eyes is to identify molecular components and their functions.
The behavioral action spectrum of sponge light sensitivity led previous authors (Leys et al., 2002) to suggest that a flavin-based molecule such as a chryptochrome, acts as the photopigment. Cryptochromes maximally absorb blue light and have light-mediated behavioral roles in multiple animal lineages. For example, Drosophila and mouse cryptochromes are involved in circadian rhythm entrainment (reviewed in Sancar, 2004), coral cryptochrome mRNA is upregulated during spawning, which occurs at the full moon (but see Hoadley et al., 2011; Levy et al., 2007), and several animal cryptochromes have been implicated in magnetoreception (Foley et al., 2011; Gegear et al., 2008; Gegear et al., 2010; Mouritsen et al., 2004). This previous research led us to examine cryptochrome as a candidate gene for use in the aneural eyes of A. queenslandica.
Here we determine gene expression patterns and the in vitro absorption spectra of sponge cryptochrome proteins for comparison with previously published behavioral action spectra. We report that A. queenslandica possesses two cryptochrome/photolyase genes with strikingly complementary expression. The mRNA of one gene (Aq-Cry2) is expressed in cells of the pigment ring. We show that the encoded protein lacks DNA photolyase activity in vitro and contains a flavin-based co-factor, and that the absorbance peak of the molecule matches the peak activity of light-mediated larval behaviors. These results suggest that Aq-Cry2 functions in an evolutionarily convergent, neuron-less, opsin-less eye.
MATERIALS AND METHODS
Bioinformatics
We first performed BLASTP searches of Amphimedon queenslandica Hooper and Van Soest 2006 gene models using known cryptochromes and, based on overall similarity, we found two putative photolyase/cryptochrome genes in the sponge. We compared the phylogenetic relationships of the two newly determined sponge genes (AqCry1 and AqCry2) to previously characterized photolyase/cryptochrome genes. First, we included in the phylogeny all previously known poriferan genes, including Aphrocallistes vastus photolyase related protein (CAD24679.1), Crateromorpha meyeri cryptochrome (CAZ66368.1) and Suberites domuncula cryptochrome (CAZ66367.1). Next, we included the cnidarian cryptochrome genes previously investigated and analyzed by Reitzel et al. (Reitzel et al., 2010), including four Nematostella vectensis genes (XP_001623146.1, XP_001631029.1, XP_001632849.1 and XP_001636303.1) and three Acropora millepora genes (ABP97099.1, ABP97098.1 and the translated EST sequence DY585180). To these, we added genes that represent the major clades of the photolyase/cryptochrome family, as defined by Brudler et al. (Brudler et al. 2003): plant cryptochromes (Arabidopsis thaliana CRY2, Q96524.2; Arabidopsis thaliana CRY1, Q43125.2); class 2 photolyases (Arabidopsis thaliana AF053365.1); DASH cryptochromes (Arabidopsis thaliana AB062926.1); class 1 photolyases (Escherichia coli P00914.1); and representatives of the major clades of the animal cryptochrome/6-4 photolyase family, including insect type-2 cryptochromes (Apis mellifera NP_001077099.1), vertebrate cryptochromes (Mus musculus Cry2 CAM15994.1), 6-4 photolyases (Drosophila melanogaster BAA12067.1) and insect type-1 cryptochromes (D. melanogaster CRY AAF55649.1).
Using each of the proteins listed above (excluding poriferan and cnidarian sequences) as ‘bait’ for database searches, we expanded the representation of genes in the phylogenetic analysis employing a pipeline of shell and perl scripts (Rivera et al., 2010; Tong et al., 2009). Each bait sequence from a major photolyase/cryptochrome clade was used to perform similarity searches using BLASTP (Altschul et al., 1997) of two non-redundant protein databases, UniRef50 and UniRef90, curated by UniProt (http://www.uniprot.org/). For each bait protein, the two most similar genes in the UniRef50 and the 15 most similar genes in the UniRef90 database were retained for phylogenetic analyses. Identical sequences, such as those obtained from both UniRef90 and UniRef50 databases, were removed from further analysis.
We next aligned together all poriferan, cnidarian, database-retained and bait sequences using MUSCLE (Edgar, 2004). Then we estimated maximum likelihood (ML) phylogenetic trees using 10 Slow ML searches in RAxML, assuming a WAG (Stamatakis, 2006) model of protein evolution. Nine of 10 searches yielded the same likelihood value, suggesting that the algorithm located the global maximum. We gauged consistency of the phylogeny using 100 fast bootstrap pseudoreplicates, also implemented in RAxML (Stamatakis et al., 2008). We visualized the resulting phylogenetic trees with TreeView (Krakauer et al., 1996) or FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Construct preparation
After finding two candidate A. queenslandica cryptochrome genes by bioinformatic analyses, we cloned both genes for subsequent in vitro expression, purification and biochemical analyses. First, we cloned Aq-Cry 1 into pCR vector (Invitrogen, Carlsbad, CA, USA) using the primers 5′-ATGGAGACTCTACAAGGCTCCTACG-3′ (forward) and 5′-CTATGGATTATCACCATATAGCTCCCT-3′ (reverse) on a cDNA library. For subcloning into an expression vector (pMALc2; New England Biolabs, Ipswich, MA, USA), we added HindIII sites to the primers and the resultant fragment was cut and cloned into the HindIII site of pMALc2 (pMALc2-CRY1). In addition, we synthesized Aq-Cry2 in pMALc4X (pMALc4X-CRY2; New England Biolabs) with GeneOnDemand (GenScript, Piscataway, NJ, USA). Both pMALc2-CRY1 and pMALc4X-CRY2 are maltose binding protein (MBP) fusions, making MBP-AqCry1 and MBP-AqCry2 proteins, respectively. MBP-AqCry1 and MBP-AqCry2 were purified from the photolyase-deficient Escherichia coli strain UNC523 and were used for in vitro photolyase activity assays. We also cloned the genes into expression plasmids constructed with His tags for Sf21 insect cell-based protein purification, which yield cleaner protein samples for spectroscopic studies. We amplified Aq-Cry1 cDNA from pMALc2-CRY1 using the oligos 5′-ACAGAATTCGCCACCATGGAGACTCTACAAGGCTCCTAC-3′ (forward) and 5′-AGCTCTAGAATAAAGTCTACAAAAAAATCCGGAA-3′ (reverse). We amplified Aq-Cry2 cDNA from pMALc4X-CRY2 using the oligos 5′-GCGAATTCGCCACCATGGACAGCTATAGCAATGGTTTC-3′ (forward) and 5′-GAATCTAGAGGGGTTTGTTTGATGCGAC-3′ (reverse), and inserted the resulting PCR products into the EcoRI and XbaI sites of pAc5.1V5/HisA (Invitrogen) to make pAc5.1AqCRY1-V5/HisA and pAc5.1AqCRY2-V5/HisA, respectively. The coding sequence from this was cut using EcoRI and PmeI and inserted into the EcoRI and StuI sites of pFast-Bac1 (Invitrogen) to make pFast-Bac1-AqCRY1-V5/HisA and pFast-Bac1-AqCRY2-V5/HisA.
Protein purification
We prepared baculovirus using the Gibco BRL Bac-to-Bac baculovirus expression system (Invitrogen). Briefly, pFast-Bac1-AqCRY2-V5/HisA was transformed into the E. coli DH10Bac, and recombinant bacmids were isolated from 1 ml of bacterial cultures grown from colonies of transformants. We transfected Sf21 cells in six-well plates with 1 μg of the bacmid using 6 μl of Cellfectin reagent (Invitrogen), and parental virus was collected after 72 h. After two more 72 h amplification steps, we obtained the third passage (P3), high titer stock, and used it to infect Sf21 cells at a large scale. We infected 1 liter of Sf21 cells (106 ml–1) growing at 27°C in spinner flasks with the P3 high titer virus at a ratio of 1:100 (v/v); the cells were harvested 2 days later. Cells were resuspended in 5 ml of 1× Tris-buffered saline (TBS; pH 7.6) containing 20 mmol l–1 imidazole, and sonicated 10×10 s with a Branson sonicator (Branson Ultrasonics, Danbury, CT, USA). The cell debris was removed by centrifugation at 4°C for 90 min in a Ti 45 rotor at 137,000 g in a Beckman L-80 centrifuge (Palo Alto, CA, USA). Cell free extract (30 ml) was incubated with 0.5 ml of Ni-NTA agarose beads (Qiagen, Valencia, CA, USA) for 3 h. Then we washed the beads four times with 15 ml of TBS containing 20 mmol l–1 imidazole to remove unbound proteins. AqCRY2V-5/HisA protein was eluted from beads by increasing the imidazole to 200 nmol l–1. We dialyzed the eluted protein against the storage buffer containing 50 mmol l–1 Tris-HCl, pH 7.5, 100 mmol l–1 NaCl, 5 mmol l–1 dithiothreitol, and 50% (v/v) glycerol and kept the eluted protein at –20°C. We analyzed the purity of recombinant protein by performing electrophoresis on a NuPAGE 4–12% Bis-Tris Gel followed by Coomassie Blue staining. The yield was 3 mg of recombinant protein from 1 liter of Sf21 culture. AqCRY1V-5/HisA protein was made in the same way.
Preparation of three photolyase controls, each with MBP fused at the N terminus, has been described previously: Arabidopsis Cry (Selby and Sancar, 2006), Xenopus laevis 6-4 photolyase (Kim et al., 1996; Zhao et al., 1997) and Caulobacter cresenteus photolyases (Ozturk et al., 2008). We purified MBP fusion proteins of Aq-Cry1 and Aq-Cry2 according to the manufacturer’s recommendations (New England Biolabs). Briefly, transformed UNC523 (photolyase deficient) bacterial cells were grown in 1 liter of Luria–Bertani (LB) medium at room temperature. At an OD600 (the optical density measured at a wavelength of 600 nm) of 0.6, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.3 mmol l–1, and growth continued for 5 h. We washed the cell pellets and resuspended them in 30 ml of column buffer (20 mmol l–1 Tris, pH 7.4, 200 mmol l–1 NaCl, 1 mmol l–1 EDTA, 10 mmol l–1 2-mercaptoethanol) before sonicating 10 times for 10 s each and pelleting. Supernatants were applied to columns containing 20 ml of amylose resin (New England Biolabs). We washed the columns with five volumes of column buffer and then eluted with 20 mmol l–1 maltose in column buffer. Fractions were analyzed on polyacrylamide gels, pooled and dialyzed against 50 mmol l–1 Tris, pH 7.5, 100 mmol l–1 NaCl, 1 mmol l–1 EDTA, 5 mmol l–1 dithiothreitol (DTT), 50% glycerol.
In vivo photolyase activity assay
We expressed Aq-Cry1 or Aq-Cry2 in bacteria that are not competent to repair UV-induced lesions in their DNA, to test whether heterologously expressed proteins rescue repair activity. We transformed competent SY2 cells [UV-sensitive strains phr-, recA- and uvrA- (Sancar et al., 1984)] with pMALc4X-CRY2, pMALc2-CRY1, pMALc4X (empty expression vector) or positive control pMS969, which expresses E. coli photylase (Sancar et al., 1983). We grew these four SY2 strains to OD600=1.0 in Luria–Bertani ampicillin (LB-Amp) medium and 1 mmol l–1 IPTG was added for 4 h to induce expression. We spun down 1.5 ml of culture and resuspended in 1 ml of 1% NaCl (saline). This was added to an additional 9 ml of saline on a Pyrex® Petri dish with the cover underneath to add an additional layer of UVC protection (Lepre et al., 1998). We then placed these on an FB-TIV 816 UV Transilluminator (Fisher Scientific, Pittsburg, PA, USA) at 20% intensity with cellulose acetate as an additional UVC filter for 1 h. Forty-microliter aliquots of each saline suspension were taken and added to 100 μl of LB-Amp + 1 mmol l–1 IPTG and plated on LB-Amp plates. We counted colonies by hand after growing overnight in ambient light. We found that positive control cells expressing E. coli Phr (a photolyase) grew to a high density, with longer UV exposures resulting in lower density of growth. Results with cells heterologously expressing each Aq-Cry varied, with each protein leading to cell growth in at least one trial (data not shown). This may be explained by the known prevalence of false positives using this method (Malhotra et al., 1995).
In vitro photolyase activity assay and photoreduction
We generated radioactive oligos with a cyclobutane thymine dimer, T<>T or (6-4) photoproduct, T[6-4]T, within an MseI cut site (TTAA). The photoproducts block digestion by MseI; photo-repair regenerates the restriction site. Single-stranded substrates were prepared as described previously (Selby and Sancar, 2006). To make double-stranded oligos, 32P-radiolabeled oligonucleotide with photoproduct and complementary oligonucleotide at 1:20 ratio were heated to 85°C for 5 min in an annealing buffer containing 50 mmol l–1 NaCl, 10 mmol l–1 Tris-HCl pH 7.5 and 10 mmol l–1 MgCl2, and then cooled slowly to 25°C. Repair reactions (20 μl) contained 50 nmol l–1 Tris-HCl, pH 7.5, 100 mmol l–1 NaCl, 1 mmol l–1 EDTA, 10 mmol l–1 DTT, 0.2 nmol l–1 substrate, and the indicated concentrations of enzyme purified from UNC523 cells. Reactions were incubated in the dark at 25°C for 15 min and then exposed to 366 nm light at a fluence rate of 2 mW cm–2 for 60 min from two black light lamps (F15J8-BLB; General Electric, Fairfield, CT, USA), filtered through a glass plate to cut out short wavelength light under 300 nm, at 4°C (dark samples were covered with aluminum foil). Following photoreactivation, the DNA was extracted with phenol, precipitated with ethanol, resuspended in 40 μl of buffer containing 10 mmol l–1 Tris-HCl, pH 7.9, 50 mmol l–1 NaCl, 10 mmol l–1 MgCl2, and 1 mmol l–1 DTT, heated to 85°C for 5 min and then cooled slowly to 25°C. To this, 40 units of MseI were added and digestion was carried out at 37°C overnight. Products were analyzed on 8% polyacrylamide sequencing gels followed by autoradiography. We quantified data from degredation and MseI digestion (repair) assays by densitometry using ImageQuant 5.0 software (Molecular Dynamics, Waukesha, WI, USA).
To quantify the absorption spectrum of Aq-Cry1 and Aq-Cry2, we grew His-tagged Aq-Cry-1 and Aq-Cry2 in insect Sf21 cells and purified them using nickel agarose beads, as described above, under yellow light to prevent photochemical modification (Song et al., 2007). We used insect cells rather than bacterial cells as they allow for purification of enzymes with their flavin adenine dinucleotide co-factor. Purified protein in storage buffer was irradiated with 366 nm light at a fluence rate of 2 mW cm–2 for 15 min using an F15T8-BLB black light (General Electric) lamp as a light source. We performed the same procedure on Aq-CRY1-His purified from Sf21 cells and purified MBP-AqCry1, which showed the same absorption spectrum. We recorded absorption spectra using a Shimadzu UV-1601 spectrophotometer (Columbia, MD, USA).
In situ hybridization
We made templates for in situ probes by designing primers against predicted coding sequences and EST sequences (Srivastava et al., 2010). We made an in situ probe for Aq-Cry1 by amplifying a 1166 bp fragment from cDNA using the primers R1261 (5′-GCAACCCACCGTAGTCTTGT-3′) and L108 (5′-CCTTCCCAATGATGCAGTTT-3′). This was cloned into TOPO-pCR II (Invitrogen) using the manufacturer’s protocol. We cloned a 1232 bp fragment of Aq-cry2 in the same way using the primers R1487 (5′-GAGATTGCCTGTCAGCCTC-3′) and L215 (5′-TAGTCTCCGTTTGGGTCCAG-3′). All clones were sequenced for verification using M13F and M13R primers. Digoxygenin (DIG)-labeled antisense RNA probes were transcribed from polymerase chain reaction (PCR) products using DIG RNA Labeling Mix (Roche Applied Sciences, Indianapolis, IN, USA) and T7 or SP6 Polymerase (Promega, Madison, WI, USA) following the manufacturer’s instructions. We obtained embryonic and larval material from Heron Island Reef according to the procedures described in Leys et al. (Leys et al., 2008). We performed in situ hybridization and subsequent processing following previously published protocols (Larroux et al., 2008). We observed whole-mount and sectioned samples under an Olympus SZH or BX60 microscope with Nomarski optics (Olympus Australia Pty Ltd, Mt Waverly, VIC, Australia) and photographed samples with a Nikon Sight DS-U1 camera (Nikon Australia Pty Ltd, Lidcombe, Australia).
RESULTS
Identification of two cryptochromes from A. queenslandica
Because A. queenslandica larvae are responsive to wavelengths of light overlapping the flavin response spectrum (Leys et al., 2002), we identified poriferan photolyase/cryptochrome orthologs as candidate genes mediating larval phototactic behaviors. Photolyases are ancient enzymes present in both prokaryotes and eukaryotes that function in blue-light-driven repair of UV-induced DNA lesions. Cryptochromes are members of this family, but lack DNA repair functions, instead acting as blue-light receptors utilizing two chromophores: flavin and pterin (Sancar, 2000; Selby and Sancar, 2006). In animals, cryptochromes are involved in light-mediated behaviors, including the entrainment of circadian rhythms.
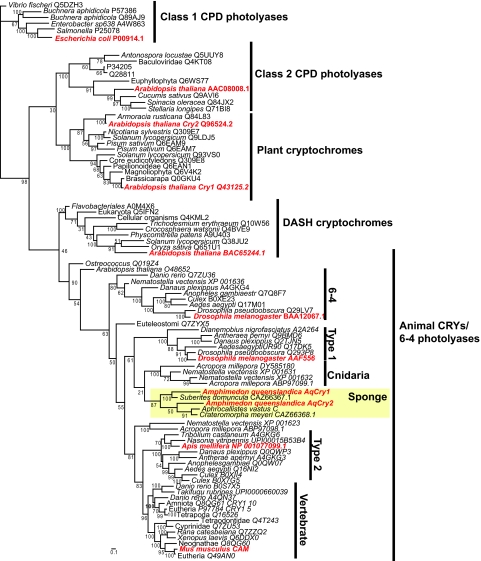
Using similarity and phylogenetic analyses, we identified and cloned two photolyase/cryptochrome genes using the fully sequenced genome of A. queenslandica (Srivastava et al., 2010) and designated them Aq-Cry1 and Aq-Cry2. These genes, Aq-Cry1 and Aq-Cry2, together with previously characterized genes from two hexactinellid sponges and another demosponge, form a clade of poriferan cryptochromes (Fig. 1). Aq-Cry1 is most closely related to another demosponge cryptochrome, suggesting that the Aq-Cry1 and Aq-Cry2 originated by duplication some 650–800 million years ago (Sperling et al., 2010), prior to the origin of demosponges, but after the divergence of hexactinellids and demosponges. The poriferan cryptochrome genes are part of a large clade of mostly animal photolyase/cryptochromes that includes the animal cryptochromes (Abushik et al., 1985) and 6-4 photolyases. Activity in response to light varies across this group of proteins with respect to proteolytic cleavage, flavin radical formation and downstream targets (Ozturk et al., 2009; Sancar, 2004; Yuan et al., 2007; Zhu et al., 2005). Although sampling is incomplete, many members, namely the 6-4 photolyases, exhibit light-mediated DNA repair activity.
Fig. 1.
Phylogenetic analysis of sponge cryptochromes (Amphimedon queenslandica Aq-Cry1 and Aq-Cry2), cnidarian cryptochromes/photolyases and previously defined major clades of the cryptochrome/photolyase family. We selected representatives of major clades (in red) and used them as ‘bait’ to find similar sequences in UniRef databases. We next aligned together all poriferan, cnidarian, database-retained and bait sequences using MUSCLE. Then we estimated maximum likelihood phylogenetic trees using 10 Slow ML searches in RAxML, assuming a WAG model of protein evolution. Numbers at nodes represent percentages of 100 fast-bootstrap pseudoreplicates containing that node. CPD, cyclobutane pyrimidine dimer.
To begin to elucidate the molecular function of sponge cryptochromes, we tested their ability to repair UV-damaged DNA, the defining feature of photolyases. Although all bona fide cryptochromes examined lack photolyase activity (Selby and Sancar, 2006), the sponge cryptochromes form a sister group to a clade of genes containing both cryptochromes and photolyases and thus may be able to act as photolyases. Furthermore, Schröder et al. (Schröder et al., 2003) concluded that the photolyase/cryptochrome protein of the hexactinellid sponge Aphrocallistes vastus possesses repair activity. We first tested for repair activity using the same assay as in Schröder et al. (Schröder et al., 2003). This assay was inconclusive, giving mixed results for both genes (data not shown; see Materials and methods for details), possibly because of the high false-positive rate of this assay (Malhotra et al., 1995).
To more rigorously test for photolyase activity in the A. queenslandica cryptochromes, we performed in vitro assays testing for the repair of a 6-4 photoproduct thymine dimer and a cyclobutane pyrimidine dimer (CPD). We found that neither sponge cryptochrome was able to repair (6-4) photoproducts, indicating that neither has (6-4) photolyase activity (Fig. 2A). Similarly, neither CRY exhibited CPD photolyase activity (Fig. 2B), demonstrating that they are cryptochromes rather than photolyases.
Fig. 2.
DNA repair ability of A. queenslandica cryptochromes. A 48-nt-long radiolabeled oligomer with photoproducts in the TTAA sequence (MseI recognition site) in the center, in single-stranded (ss) or double-stranded (ds) form, was mixed with the appropriate enzyme and exposed to photoreactivating light (l) or kept in the dark (d). The level of repair was determined by the susceptibility of the DNA to cleavage by MseI endonuclease. Repaired products were annealed with the complementary strand (1:100 fold) before digestion with MseI. The reaction products were separated on 8% polyacrylamide gels. (A) Single- and double-stranded oligos containing CPD dimers were incubated in 100 nmol l–1 of Aq-Cry1, Aq-Cry2, and positive controls Arabidopsis DASH photolyase (ss substrate) and Caulobacter cresenteus photolyase (ds substrate) for 90 min in the dark or 2 mW cm–2 black light. Oligos with repaired photoproducts became sensitive to MseI digestion, resulting in a shorter oligo. In these conditions, Aq-Cry1 and Aq-Cry2 did not repair photoproducts, indicating that they do not have any photolyase activity in vitro. (B) Single- and double-stranded oligos containing [6-4] photoproducts were incubated in 100 nmol l–1 of Aq-Cry1, Aq-Cry2, or positive control Xenopus 6-4 photolyase (ss and ds substrate) for 90 min in the dark or 2 mW cm–2 black light. Oligos with repaired photoproducts became sensitive to MseI digestion, resulting in a shorter oligonucleotide. In these conditions, Aq-Cry1 and Aq-Cry2 did not repair photoproducts, indicating that they do not have any photolyase activity in vitro. (C) Analysis of purified protein by 4–12% gradient SDS-PAGE. The numbers on the left indicate size markers. Lane 1, purified MBP-AqCRY1; lane 2, purified MBP-AqCRY2 through amylose resin. The positions of full-length CRYs are indicated by arrows. Extensive washing of the amylose resin by high-salt buffer causes considerable loss of the flavin co-factor. Therefore, the proteins bound to amylose resin were washed with two column volumes of buffer with moderate ionic strength before elution with maltose, and the preparation contains contaminating proteins at a considerable level. However, because the recombinant proteins were expressed in the photolyase-deficient E. coli strain UNC523 (phr::kan uvrA::Tn10), the contaminating proteins do not interfere with subsequent in vitro photoreactivation assays.
Expression of Aq-Cry1 and Aq-Cry2 show complementary patterns
Having established that the Aq-Cry1 and Aq-Cry2 are indeed cryptochromes, by phylogenetic position and lack of photolyase activity, we next examined whether either gene was expressed in a pattern suggesting involvement with phototactic behaviors. To this end, we performed in situ hybridizations across developmental stages to reveal patterns of gene expression.
The two A. queenslandica cryptochromes show strikingly complementary expression patterns in in situ hybridization experiments with Aq-Cry2, but not Aq-Cry1, expressed around the pigment ring (Figs 3, 4). Early expression of both genes is similar and is seen in the medium-sized cells scattered throughout the embryo, and Aq-Cry1 transcripts are found in maternal follicle cells around the developing embryo (Fig. 3A,B, Fig. 4A,B). For descriptions of A. queenslandica development, see Adamska et al., Leys et al., and Leys and Degnan (Adamska et al., 2007; Leys et al., 2002; Leys and Degnan, 2001). During the ‘spot stage’ of embryonic development, pigment cells coalesce to form a pigmented spot at the posterior pole of the embryo. At this stage, Aq-Cry2 transcripts are strongly enriched directly adjacent to and around the pigmented cells (Fig. 4C–E). In contrast, during this spot stage, we observed Aq-Cry1 expression in cells restricted to the outer layer of the embryo, with no expression detected in the vicinity of the pigment ring (Fig. 3C,D). Later during development, as the pigment cells form the ring-eye, Aq-Cry2 expression remains largely restricted to cells adjacent to, underneath and inside the ring, although transcripts are also detected in cells comprising an epithelial-like layer (Fig. 4F,G). In contrast, during ring formation, Aq-Cry1 expression is weaker in the future photosensitive cells around the pigment ring (Fig. 3E–G). These expression patterns are maintained until larval hatching (Fig. 3H,I, Fig. 4H–J). After hatching, the expression of both cryptochrome genes becomes more diffuse and Aq-Cry2 is no longer enriched around the pigment ring, though it still shows expression in pigment cells (Fig. 3J–M, Fig. 4K,L).
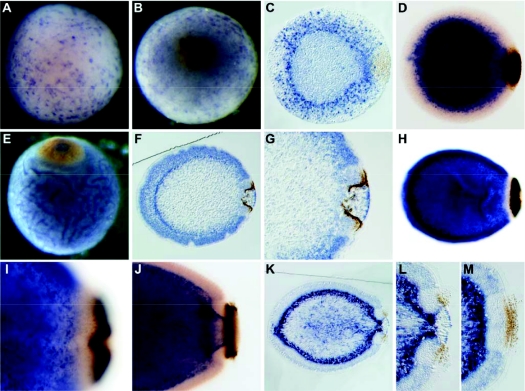
Fig. 3.
Developmental expression of Aq-Cry1. (A,B,E) Whole-mounts; (C,F,G,K,L,M) sections; (D,H,I,J) cleared whole-mounts; (B,E) posterior pole facing up; (C,D,F–M) posterior pole to right. (A,B) In early gastrula and spot stages, Aq-Cry1 expression is evident in maternal follicle cells on the surface of the embryo. (C,D) After gastrulation, at the spot stage, embryonic Aq-Cry1 expression is strongest in a population of cells scattered throughout the outer region of the embryo. (E,F) During ring formation, the gene is upregulated in presumptive epithelial-like cells and in a population of cells inside the pigment ring. (G) Higher magnification of the embryo shown in F, showing expression diminished in cells adjacent to the outer part of the ring. (H) Excluding cells adjacent to the pigment ring, expression in the outer layer continues in early larvae (pre-release). (I) Higher magnification of the embryo shown in H. (J,K) In hatched, swimming larvae, Aq-Cry1 is expressed in sub-epithelial cells and cells located inside the ring, and in the epithelial-like cells a in a pattern similar to that observed at earlier stages. (L,M) Higher magnifications of the embryo shown in K, sectioned through the middle of the ring (L) or the edge of the ring (M).
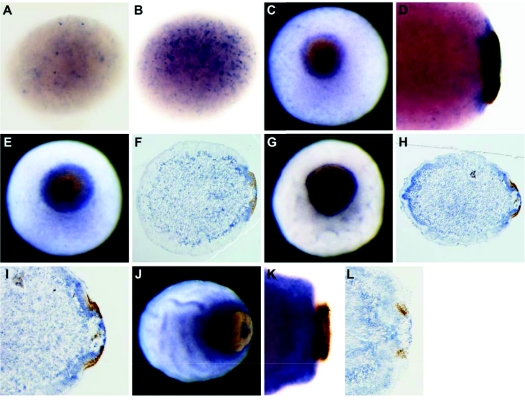
Fig. 4.
Developmental expression of Aq-Cry2. (A,B,D,K) Cleared whole-mounts; (C,E,G,J) whole-mounts; (F,H,I,L) sections; (C,E,G) posterior pole facing up; (D,F,H–L) posterior pole to right. (A,B) Aq-Cry2 expressing cells are scattered throughout the embryo during cleavage. (C–E) In later stages, Aq-Cry2 transcripts are predominantly detected in the vicinity of the pigment spot and early ring. (F,G) During ring formation, Aq-Cry2 expression continues outside, underneath and inside the ring, and also becomes evident in a population of cells located beneath the outer-layer, putative sub-epithelial cells. (H–J) As the embryo develops into an early larva, expression of the gene increases in the outer layer, appearing strongest at the anterior and posterior poles. (I) Higher magnification of the embryo shown in H. (K,L) In swimming larvae, Aq-Cry2 is expressed in sub-epithelial cells, epithelial-like cells, pigment cells, cells with long cilia and in a subset of cells located inside the pigment ring.
Aq-Cry2 is responsive to blue light but does not undergo proteolysis
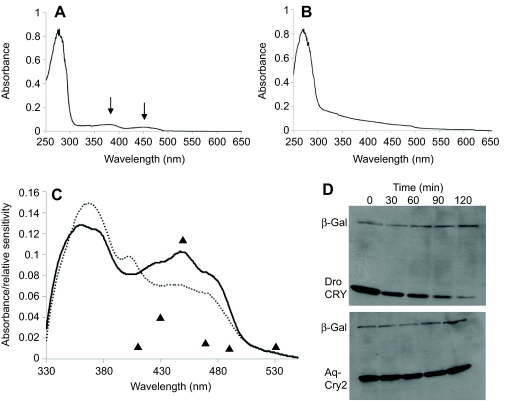
We next examined the in vitro absorption spectrum of Aq-Cry2 and found similarities to the action spectrum of A. queenslandica larval photo-behaviors. The absorption spectrum of fully oxidized, purified Aq-Cry2 ranges between 366 and 450 nm and is typical of enzyme-bound oxidized flavin adenine dinucleotide (FAD) (Fig. 5A,C), and the ratio of co-factor to apoprotein shows that recombinant Aq-Cry2 contained stoichiometric flavin (Fig. 5A) (Worthington et al., 2003). Upon exposure to UV light, Aq-Cry2 showed a typical type-1 cryptochrome reduction pattern (Fig. 5C), being converted to FAD ionic radical (Ozturk et al., 2009). The absorption peak of Aq-Cry2 near 450 nm corresponds well with the primary, dominant peak of the A. queenslandica behavioral action spectrum (Leys et al., 2002). The lack of activity at 400 nm in the behavioral action spectrum could be due to filtering by another pigment, but this explanation does not account for an additional, minor behavioral response to longer wavelengths near 600 nm. In contrast to the Aq-Cry2 gene with pigment ring expression, Aq-Cry1 did not show measurable absorption in the near-UV/visible wavelength range, possibly because it lacks flavin and therefore lacks a light response, meaning its function remains unknown (Fig. 5B). Flavin is not covalently bound to photolyases/cryptochromes, so it is not unusual to purify a protein without stoichiometric flavin (Zhao et al., 1997).
Fig. 5.
Spectroscopic properties and proteolytic activity of A. queenslandica cryptochromes. (A,B) Absorption spectra of purified Aq-Cry2 (A) and Aq-Cry1 (B). Arrows in A point to peaks, indicating purification of the protein along with stoichiometric flavin. The lack of measurable absorption in the 350–350 nm range in B suggests that Aq-Cry1 (AqCRY1-V-5/HisA) did not contain substantial flavin after purification. MBP-AqCry1 showed the same result (data not shown). In contrast, the ratio of absorbance at 280 nm of apoprotein to the 450 nm flavin adenine dinucleotide absorption peaks suggests that the recombinant MBP-AqCry2 contained stoichiometric flavin. (C) Photoreduction of Aq-Cry2 by blue light. Solid line: spectrum of Aq-Cry2 purified under yellow light. Dashed line: after exposure to UV light (366 nm at 2 mW cm–2 for 15 min). Aq-Cry2 showed the typical type-1 cryptochrome reduction pattern, with an increase of the 366 nm peak and the formation of a new peak at 405 nm and concomitant decrease of the 450 nm peak. Triangles indicate the behavioral response in living larvae (Leys et al., 2002); the minor behavioral response to 600 nm light not shown (units for behavioral response are arbitrary). (D) Light-induced proteolysis of cryptochromes. Although Aq-Cry2 displays spectroscopic properties similar to animal type-1 cryptochromes, it does not undergo photo-induced proteolysis. Drosophila CRY (Dro CRY) and Aq-Cry2 were transfected into S2 cells alongside with beta-galactosidase (β-Gal) as an internal control, and tested for light-induced proteolysis after 36 h. Although the Drosophila CRY protein was degraded via light-induced proteolysis (upper panel), levels of Aq-Cry2 remained high even after 2 h of light activation (lower panel).
Unlike several previously studied animal Type 1 cryptochromes (Ozturk et al., 2009), we found that Aq-Cry2 lacked photo-induced proteolysis (Fig. 5E,F). When expressed in insect S2 cells and exposed to blue light, Drosophila CRY was degraded after 90 min whereas Aq-Cry2 remained intact (Fig. 5E,F). This key difference between Aq-Cry2 and well-characterized cryptochromes such as Drosophila CRY indicates that, despite similar spectral properties, Aq-Cry2 must mediate its activation through a different, as-yet-undetermined mechanism.
DISCUSSION
Pigment ring eyes of sponges such as A. queenslandica provide a fascinating case study in convergent evolution. Although all eyes of eumetazoan animals examined to date utilize phototransduction cascades initiated when photons strike homologous opsin proteins, A. queenslandica provides an exception to this rule (Plachetzki et al., 2007). Without neurons, and with a genome lacking opsin, this sponge’s eye ring must utilize different and evolutionarily convergent molecular machinery compared with other animal eyes. An important candidate gene for involvement in these pigment ring eyes is cryptochrome (Leys et al., 2002).
Here we present correlative evidence for the function of cryptochrome in the A. queenslandica eye. After finding candidate genes based on sequence similarity, we showed that one of two genes (Aq-Cry2) lacks DNA repair activity, and is therefore a true cryptochrome, not a photolyase. We then showed that Aq-Cry2 is expressed in the cells thought to mediate the light response (Leys and Degnan, 2001). Further, its absorption pattern shows a prominent peak corresponding to peak phototactic behaviors in A. queenslandica (Leys et al., 2002). This peak absorption disappears after photoreduction of Aq-Cry2, demonstrating photoresponsiveness of Aq-Cry2 itself. These results are consistent with the hypothesis that opsin-less sponge eyes utilize cryptochrome, along with other proteins, to direct or act in eye-mediated phototactic behavior.
Although our expression and in vitro biochemical results suggest that Aq-Cry2 functions by responding to light in the pigment ring eye, we cannot yet determine the precise nature of this function in the organism. There are multiple possibilities, including acting as the primary photoreceptor effecting ciliary rudders or acting in the entrainment of timing cues for swimming and settlement. The first possibility, Aq-Cry2 is the primary photoreceptor, is supported by our finding that the peak sensitivity of the behavioral response is similar to the peak of the Aq-Cry2 absorption spectrum. However, a minor behavioral response to light near 600 nm may require either a second Aq-Cry2 co-factor (besides flavin), or a second photoresponsive protein mediating the long-wavelength response. As we see no evidence for a second co-factor in our study or in other cryptochromes in the literature that could explain a response to 600 nm light, Aq-Cry2 may not be the primary photoreceptive molecule, or may not be acting alone.
A second possibility, that Aq-Cry2 is involved as a timing and/or entrainment molecule, is supported by the involvement of other animal cryptochromes in entrainment (Emery et al., 1998; Stanewsky et al., 1998; Thresher et al., 1998). In this case, Aq-Cry2 could be acting as an upstream signaling factor indicating the presence of light in general or as a developmental timing cue. As animal cryptochromes are known to function as transcription factors in signaling transduction pathways (Reppert and Weaver, 2002) as well as modifiers of protein function (Ceriani et al., 1999), Aq-Cry2 could be activating, via transcription or protein modification, other downstream factor(s) responsible for the light response. If Aq-Cry2 is indeed acting as a cue, rather than an effector, then it only needs to be tuned to the major wavelengths of light associated with the behavior (420–460 nm). Future studies into genes involved in entrainment could give insight into the function of sponge cryptochromes. For example, if other known entrainment molecules are expressed in the developing pigment ring, then a role for Aq-Cry2 in entrainment could be supported. If so, a different and unknown photoreceptive molecule could be the directional indicator needed to guide swimming behavior.
Other candidate genes for the primary photoreceptive molecule, beside cryptochrome, are also worthy of consideration. The short-wavelength receptor lite-1 was recently characterized in Caenorhabditis elegans (Edwards et al., 2008) and D. melanogaster (Xiang et al., 2010). However, lite-1, like opsin, acts in neurons and would therefore require a different mechanism in sponges. Another candidate is the mitochondrial gene cytochrome oxidase I (CO1). The absorption spectrum for CO1 closely matches the entire behavioral action spectrum for A. queenslandica, including the minor long-wavelength behavioral response (Bjorn and Rasmusson, 2009). Although CO1 is not generally thought of as a light-responding molecule, it has been shown to absorb light in various biological contexts (Hallén and Brzezinski, 1994; Karu, 1999; Karu et al., 2004). However, a mechanism for how CO1 could mediate ciliary responses is unknown. An additional difficulty is that testing whether CO1 functions as a photoreceptor is challenging. A search of the A. queenslandica genome did not uncover a nuclear copy of CO1, and expression studies of a mitochondrial gene would be uninformative, in part because increased concentration could represent increased numbers of mitochondria for energetic needs that may be required by active ciliated cells, rather than indicating localization of a photoreceptive gene. Furthermore, knockdown of a mitochondrial gene such as CO1 would cause numerous side effects, and preclude testing its function as a photoreceptor.
Given our expression and biochemical results, elucidation of the exact function of A. queenslandica cryptochromes provides rich prospects for future research that will depend on multiple complementary approaches. Although a gene knockdown approach (Rivera et al., 2011) could confirm that Aq-Cry2 plays some role in larval phototaxis, it may not be able to distinguish between a specific role as the photoreceptor regulating the ciliary rudder, an ongoing role in settlement timing, or both. Indeed, both duplicate sponge cryptochromes are expressed throughout development and it would not be surprising if they had multiple functions at different developmental time points. Another hurdle is that techniques for gene knockdown in sponges are limited to feeding adults and have not yet been developed for embryos or larvae, the stage when the animals are phototactic (Rivera et al., 2011). In light of this, experiments manipulating magnetic fields (Henbest et al., 2008) or examining a potential role for cryptochromes in the regulation of ciliary movements via calcium signaling (Liscum et al., 2003; Tamm and Terasaki, 1994) might be used to provide mechanistic insight into Aq-Cry2 function.
This study explored the molecular basis of sponge vision, which likely evolved in the absence of neurons and opsins. Although eyes have evolved up to 60 times in eumetazoan animals (Salvini-Plawen and Mayr, 1977), all of these share many genetic components (Kozmik, 2008; Mutsuddi et al., 2005). In particular, they are opsin-based (Spudich et al., 2000; Suga et al., 2008; Vopalensky and Kozmik, 2009), illustrating an evolutionary continuity that runs through all non-poriferan animal eyes examined to date. Of particular interest for future study will be to elucidate multiple molecular components of sponge phototaxis, and to determine whether any are shared with non-poriferan eyes, or whether the sponge eyes are instead completely convergent at the genetic level.
Acknowledgments
ACKNOWLEDGEMENTS
We thank S. Leys for comments and discussion, especially regarding Amphimedon morphology. D. Speiser and M. Alexandrou provided helpful comments on earlier drafts.
FOOTNOTES
FUNDING
This work was supported by funds from the National Science Foundation [DEB-0643840 to T.H.O.], the University of California, Santa Barbara [to T.H.O.], the National Institutes of Health [GM31082 to A.S.] and the Australian Research Council [to B.M.D.]. Deposited in PMC for release after 12 months.
REFERENCES
- Abushik A. F., Berger A. Y., Koren T. N., Modzalevskaya T. L., Nikiforova O. I., Predtechensky N. N. (1985). The fourth series of the Silurian System in Podolia. Lethaia 18, 125–146 [Google Scholar]
- Adachi Y., Hauck B., Clements J., Kawauchi H., Kurusu M., Totani Y., Kang Y. Y., Eggert T., Walldorf U., Furukubo-Tokunaga K., et al. (2003). Conserved cis-regulatory modules mediate complex neural expression patterns of the eyeless gene in the Drosophila brain. Mech. Dev. 120, 1113–1126 [DOI] [PubMed] [Google Scholar]
- Adamska M., Degnan S. M., Green K. M., Adamski M., Craigie A., Larroux C., Degnan B. M. (2007). Wnt and TGF-β expression in the sponge Amphimedon queenslandica and the origin of metazoan embryonic patterning. PLoS ONE 2, e1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D., Wittbrodt J. (2001). Reconstructing the eyes of Urbilateria. Philos. Trans. R. Soc. Lond. B 356, 1545–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorn L. O., Rasmusson A. G. (2009). Photosensitivity in sponge due to cytochrome c oxidase? Photochem. Photobiol. Sci. 8, 755–757 [DOI] [PubMed] [Google Scholar]
- Brudler R., Hitomi K., Daiyasu H., Toh H., Kucho K., Ishiura M., Kanehisa M., Roberts V. A., Todo T., Tainer J. A., et al. (2003). Identification of a new cryptochrome class: structure, function, and evolution. Mol. Cell 11, 59–67 [DOI] [PubMed] [Google Scholar]
- Ceriani M. F., Darlington T. K., Staknis D., Más P., Petti A. A., Weitz C. J., Kay S. A. (1999). Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 285, 553–556 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. L., Charlie N. K., Milfort M. C., Brown B. S., Gravlin C. N., Knecht J. E., Miller K. G. (2008). A novel molecular solution for ultraviolet light detection in Caenorhabditis elegans. PLoS Biol. 6, e198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P., So W. V., Kaneko M., Hall J. C., Rosbash M. (1998). CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679 [DOI] [PubMed] [Google Scholar]
- Foley L. E., Gegear R. J., Reppert S. M. (2011). Human cryptochrome exhibits light-dependent magnetosensitivity. Nat. Commun. 2, 356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegear R. J., Casselman A., Waddell S., Reppert S. M. (2008). CRYPTOCHROME mediates light-dependent magnetosensitivity in Drosophila. Nature 454, 1014–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegear R. J., Foley L. E., Casselman A., Reppert S. M. (2010). Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature 463, 804–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallén S., Brzezinski P. (1994). Light-induced structural changes in cytochrome c oxidase: implication for the mechanism of electron and proton gating. Biochim. Biophys. Acta 1184, 207–218 [DOI] [PubMed] [Google Scholar]
- Henbest K. B., Maeda K., Hore P. J., Joshi M., Bacher A., Bittl R., Weber S., Timmel C. R., Schleicher E. (2008). Magnetic-field effect on the photoactivation reaction of Escherichia coli DNA photolyase. Proc. Natl. Acad. Sci. USA 105, 14395–14399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley K. D., Szmant A. M., Pyott S. J. (2011). Circadian clock gene expression in the coral Favia fragum over diel and lunar reproductive cycles. PLoS ONE 6, e19755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karu T. I. (1999). Primary and secondary mechanisms of action of visible to near-IR radiation on cells. J. Photochem. Photobiol. B 49, 1–17 [DOI] [PubMed] [Google Scholar]
- Karu T. I., Pyatibrat L. V., Afanasyeva N. I. (2004). A novel mitochondrial signaling pathway activated by visible-to-near infrared radiation. Photochem. Photobiol. 80, 366–372 [DOI] [PubMed] [Google Scholar]
- Kim S. T., Malhotra K., Taylor J. S., Sancar A. (1996). Purification and partial characterization of (6-4) photoproduct DNA photolyase from Xenopus laevis. Photochem. Photobiol. 63, 292–295 [DOI] [PubMed] [Google Scholar]
- Koyanagi M., Takano K., Tsukamoto H., Ohtsu K., Tokunaga F., Terakita A. (2008). Jellyfish vision starts with cAMP signaling mediated by opsin-G(s) cascade. Proc. Natl. Acad. Sci. USA 105, 15576–15580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmik Z. (2008). The role of Pax genes in eye evolution. Brain Res. Bull. 75, 335–339 [DOI] [PubMed] [Google Scholar]
- Kozmik Z., Ruzickova J., Jonasova K., Matsumoto Y., Vopalensky P., Kozmikova I., Strnad H., Kawamura S., Piatigorsky J., Paces V., et al. (2008). Assembly of the cnidarian camera-type eye from vertebrate-like components. Proc. Natl. Acad. Sci. USA 105, 8989–8993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer D. C., Pagel M., Southwood T. R., Zanotto P. M. (1996). Phylogenesis of prion protein. Nature 380, 675 [DOI] [PubMed] [Google Scholar]
- Larroux C., Fahey B., Adamska M., Richards G. S., Gauthier M., Green K., Lovas E., Degnan B. M. (2008). Whole-mount in situ hybridization in Amphimedon. Cold Spring Harb. Protoc. 2008, prot5096 [DOI] [PubMed] [Google Scholar]
- Lepre A. M., Sutherland J. C., Trunk J. G., Sutherland B. M. (1998). A robust, inexpensive filter for blocking UVC radiation in broad-spectrum ‘UVB’ lamps. J. Photochem. Photobiol. 43, 34–40 [DOI] [PubMed] [Google Scholar]
- Levy O., Appelbaum L., Leggat W., Gothlif Y., Hayward D. C., Miller D. J., Hoegh-Guldberg O. (2007). Light-responsive cryptochromes from a simple multicellular animal, the coral Acropora millepora. Science 318, 467–470 [DOI] [PubMed] [Google Scholar]
- Leys S. P., Degnan B. M. (2001). Cytological basis of photoresponsive behavior in a sponge larva. Biol. Bull. 201, 323–338 [DOI] [PubMed] [Google Scholar]
- Leys S. P., Cronin T. W., Degnan B. M., Marshall J. N. (2002). Spectral sensitivity in a sponge larva. J. Comp. Physiol. A 188, 199–202 [DOI] [PubMed] [Google Scholar]
- Leys S. P., Larroux C., Gauthier M., Adamska M., Fahey B., Richards G. S., Degnan S. M., Degnan B. M. (2008). Isolation of Amphimedon developmental material. Cold Spring Harb. Protoc. 2008, prot5095 [DOI] [PubMed] [Google Scholar]
- Liscum E., Hodgson D. W., Campbell T. J. (2003). Blue light signaling through the cryptochromes and phototropins. So that’s what the blues is all about. Plant Physiol. 133, 1429–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra K., Kim S. T., Batschauer A., Dawut L., Sancar A. (1995). Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry 34, 6892–6899 [DOI] [PubMed] [Google Scholar]
- Mouritsen H., Janssen-Bienhold U., Liedvogel M., Feenders G., Stalleicken J., Dirks P., Weiler R. (2004). Cryptochromes and neuronal-activity markers colocalize in the retina of migratory birds during magnetic orientation. Proc. Natl. Acad. Sci. USA 101, 14294–14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsuddi M., Chaffee B., Cassidy J., Silver S. J., Tootle T. L., Rebay I. (2005). Using Drosophila to decipher how mutations associated with human branchio-oto-renal syndrome and optical defects compromise the protein tyrosine phosphatase and transcriptional functions of eyes absent. Genetics 170, 687–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk N., Kao Y.-T., Selby C. P., Kavakli I. H., Partch C. L., Zhong D., Sancar A. (2008). Purification and characterization of a type III photolyase from Caulobacter crescentus. Biochemistry 47, 10255–10261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk N., Selby C. P., Song S.-H., Ye R., Tan C., Kao Y.-T., Zhong D., Sancar A. (2009). Comparative photochemistry of animal type 1 and type 4 cryptochromes. Biochemistry 48, 8585–8593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachetzki D. C., Degnan B. M., Oakley T. H. (2007). The origins of novel protein interactions during animal opsin evolution. PLoS ONE 2, e1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel A. M., Behrendt L., Tarrant A. M. (2010). Light entrained rhythmic gene expression in the sea anemone Nematostella vectensis: the evolution of the animal circadian clock. PLoS ONE 5, e12805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert S. M., Weaver D. (2002). Coordination of circadian timing in mammals. Nature 418, 935–941 [DOI] [PubMed] [Google Scholar]
- Rivera A., Pankey M., Syme A., Villacorta C., Serb J. M., Plachetzki D. C., Omilian A., Oakley T. H. (2010). Gene duplication and the origins of morphological complexity in pancrustacean eyes, a genomic approach. BMC Evol. Biol. 10, 123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera A. S., Hammel J. U., Haen K. M., Danka E. S., Cieniewicz B., Winters I. P., Posfai D., Worheide G., Lavrov D. V., Knight S. W., et al. (2011). RNA interference in marine and freshwater sponges: actin knockdown in Tethya wilhelma and Ephydatia muelleri by ingested dsRNA expressing bacteria. BMC Biotechnol. 11, 67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvini-Plawen L. V., Mayr E. (1977). On the Evolution of Photoreceptors and Eyes. New York: Plenum Press; [Google Scholar]
- Sancar A. (2000). Cryptochrome: The second photoactive pigment in the eye and its role in circadian photoreception. Annu. Rev. Biochem. 69, 31–67 [DOI] [PubMed] [Google Scholar]
- Sancar A. (2004). Regulation of the mammalian circadian clock by cryptochrome. J. Biol. Chem. 279, 34079–34082 [DOI] [PubMed] [Google Scholar]
- Sancar A., Smith F. W., Sancar G. B. (1984). Purification of Escherichia coli DNA photolyase. J. Biol. Chem. 259, 6028–6032 [PubMed] [Google Scholar]
- Sancar G. B., Smith F. W., Sancar A. (1983). Identification and amplification of the E. coli phr gene product. Nucleic Acids Res. 11, 6667–6678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder H. C., Krasko A., Gundacker D., Leys S. P., Müller I. M., Müller W. E. G. (2003). Molecular and functional analysis of the (6-4) photolyase from the hexactinellid Aphrocallistes vastus. Biochim. Biophys. Acta 1651, 41–49 [DOI] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. (2006). A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA 103, 17696–17700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.-H., Ozturk N., Denaro T. R., Arat N. O., Kao Y.-T., Zhu H., Zhong D., Reppert S. M., Sancar A. (2007). Formation and function of flavin anion radical in cryptochrome 1 blue-light photoreceptor of monarch butterfly. J. Biol. Chem. 282, 17608–17612 [DOI] [PubMed] [Google Scholar]
- Sperling E. A., Robinson J. M., Pisani D., Peterson K. J. (2010). Where’s the glass? Biomarkers, molecular clocks, and microRNAs suggest a 200-Myr missing Precambrian fossil record of siliceous sponge spicules. Geobiology 8, 24–36 [DOI] [PubMed] [Google Scholar]
- Spudich J. L., Yang C. S., Jung K. H., Spudich E. N. (2000). Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16, 365–392 [DOI] [PubMed] [Google Scholar]
- Srivastava M., Simakov O., Chapman J., Fahey B., Gauthier M. E., Mitros T., Richards G. S., Conaco C., Dacre M., Hellsten U., et al. (2010). The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771 [DOI] [PubMed] [Google Scholar]
- Stanewsky R., Kaneko M., Emery P., Beretta B., Wager-Smith K., Kay S. A., Rosbash M., Hall J. C. (1998). The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692 [DOI] [PubMed] [Google Scholar]
- Suga H., Schmid V., Gehring W. J. (2008). Evolution and functional diversity of jellyfish opsins. Curr. Biol. 18, 51–55 [DOI] [PubMed] [Google Scholar]
- Tamm S. L., Terasaki M. (1994). Visualization of calcium transients controlling orientation of ciliary beat. J. Cell Biol. 125, 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thresher R. J., Vitaterna M. H., Miyamoto Y., Kazantsev A., Hsu D. S., Petit C., Selby C. P., Dawut L., Smithies O., Takahashi J. S., et al. (1998). Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 282, 1490–1494 [DOI] [PubMed] [Google Scholar]
- Tong D., Rozas N. S., Oakley T. H., Mitchell J., Colley N. J., McFall-Ngai M. J. (2009). From the Cover: Evidence for light perception in a bioluminescent organ. Proc. Natl. Acad. Sci. USA 106, 9836–9841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vopalensky P., Kozmik Z. (2009). Eye evolution: common use and independent recruitment of genetic components. Philos. Trans. R. Soc. Lond. B 364, 2819–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington E. N., Kavakli I. H., Berrocal-Tito G., Bondo B. E., Sancar A. (2003). Purification and characterization of three members of the photolyase/cryptochrome family blue-light photoreceptors from Vibrio cholerae. J. Biol. Chem. 278, 39143–39154 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Yuan Q., Vogt N., Looger L. L., Jan L. Y., Jan Y. N. (2010). Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature 468, 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q., Metterville D., Briscoe A. D., Reppert S. M. (2007). Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol. Biol. Evol. 24, 948–955 [DOI] [PubMed] [Google Scholar]
- Zhao X., Liu J., Hsu D. S., Zhao S., Taylor J. S., Sancar A. (1997). Reaction mechanism of (6-4) photolyase. J. Biol. Chem. 272, 32580–32590 [DOI] [PubMed] [Google Scholar]
- Zhu H., Yuan Q., Briscoe A. D., Froy O., Casselman A., Reppert S. M. (2005). The two CRYs of the butterfly. Curr. Biol. 15, R953–R954 [DOI] [PubMed] [Google Scholar]